ABSTRACT
The global pandemic of COVID-19 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in widespread social and economic disruption. Effective interventions are urgently needed for the prevention and treatment of COVID-19. Neutralizing monoclonal antibodies (mAbs) have demonstrated their prophylactic and therapeutic efficacy against SARS-CoV-2, and several have been granted authorization for emergency use. Here, we discover and characterize a fully human cross-reactive mAb, MW06, which binds to both SARS-CoV-2 and SARS-CoV spike receptor-binding domain (RBD) and disrupts their interaction with angiotensin-converting enzyme 2 (ACE2) receptors. Potential neutralization activity of MW06 was observed against both SARS-CoV-2 and SARS-CoV in different assays. The complex structure determination and epitope alignment of SARS-CoV-2 RBD/MW06 revealed that the epitope recognized by MW06 is highly conserved among SARS-related coronavirus strains, indicating the potential broad neutralization activity of MW06. In in vitro assays, no antibody-dependent enhancement (ADE) of SARS-CoV-2 infection was observed for MW06. In addition, MW06 recognizes a different epitope from MW05, which shows high neutralization activity and has been in a Phase 2 clinical trial, supporting the development of the cocktail of MW05 and MW06 to prevent against future escaping variants. MW06 alone and the cocktail show good effects in preventing escape mutations, including a series of variants of concern, B.1.1.7, P.1, B.1.351, and B.1.617.1. These findings suggest that MW06 recognizes a conserved epitope on SARS-CoV-2, which provides insights for the development of a universal antibody-based therapy against SARS-related coronavirus and emerging variant strains, and may be an effective anti-SARS-CoV-2 agent.
Introduction
COVID-19, caused by SARS-CoV-2, is currently spreading globally, threatening human health and economic development. As of June 4, 2021, SARS-CoV-2 has caused over 183 million infections in more than 200 countries/regions and resulted in over 3.9 million deaths, according to data from the Johns Hopkins Coronavirus Resource Center. Interventions for the prevention or treatment of COVID-19 are crucial for the ongoing outbreak.
Both SARS-CoV-2 and SARS-CoV belong to Betacoronaviruses, Sarbecovirus (lineage B), and they share ~79.6% sequence identity.Citation1,Citation2 Cellular entry of SARS-CoV-2 and SARS-CoV is mediated by the viral spike glycoprotein, which forms trimeric spikes on the viral surface. Like SARS-CoV, the receptor-binding domain (RBD) of SARS-CoV-2 spike protein is responsible for engaging the angiotensin-converting enzyme 2 (ACE2) receptor on the host cell surface and mediating cell-virus membrane fusion by the class I fusion mechanism.Citation3,Citation4 Therefore, spike protein, especially the RBD of SARS-CoV-2, is the primary target for neutralizing antibody and vaccine development. SARS-CoV and SARS-CoV-2 share ~76% amino acid identity in their spike proteins, raising the possibility of conserved epitopes on these antigens.Citation1 Remarkably, the essential SARS-CoV contact residues that interact with ACE2 were highly conserved in SARS-CoV-2 as well as in SARS-related coronaviruses, indicating the possibility of developing broadly neutralizing mAbs for potential future diseases caused by other emerging SARS-related viruses.Citation5,Citation6
Vaccines and neutralization antibodies are the best strategies to control the worldwide pandemic of SARS-CoV-2. After the outbreak, multiple vaccine candidates derived from different platforms were developed, and vaccines based on inactivated virus and mRNA-encoded viral proteins, as well as an adenovirus-vectored vaccine, have been authorized for use. During the SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) outbreaks, neutralizing mAbs were developed and proved their potential therapeutic uses for the treatment of coronavirus infections.Citation7,Citation8 Likewise, several highly potent neutralizing mAbs targeting SARS-CoV-2 spike protein, especially RBD, have been identified and evaluated in clinical trials. Neutralizing mAbs developed by Regeneron and Lilly/AbCellera were granted emergency use authorization by the Food and Drug Administration. Many other neutralizing mAbs are in preclinical and clinical development and show good antiviral activities both in vitro and in vivo.Citation9–11
We recently reported the isolation and characterization of a panel of SARS-CoV-2 RBD-binding mAbs from a COVID-19 convalescent patient using a single B cell cloning strategy.Citation9 Among these, MW05 (drug code: MW33), which showed the most potent neutralizing activity in an authentic virus neutralization assay without cross-reactivity with SAR-CoV, was selected for further development; an engineered version is in a Phase 2 clinical trial (NCT04627584). Here, we describe a novel SARS-CoV-2 spike RBD-targeting mAb, MW06, which shows strong cross-binding activity and high neutralizing potency with both SARS-CoV-2 and SARS-CoV. SARS-CoV-2 RBD/MW06 Fab complex structure determination revealed that the epitope of MW06 is highly conserved among SARS-related coronaviruses, indicating the broad antiviral activities of MW06. No ADE activity was detected for MW06, indicating the safety of MW06 in potential clinical use. MW06 and MW05 recognize different epitopes on RBD of SARS-CoV-2, supporting the development of the cocktail of MW05 and MW06 to prevent against future escape variants.
Results
Cross-reactivity of MW06 with SARS-CoV-2 and SARS-CoV
Single B cell strategy is an efficient way to obtain fully human SARS-CoV-2 neutralizing mAbs. Since the spike (S) protein on the surface of SARS-CoV-2 is the major molecular determinant for viral infection, RBD or trimeric prefusion ectodomain of S protein can be used as bait to isolate specific memory B cells.Citation9,Citation12,Citation13 We have previously developed and characterized a panel of fully human mAbs that target the RBD of SARS-CoV-2 spike protein from a COVID-19 convalescent patient using single B cell cloning strategy.Citation9 One mAb, MW06, was identified to have cross-reactivity with recombinant RBDs of both SARS-CoV-2 and SARS-CoV. MW06 exhibited strong binding ability to SARS-CoV-2 and SARS-CoV RBD recombinant proteins in ELISA, with the EC50 of 0.004 μg/mL and 0.003 μg/mL, respectively (). Bio-layer interferometry (BLI) assay was then used to measure the binding affinity of MW06. The equilibrium dissociation constant (KD) of MW06 to SARS-CoV-2 and SARS-CoV RBD recombinant proteins were 5.48 nM and 12.3 nM, respectively ().
Figure 1. The cross-reactivity of MW06 to SARS-CoV-2 and SARS-CoV. (a-b) The binding ability of MW06 to SARS-CoV-2 and SARS-CoV RBD recombinant proteins was assessed by ELISA. Irrelative hIgG1 was used as a control. EC50 was labeled accordingly. (c-d) The binding kinetics of MW06 to SARS-CoV-2 and SARS-CoV RBD recombinant proteins was measured by BLI. The KD was labeled accordingly. (e-f) The ability of MW06 to block SARS-CoV-2 and SARS-CoV RBD interaction with ACE2 was evaluated by competition ELISA. Irrelative hIgG1 was used as a control. IC50 was labeled accordingly
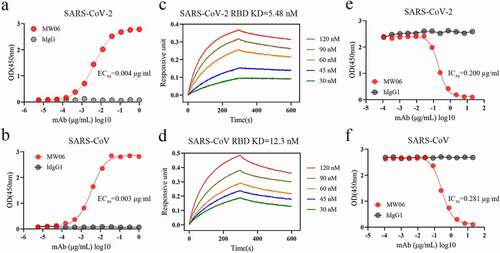
To further evaluate whether the binding of MW06 to RBD could block the interaction of RBD with ACE2 or not, a competitive assay was performed. The results showed that MW06 could effectively block the interaction of ACE2 with both SARS-CoV-2 and SARS-CoV RBD recombinant proteins, with the IC50 of 0.200 μg/mL for SARS-CoV-2 RBD and 0.281 μg/mL for SARS-CoV RBD (). In summary, MW06 showed cross-binding ability and ACE2/RBD blocking activity for both SARS-CoV-2 and SARS-CoV.
Neutralization activity of MW06 against SARS-CoV-2 and SARS-CoV
To evaluate the cross-neutralization potency of MW06, both pseudovirus and authentic virus assays were performed. MW06 effectively inhibited the transduction of SARS-CoV-2 pseudovirus into both Huh-7 and Vero cells, with NT50 of 0.338 μg/mL and 1.317 μg/mL, respectively (). Further, the neutralization potency of MW06 against SARS-CoV-2 authentic virus was evaluated on Vero E6 and ACE2 overexpression Huh-7 (Huh-7/ACE2) cells using cytopathic effect inhibition (CPE) and plaque reduction assays. MW06 was shown to significantly inhibit SARS-CoV-2 replication in both cell lines, with an NT50 of 0.248 μg/mL for Huh-7/hACE2 cells and 0.252 μg/mL for Vero E6 cells in CPE assay (). In line with the CPE assay, MW06 exhibited strong neutralization activity against SARS-CoV-2 authentic virus on Vero E6 cells in plaque reduction assay, with an NT50 of 0.654 μg/mL ().
Figure 2. Neutralization activity of MW06 against SARS-CoV-2 and SARS-CoV. (a) Neutralization potency of MW06 against SARS-CoV-2 pseudovirus on Huh-7 and Vero cells, 50% neutralization titer (NT50) were labeled. (b-c) Neutralization potencies of MW06 against SARS-CoV-2 authentic virus were evaluated by CPE assay on Huh-7/hACE2 and Vero E6 cells, and plaque reduction assay on Vero E6 cells. (d) Neutralization potency of MW06 against SARS-CoV pseudovirus on Huh-7 and Vero cells. 50% neutralization titer (NT50) was labeled, accordingly
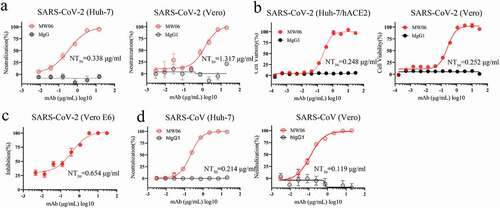
We confirmed that the cross-binding activity of MW06, the SARS-CoV pseudovirus system was further used to evaluate the potential neutralization activity of MW06. As expected, a potent neutralization activity against SARS-CoV was observed for MW06. The NT50 was 0.214 μg/mL on Huh-7 cells and 0.119 μg/mL on Vero cells (). Taken together, the results indicate that MW06 could effectively neutralize both SARS-CoV-2 and SARS-CoV in vitro.
Structural basis of MW06 binding to SARS-CoV-2 RBD
To explore the mechanism of anti-viral activity of MW06 against SARS-CoV-2, we solved the crystal structures of MW06 Fab in complex with SARS-CoV-2 RBD at a resolution of 3.3 Å (). The variable domain of MW06 was bound to the RBD mainly through its core subdomain (). The binding buried ~1555 Å of surface area, of which ~1065 Å involves heavy-chain residues. However, only HCDR3, which was significantly longer than that of most antibodies and rich in aromatic amino acid residues, contributed to the recognition by the heavy chain. Specifically, the main-chain atoms of Ser105, Tyr107, and Arg110 in HCDR3 formed several hydrogen bonds with the main-chain atoms of Cys379, Phe377, and Ser375 of spike RBD. The side chains of Tyr101, Tyr102, Tyr107, Tyr108, and Phe109 contacted spike RBD through hydrophobic interactions and hydrogen bonds (). The LCDR1, LCDR2, and LCDR3 of the light chain were all involved in recognition. Specifically, the side chains of Tyr32 in LCDR1, Ser53 in LCDR2, and Asn92 in LCDR3 formed hydrogen bonds with main chains of Phe374, Val503, and Ala372 of SARS-CoV-2 RBD, respectively (). Superimposition of RBD/MW06 complex structure with RBD/hACE2 complex structureCitation14 showed that the recognition site of MW06 slightly overlapped with the hACE2 binding site (), suggesting that MW06 competes for ACE2 binding.
Figure 3. Determination of the complex structure of SARS-CoV-2 RBD/MW06. (a) The overall structure of MW06 Fab in complex with SARS-CoV-2 Spike RBD. The heavy chain and light chain of MW06 are shown as blue and light cyan cartoons, and the SARS-CoV-2 Spike RBD is shown as gray surface. (b) The epitope of MW06. The residues contacted by heavy chain, light chain, and both chains were colored in blue, light cyan, and yellow, respectively. (c) The detail of interactions between SARS-CoV-2 Spike RBD and heavy chain of MW06. (d) The detail of interactions between SARS-CoV-2 Spike RBD and light chain of MW06. The involved residues are shown as sticks with the same colors in Figure 3a. The hydrogen bonds and salt bridges are indicated as blue dashed lines. (e) Superimposition of SARS-CoV-2 RBD/MW06 with RBD/hACE2 complex (PDB code: 6LZG). The heavy chain and light chain of MW06 Fab, and the hACE2 are shown as blue, light cyan, and salmon cartoons, respectively. SARS-CoV-2 Spike RBD is represented as gray surface. (f) The MW06 and hACE2 binding surface of SARS-CoV-2 Spike RBD. The residues contacting MW06, hACE2 are colored in blue and red, respectively; the residues contacting both are colored in yellow. (g) Superimposition of SARS-CoV-2 RBD/MW06 with SARS-CoV-2 Spike trimer in “close” state (PDB code: 6VXX). One MW06 Fab is superposed to one “down” RBD (wheat) in the “close” SARS-CoV-2 Spike trimer. The binding of MW06 Fab introduces clashes (enclosed with red dashed cycle) with another RBD (light yellow). (h) Superimposition of RBD/MW06 with SARS-CoV-2 Spike trimer in “open” state (PDB code: 6VYB). The MW06 Fab is superposed to the “up” RBD (wheat). Binding of MW06 Fab introduces clashes (enclosed with red dashed cycle) with one of the “down” RBDs (light yellow). (i) Superimposition of RBD/MW06 with SARS-CoV-2 Spike trimer with 3-“up” RBDs (PDB code: 7A98). Three MW06 Fabs are superposed to three “up” RBDs in SARS-CoV-2 Spike trimer. The heavy chain and light chain of MW06 Fab are shown as blue and light cyan cartoons, respectively, and the three chains of SARS-CoV-2 Spike trimer are shown as wheat, gray, and yellow surface
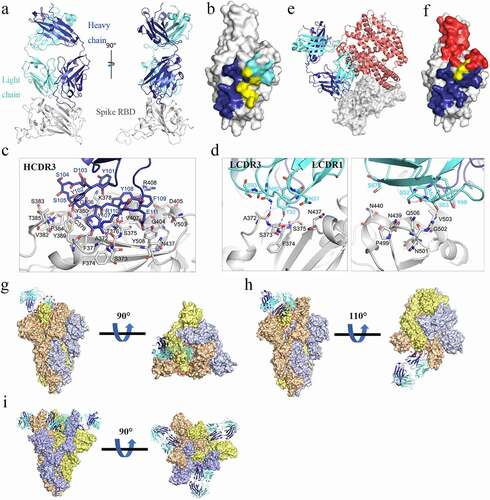
To better understand the binding model of MW06 to SARS-CoV-2 spike trimer, RBD/MW06 complex structure was further superimposed into spike trimer with different conformations. Superimposition of the RBD/MW06 complex structure into a spike trimer structure in “open” state, which include 2-“down” RBDs and 1-“up” RBD,Citation15,Citation16 revealed that MW06 could not bind to either “down” or “up” RBDs. The superimposition of the RBD/MW06 complex structure with spike trimer structure in “close” state, which include 3-“down” RBDs, showed that MW06 could not bind to the “down” RBDs. Binding with any one of “up” or “down” RBDs in the spike trimer in “close” or “open” conformation could induce clashes (). Recently, a spike trimer structure with 3-“up” RBDs was published.Citation17 We further superimposed RBD/MW06 complex structure with the spike trimer with 3-“up” RBDs. The result showed that three “up” RBDs in spike trimer could bind to three MW06 Fabs without any clashes (). These results suggested that MW06 only binds SARS-CoV-2 spike trimer in a specific conformation, indicating that MW06 might neutralize SARS-CoV2 through a unique way or in a unique stage of viral infection.
Epitope conservation of SARS-related coronavirus recognized by MW06
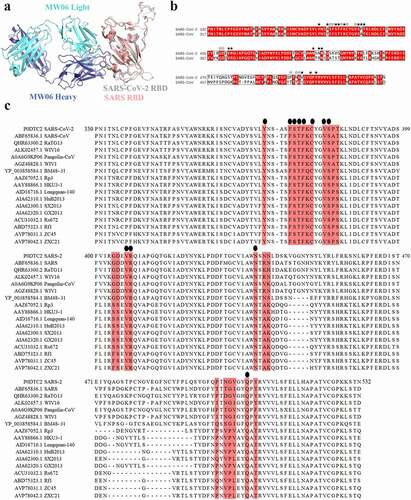
To explore the molecular mechanism of SARS-CoV-2 and SARS-CoV cross-reactivity of MW06, SARS-CoV-2 RBD/MW06 complex structure was superimposed on the structure of SARS-CoV RBD. The model revealed that the structure of SARS-CoV spike RBD was quite similar to that of SARS-CoV-2 spike RBD (). Furthermore, the sequence alignment of these two RBDs showed that the amino acid residues of SARS-CoV-2 spike RBD involving the interaction with MW06 Fab were highly conserved (). Therefore, the structural similarity and epitope conservation of the two SARS-related viruses contribute to the cross-reactivity of MW06. In addition, we included another 15 SARS-related coronavirus strains into the alignment analysis. Of the 24 amino acid residues in the epitope recognized by MW06, 12 residues are identical in all coronavirus strains (). This result indicates a highly conserved epitope recognized by MW06 in most of these SARS-related coronavirus strains, which implies broad coverage of coronavirus by MW06.
Figure 4. Conserved epitope recognized by MW06. (a) Superposition of SARS-CoV-2 Spike RBD/MW06 complex structure with SARS-CoV Spike RBD. The heavy chain and light chain of MW06 are shown as blue and light cyan cartoons; the SARS-CoV-2 Spike RBD is shown as gray cartoon, and the SARS-CoV Spike RBD is shown as red cartoon. (b) Sequence alignment of the Spike RBD of SARS-CoV-2 and SARS-CoV. The amino acid residues involved in interaction with MW06 Fab was indicated with pentangle. The solid pentangle indicates that the side-chains of the residue participated in the interaction while the hollow pentangle indicates main-chains. The red boxes indicated the identical amino acid residues. (c) Amino acid sequence alignment of RBDs from SARS-CoV-2, SARS-CoV and other SARS-related coronavirus strains. Epitope residues recognized by MW06 are highlighted in red. Conserved residues are indicated by small black dots on the top of the alignment
No observed ADE activity of MW06
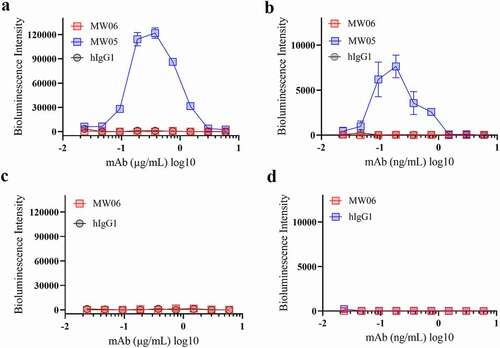
Neutralizing antibodies against spike protein play a critical role in host defense against the SARS-CoV-2 virus. However, antibody-dependent enhancement (ADE) is a substantial concern for antibody-based therapeutics development. ADE has been observed for coronaviruses, and several publications have shown that sera induced by SARS-CoV spike protein enhanced viral entry into immune cells and exacerbated inflammation.Citation18–21 We have previously reported that MW05 with wild-type IgG1 format (MW05/IgG1) enhances SARS-CoV-2 infection on Raji cells due to the interaction of Fc with FcγRIIB.Citation9 In order to investigate whether MW06/IgG1 could enhance the infection of SAR-CoV-2 and SARS-CoV on FcγRIIB expressing B cells, Raji and Daudi cells were used to evaluate the ADE activity of MW06. As shown before, mAb MW05/IgG1 enhanced the infection of SARS-CoV-2 on both Raji and Daudi cells (). Instead, no ADE of SARS-CoV-2 or SARS-CoV infection in immune cells was observed for MW06 (). As ADE is one of the main concerns for the development of anti-SARS-CoV-2 neutralizing monoclonal antibodies, these results support that MW06 is a good therapeutic antibody candidate for further clinical development.
Figure 5. ADE activity of MW06. ADE of SARS-CoV-2 pseudovirus infection of nonpermissive Raji cells (a) and Daudi cells (b) by MW06 were evaluated in a luciferase assay system. SARS-CoV-2 pseudovirus were incubated with 2-fold serially diluted mAbs. The mixtures were then added into Raji cells. After 24 hours of incubation, luciferase activity was measured. MW05/IgG1 was used as positive control. (c-d) ADE of SARS-CoV pseudovirus infection of nonpermissive Raji cells and Daudi cells by MW06
Cocktail of MW06 with MW05
Over the course of the pandemic, the spike protein of SARS-CoV-2 has been mutating. Antibody cocktails that include two or more mAbs recognizing distinct epitopes could target different mutants and prevent escape mutants. We therefore evaluated the possibility of developing a cocktail of MW06 and MW05, as MW05 shows high neutralization activity by disrupting the interaction of RBD with ACE2 receptorCitation9 and has been in a Phase 2 clinical trial. We first compared the epitope of MW05 and MW06. The structure of SARS-CoV-2 RBD in complex with MW05 has been resolved (PDB accession number: 7DK0). We then superimposed this complex structure with that of SARS-CoV-2 RBD/MW06. MW06 and MW05 could bind to SARS-CoV-2 RBD simultaneously without inducing clashes (). A competition assay was performed on the Octet system to confirm whether MW06 and MW05 bind to distinct epitopes on RBD of SARS-CoV-2. As shown in , binding of MW05 on SARS-CoV-2 RBD did not interfere with the binding of MW06. Binding of MW06 on SARS-CoV-2 RBD also did not affect the binding of MW05. Altogether, these results suggested that the cocktail of MW05 and MW06 may have complementary effects in terms of binding and neutralizing different SARS-CoV-2 mutants.
Figure 6. Cocktail of MW05 with MW06. (a) The overall structure of MW06 Fab and MW05 Fab in complex with SARS-CoV-2 Spike RBD. The heavy chain and light chain of MW06 are shown as blue and light cyan cartoons, the heavy chain and light chain of MW05 are shown as dark green and light green cartoons, and the SARS-CoV-2 Spike RBD is shown as gray surface. (b) Epitope competition assay was performed between MW05 and MW06. (c) Heatmap showing the ACE2/RBD blocking activity of MW05, MW06 and MW05+ MW06 were assessed by ELISA. The IC50 (μg/mL) value for each mutant is shown with red, light blue and blue indicating strong, intermediate, weak or non-detectable binding, respectively. “>20” means non-detectable activity. (d) The ACE2/RBD blocking curves of MW05 (left), MW06 (middle) and MW05+ MW06 (right). (e) The blocking ability of MW05, MW06 and the cocktail to block RBD recombination protein of SARS-CoV-2 variants strains interaction with ACE2 was evaluated by competition ELISA. (f) Neutralization potency of MW05, MW06 and the cocktail against pseudovirus of SARS-CoV-2 variants strains on Huh-7/ACE2 cells
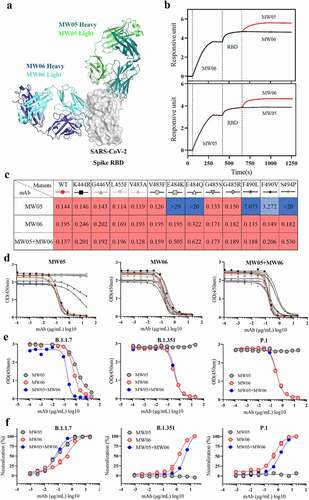
After the outbreak, several SARS-CoV-2 RBD mutations, which are located on the binding surface of MW05, have been reported. All these mutated RBD recombinant proteins were prepared, and we tested their binding with MW05, MW06, and the cocktail of MW05 with MW06. As shown in and d, MW05 showed decreased RBD/ACE2 blocking activity to F490L and F490V, and totally lost the blocking activity to E484K, E484Q, and S494P mutations. However, MW06 and the cocktail could effectively block the interaction of all these RBD mutants with ACE2. Consistent with the blocking data, MW06 and the cocktail demonstrate binding to all these RBD mutants, with EC50 all lower than 0.02 μg/ml (Figure S1 a,b).
Novel variants with combinations of mutations and deletions in RBD have recently appeared in multiple countries. The emergence of variants B.1.1.7, B.1.351, and P.1 marked the beginning of SARS-CoV-2 antigenic drift, among which, both B.1.1.7 and B.1.351 have been reported to be resistant to neutralization by mAbs.Citation22,Citation23 Notably, B.1.351 is resistant to a major group of potent mAbs that target the RBD of SARS-CoV-2, including LY-CoV555, LY-CoV55 combined with CB6, and REGN10933, which are regimens for emergency use.Citation23 We further assessed the antiviral activity of the MW05, MW06, and the cocktail of these SARS-CoV-2 variant strains. As shown in and Figure S1c, MW05 retains its activity against variant B.1.1.7, while its activities against variant B.1.351 and P.1 are completely abolished. However, MW06 and the cocktail could effectively disrupt the interaction of ACE2 with RBD mutants and also can block the entry of B.1.1.7, B.1.351, and P.1 pseudoviruses into Huh-7/ACE2 cells. Furthermore, the binding and blocking activities against a newly emerged variant, B.1.617.1, have been tested. MW06 and the cocktail show good binding and blocking activities, while MW05 does not (Figure S1d,e).
Taken together, these results support the potential clinical use of the combination of MW05 and MW06 with a strong resistance potential for diverse SARS-CoV-2 mutation strains.
Discussion
Due to the rapid spread and high mortality rate of SARS-CoV-2, therapeutic strategies for COVID-19 are urgently needed. A promising approach to combat the COVID-19 pandemic involves the development of neutralizing antibodies targeting the spike protein of SARS-CoV-2. The spike protein is a key mediator of viral infectivity required for attachment and entry into target cells by binding to the ACE2 receptor. To date, over 190 antibody-based therapeutics against SARS-CoV-2 are being explored.Citation24 This study reports a novel SARS-CoV-2 neutralizing antibody, MW06. MW06 exhibited good performance in blocking ACE2/RBD interaction, and showed strong neutralizing activities against pseudo and authentic SARS-CoV-2 in vitro. Interestingly, MW06 could also bind and neutralize SARS-CoV with high potency ().
SARS-CoV-2 has undergone several mutations and is rapidly evolving since its emergence, which could affect the efficacy of antibody-based therapeutics. Neutralizing mAbs targeting highly conserved regions on RBD of SARS-CoV-2 may provide better performance in clinical trials.Citation25 In this study, we demonstrated that epitope residues recognized by MW06 are quite conserved in SARS-CoV-2, SARS-CoV, and other SARS-related coronaviruses (). Unsurprisingly, MW06 shows a cross-activity against spike RBD of both SARS-CoV-2 and SARS-CoV. As expected, MW06 could neutralize both SARS-CoV-2 and SARS-CoV pseudovirus effectively in in vitro neutralizing assays using different cell lines. Although further verification is needed to confirm whether MW06 can neutralize SARS-CoV authentic virus, it is a reasonable assertion that MW06 has a potentially wide spectrum of antiviral activities against SARS-related coronavirus.
Other spike RBD-targeting antibodies with cross-neutralization activity, such as CR3022,Citation26 S309,Citation27 and COVA1-16,Citation28 have been reported. Like MW06, CR3022, and COVA1-16 also only bind the spike trimer with 3-“up” RBDs.Citation26,Citation28 This common feature seems to be related to the epitope residues. Indeed, MW06 shares several common epitope residues in SARS-COV-2 RBD with CR3022 and COVA1-16, especially in the regions recognized by HCDR3 of MW06.Citation26,Citation28 This evidence indicates that immunogens based on this region may elicit cross-neutralization antibodies to SARS-related coronaviruses.
A main concern of neutralizing mAbs against coronavirus is the ADE of virus infection in immune cells. Several antibodies against SARS-CoV or SARS-CoV-2 Spike have been reported to exhibit ADE activities in vitro or exacerbate disease in animal models.Citation9,Citation18,Citation29,Citation30 Studies indicate that the ADE of neutralizing mAbs is mediated by the interaction of Fc domain with Fc receptors.Citation9,Citation31,Citation32 Some neutralizing antibodies against SARS-CoV-2 that are under clinical development include LALA mutations to the Fc to diminish the interaction with Fc receptors and eliminate the ADE effect.Citation9,Citation11 No ADE of MW06 was observed on nonpermissive Raji and Daudi cells (). Compared with MW05, MW06 has the same Fc region, but a different Fab region, which leads to different binding epitopes and recognition of different conformations of the spike trimer. These results indicate that epitope and binding models also contribute to the mechanism of ADE. The correlation between antibody epitope and ADE activity needs further study.
An antibody cocktail approach has provided profound survival benefit for patients infected with the Ebola virus.Citation33 Similar findings were reported recently regarding cocktails for COVID-19. REGN-COV2, an antibody cocktail that consists of two anti-SARS-CoV-2 antibodies, REGN10933 and REGN10987, enhanced viral clearance and led to improved clinical outcomes.Citation34 More importantly, REGN10933 and REGN10987, targeting distinct structural epitope, can target more mutants than either single antibody.Citation35 Pairs of antibodies that bind to different epitopes of the SARS-COV-2 RBD spike protein may reduce the risk of treatment-resistant mutants emergence. In this study, our group previously resolved the SARS-CoV-2 RBD/MW05 complex, and we determined the structure of SARS-CoV-2 RBD/MW06 in this study, which encourages us to try a cocktail of MW05 with MW06. No epitope competition between MW05 and MW06 was observed in molecular docking and was further confirmed by BLI assays (). For several epidemic variants we tested, the cocktail shows similar performance with MW06, in both binding and blocking activity ( and Figure S1). Considering that the spike protein of SARS-COV-2 is continuously mutating, the cocktail of MW05 and MW06 could prevent against future escape variants more efficiently. In addition, as MW05/IgG1 shows ADE activity, LALA mutations were introduced in MW05 (MW05/LALA; MW33)Citation9 in anticipation of the future clinical development of cocktails. Animal studies are needed to further confirm the prophylactic and therapeutic efficacy of the cocktail of MW05/LALA with MW06.
In summary, MW06 has the potential to be a good therapeutic antibody candidate for COVID-19 treatment due its high neutralization activity, broad anti-SARS-related coronaviruses activity, and lack of in vitro ADE effects. Moreover, the cocktail of MW06 and MW05/LALA is also valuable for further development to prevent outbreaks that may be caused by potential viral mutations in the future.
Materials and methods
Cells and viruses
Huh-7 (Institute of Basic Medical Sciences CAMS, 3111C0001CCC000679) cells and Vero (ATCC, CCL-81) cells were cultured at 37°C in Dulbecco’s Modified Eagle medium (DMEM, Hyclone SH30243.01) supplemented with 10% fetal bovine serum (FBS). CHO-K1 (ATCC, CCL-61) cells were cultured at 37°C in Dynamis Medium (GIBCO A2661501). Raji (ATCC, CCL-86) and Daudi (National collection of Authenticated Cell Cultures, TCHu140) cells were cultured at 37°C in RPMI 1640 Medium (BasalMedia L220KJ) with 10% FBS. SARS-CoV-2 (Wild type, B.1.1.7, B.1.351, and P.1 variant strains) and SARS-CoV pseudoviruses were prepared and provided by the Institute for Biological Product Control, National Institutes for Food and Drug Control.
Recombinant protein generation
The SARS-CoV-2 RBD (319–533aa, accession number: QHD43416.1) tagged with C-terminal 6 × His or mouse Fc fragment, SARS-CoV-2 RBD mutants recombinant proteins (Supplementary Table 1) tagged with C-terminal 6 × His, and human ACE2 (1–615 aa, accession number: NP_068576.1) tagged with human Fc fragment were cloned into expression vector. HEK293 cells were transiently transfected with plasmids using 293fectinTM Transfection Reagent (Cat. 12347019, Life Technologies) when the cell density reached 1 × 106 cells/mL. Four days after transfection, the conditioned media was collected by centrifugation followed by purification using HisTrapTM HP (Cat. 17–5248-01, GE Healthcare), or MabSelect SuRe antibody purification resin (Cat: 29–0491-04, GE Healthcare). SEC-HPLC and SDS-PAGE were used to check the size and purity of these recombinant proteins.
ELISA
To access the binding of mAbs to recombinant proteins (SARS-CoV-2 RBD, SARS-CoV-2 RBD mutants, SARS-CoV RBD), recombinant proteins were coated on 96-well ELISA plates at 10 µg/ml in 100 µL at 4°C for overnight. After blocking with 5% bovine serum albumin in phosphate-buffered saline (PBS), serially diluted mAbs were added to the plates and incubated at 37°C for 1 hour. Plates were washed and secondary Ab Goat Anti-Human IgG Fc-HRP (Cat. 109–035-098, Jackson ImmunoResearch) was added. 3,3′,5,5′-tetramethylbenzidine (TMB) was used for color development and absorbance at 450 nm was measured using a microplate reader. For the RBD/ACE2-hFc blocking assay, ACE2-hFc recombinant protein was coated on a 96-well ELISA plate at 0.75 µg/ml in 100 µL at 4°C for overnight. The equivalent volumes (100 µL + 100 µL) of pre-incubated RBD-mFc/mAb complex (RBD-mFc concentration: 100 ng/ml, mAb concentrations between 0.00023 and 40 µg/ml) were added to the plates and incubated for 1 hour at 37°C. Plates were washed and secondary Ab Goat Anti-Mouse IgG Fc-HRP (Cat. 115–035-071, Jackson ImmunoResearch) was added. TMB was used for color development and absorbance at 450 nm was measured using a microplate reader (Multiskan MK3, ThermoFisher).
Affinity measurement and epitope competition assays
Affinity measurement and epitope competition assays were performed on Octet Red 384 system (Pall ForteBio, USA) using BLI strategy. For MW06 affinity measurement, antibody (4 µg/ml) was loaded onto the AHC biosensors (Cat. 18–5060, FORTEBIO). Following a short baseline in kinetics buffer, the loaded biosensors were exposed to a series of recombinant RBD concentrations (30–120 nM) and background subtraction was used to correct for sensor drifting. Background wavelength shifts were measured from reference biosensors that were loaded only with antibodies. ForteBio’s data analysis software was used to fit the data to a 1:1 binding model to extract the association and dissociation rates. The KD was calculated using the ratio koff/kon.
For MW05 and MW06 epitope competition assays, biotinylated MW05 or MW06 were loaded on SA biosensors (Cat. 18–0009, FORTEBIO). Then, SARS-CoV-2 RBD recombinant proteins were loaded at a concentration of 200 nM. Finally, the biosensors were exposed to the secondary antibody at a concentration of 60 nM to detect the binding. If no increased binding signal was observed with the second antibody over the binding signal of the first antibody; then, these two antibodies recognized the same epitope. In contrast, if an increased binding signal was observed, these two antibodies recognized two distinct epitopes.
Neutralization assay
For the pseudovirus neutralization assay, 100 µL of mAbs at different concentrations were mixed with 50 µL supernatant containing ~1000 TCID50 SARS-CoV-2 or SARS-CoV pseudovirus. The mixture was incubated at 37°C for 1 hour, supplied with 5% CO2. 100 µL of Huh-7 cell or Vero cell suspension (2 × 105 cells/mL) was then added to the mixtures of pseudoviruses and mAbs for an additional 24-hour incubation at 37°C. Then, 150 μL of supernatant was removed, and 100 μL luciferase detecting regents (Cat. 6066769, PerkinElmer) was added to each well. After 2 minutes of incubation, each well was mixed 6–8 times by pipetting, and 150 μL of the mixture was transferred to a new microplate. Luciferase activity was measured using a microplate luminometer (SpectraMax L, Molecular Devices). The 50% neutralization titer (NT50) was calculated using GraphPad Prism 8.2.1.
For authentic virus neutralization assays, Vero E6 and Huh-7/ACE2 cells in logarithmic growth phase were digested with trypsin to prepare a cell suspension with a cell density of 4 × 105 cells/ml (Vero E6) and 2 × 105 cells/ml (Huh-7/ACE2), then seeded into 96-well plates with 100 μL per well (4 × 104 cells per well). 60 μL of the serially diluted test samples were mixed with 60 μL of the SARS-CoV-2 virus with a titer of 100 TCID50/50 μL. The mixture was then incubated at 37°C for 1 hour. Media in the cell plates were discarded. 100 μL of test antibody-virus mixture was then added into the cell culture plates, and cultured at 37°C for 1 hour for viral infection. After viral infection, liquid supernatant in the culture plate was aspirated. The plate was washed twice with 1x PBS. The maintenance medium (DMEM medium containing 2% of FBS for CPE assay, additional 1% of methylcellulose was added for plaque reduction assay) was added into the samples followed by culturing at 37°C for 72–96 hours. After fixation and stained by crystal violet, plaque counts were recorded and analyzed. CCK-8 reagent (10%, 50 μL per well) was used to measure the cell viability for each well. The 50% neutralization titer (NT50) was calculated using GraphPad Prism 8.2.1.
Fab generation
The variable region of the heavy chain coding sequence of MW06 was cloned into the upstream of a human IgG1 CH1 domain sequence to produce VH-CH1 expression vector. The light chain expression vector was generated by inserting the variable region of light chain into the upstream of the human Kappa light chain constant region. For Fab generation, VH-CH1 and light-chain expression plasmids were transiently co-transfected into HEK293 cells. Four days after transfection, the conditioned media was collected by centrifugation followed by purification using Protein G affinity column (Cat. 17–5248-01, Cytiva). The purified protein was buffer exchanged into PBS using a Vivacon 500 concentrator (Cat. VS0122, Sartorius Stedim). SEC-HPLC and SDS-PAGE were used to check the size and purity of MW06 Fab.
Generation of Fab/RBD complex
For the preparation of RBD/MW06 Fab complex, SARS-CoV-2 RBD recombinant protein tagged with an N-terminal 6 × His tag together with MW06 Fab were mixed at a molar ratio of 1:1 in a 20 mM Tris,150 mM NaCl buffer. After incubation at room temperature for 2 hours, the mixture was concentrated by an ultrafiltration concentrator (Cat. VS2002, Sartorius Stedim) to a concentration of 20 mg/mL in a volume of 0.3 mL. SEC-HPLC and SDS-PAGE were used to check their size and purity.
Crystallization, data collection, structure determination, and spike trimer-binding modeling
The crystals of SARS-CoV-2 RBD/MW06 Fab complex were grown in a buffer composed of 0.1 M citric acid, pH 3.5, 12% (v/v) ethylene glycol, and 14% (w/v) polyethylene glycol 6000 using the sitting-drop vapor-diffusion method at 16°C. The crystals were soaked in reservoir solutions with additional 10% glycerol for several seconds and flash-frozen in liquid nitrogen before diffraction data collection. The X-ray diffraction data were collected at beamline BL19U1Citation36 of the National Facility for Protein Science in Shanghai (NFPS) at the Shanghai Synchrotron Radiation Facility (SSRF) with a wavelength of 0.9785 Å, using a Pilatus 6 M detector. The data were indexed, integrated, and scaled with HKL3000 software.Citation37 The structure was solved by the molecular replacement method with PHASERCitation38 in the CCP4 suite,Citation39 using the Spike RBD structure in the SARS-CoV2 Spike RBD/hACE2 complex (PDB code 6LZG)Citation14 and the Fab structure in the MW317-PD1 complex (PDB code 6JJP)Citation9 as the search models. The model was rebuilt with COOTCitation40 and refined with PHENIX.Citation41 Coordinates and structure factors have been deposited into the Protein Data Bank (PDB) under an accession number of 7DPM. Detailed statistics for data collection and structure determination are summarized in Supplementary Tables 2 and 3. The models of MW06 Fab bound to “close”, “open”, and “3 up” Spike trimers were generated by superimposing the RBD in the RBD/MW06 Fab complex with RBDs in the “close” Spike trimer (PDB code 6VXX),Citation16 “open” Spike trimer (PDB code 6VYB),Citation16 and “3-up” Spike trimer-hACE2 complex (PDB code 7A98)Citation17 structures, respectively.
Sequence conservation analysis
RBD amino acid sequences from SARS-CoV-2, SARS-CoV, and SARS-related coronavirus (SARSr-CoV) strains were retrieved from the following accession codes:
UniProt P0DTC2 (SARS-CoV-2), GenBank ABF65836.1 (SARS-CoV), GenBank ALK02457.1 (Bat SARSr-CoV WIV16), GenBank AGZ48828.1 (Bat SARSr-CoV WIV1), GenBank ACU31032.1 (Bat SARSr-CoV Rs672), GenBank AIA62320.1 (Bat SARSr-CoV GX2013), GenBank AAZ67052.1 (Bat SARSr-CoV Rp3), GenBank AIA62300.1 (Bat SARSr-CoV SX2013), GenBank ABD75323.1 (Bat SARSr-CoV Rf1), GenBank AIA62310.1 (Bat SARSr-CoV HuB2013), GenBank AAY88866.1 (Bat SARSr-CoV HKU3-1), GenBank AID16716.1 (Bat SARSr-CoV Longquan-140), GenBank AVP78031.1 (Bat SARSr-CoV ZC45), GenBank AVP78042.1 (Bat SARSr-CoV ZXC21), GenBank QHR63300.2 (Bat CoV RaTG13), UniProt A0A6G9KP06 (Pangolin BetaCoV Guandong2019), NCBI Reference Sequence YP_003858584.1 (Bat SARSr-CoV BM48-31). Multiple sequence alignment of the RBD sequences was performed by Vector NTI Advance version 11.5.1.
Antibody-dependent enhancement
The ADE assays were performed using Raji and Daudi cell lines. 25 µL of serially diluted mAbs were mixed with 25 µL supernatant containing 750 TCID50 SARS-CoV-2 or SARS-CoV pseudovirus. The mixture was incubated for 1 h at 37°C. 50 µL cells at the density of 2 × 106 cells/mL were added to the mixtures of pseudoviruses and mAbs for an additional 24-h incubation. Then, the same volume of luciferase detection reagents (Cat. 6066769, PerkinElmer) was added to each well. After 2 minutes of incubation, the luciferase activity was measured using a microplate luminometer (SpectraMax i3x, Molecular Devices).
Supplemental Material
Download Zip (467.6 KB)Acknowledgments
We thank Dr. Cuicui Guo for reviewing and editing this manuscript. This work was supported by the National Key R&D Program (grant number 2020YFC0848600).
Disclosure statement
X.G., S.W., R.W., S.J., and J.Z. are listed as inventors on the filing patent for MW06. W.J., S.J., B.C., R.W., H.C., A.W., G.L., J.Z., and X.G. are employees of Mabwell (Shanghai) Bioscience Co., Ltd. and may hold shares in Mabwell (Shanghai) Bioscience Co., Ltd. The other authors declare no competing interests.
Data availability statement
The crystal structure for the complex of SARS-CoV-2 RBD/MW06 has been deposited in the Protein Data Bank (www.rcsb.org), under the accession no. 7DPM. All other data to support the conclusions are in the main paper or supplementary materials.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
Additional information
Funding
References
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–12. doi:10.1038/s41586-020-2012-7.
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–69.
- Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11(1):1620. doi:10.1038/s41467-020-15562-9.
- Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–80 e8. doi:10.1016/j.cell.2020.02.052.
- Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–20. doi:10.1038/s41586-020-2180-5.
- Li F. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–68. doi:10.1126/science.1116480.
- Sui J, Li W, Murakami A, Tamin A, Matthews LJ, Wong SK, Moore MJ, Tallarico ASC, Olurinde M, Choe H, et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci U S A. 2004;101(8):2536–41. doi:10.1073/pnas.0307140101.
- ter Meulen J, Bakker AB, En EN, Weverling GJ, Martina BE, Haagmans BL, Kuiken T, de Kruif J, Preiser W, Spaan W, et al. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet. 2004;363(9427):2139–41. doi:10.1016/S0140-6736(04)16506-9.
- Wang S, Peng Y, Wang R, Jiao S, Wang M, Huang W, Shan C, Jiang W, Li Z, Gu C, et al. Characterization of neutralizing antibody with prophylactic and therapeutic efficacy against SARS-CoV-2 in rhesus monkeys. Nat Commun. 2020;11(1):5752. doi:10.1038/s41467-020-19568-1.
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–33. doi:10.1056/NEJMoa2001017.
- Shi R, Shan C, Duan X, Chen Z, Liu P, Song J, Song T, Bi X, Han C, Wu L, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584(7819):120–24. doi:10.1038/s41586-020-2381-y.
- Zost SJ, Gilchuk P, Chen RE, Case JB, Reidy JX, Trivette A, Nargi RS, Sutton RE, Suryadevara N, Chen EC, et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat Med. 2020;26:1422–27. doi:10.1038/s41591-020-0998-x.
- Wu Y, Wang F, Shen C, Peng W, Li D, Zhao C, Li Z, Li S, Bi Y, Yang Y, et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–78. doi:10.1126/science.abc2241.
- Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen K-Y, et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181(4):894–904 e9. doi:10.1016/j.cell.2020.03.045.
- Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler VD. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181(2):281–92 e6. doi:10.1016/j.cell.2020.02.058.
- Ke Z, Oton J, Qu K, Cortese M, Zila V, McKeane L, Nakane T, Zivanov J, Neufeldt CJ, Cerikan B, et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588(7838):498–502. doi:10.1038/s41586-020-2665-2.
- Benton DJ, Wrobel AG, Xu P, Roustan C, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588(7837):327–30. doi:10.1038/s41586-020-2772-0.
- Jaume M, Yip MS, Cheung CY, Leung HL, Li PH, Kien F, Dutry I, Callendret B, Escriou N, Altmeyer R, et al. Anti-Severe Acute Respiratory Syndrome Coronavirus Spike Antibodies Trigger Infection of Human Immune Cells via a pH- and Cysteine Protease-Independent Fc R Pathway. J Virol. 2011;85(20):10582–97. doi:10.1128/JVI.00671-11.
- Jaume M, Yip MS, Kam YW, Cheung CY, Kien F, Roberts A, Li PH, Dutry I, Escriou N, Daeron M, et al. SARS CoV subunit vaccine: antibody-mediated neutralisation and enhancement. Hong Kong Med J. 2012;18(Suppl 2):31–36.
- Liu L, Wei Q, Lin Q, Fang J, Wang H, Kwok H, Tang H, Nishiura K, Peng J, Tan Z, et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4(4):e123158. doi:10.1172/jci.insight.123158.
- Wan Y, Shang J, Sun S, Tai W, Chen J, Geng Q, He L, Chen Y, Wu J, Shi Z, et al. Molecular Mechanism for Antibody-Dependent Enhancement of Coronavirus Entry. J Virol. 2020;94(5):e02015-19.
- Planas D, Bruel T, Grzelak L, Guivel-Benhassine F, Staropoli I, Porrot F, Planchais C, Buchrieser J, Rajah MM, Bishop E, et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med. 2021;27(5):917–24. doi:10.1038/s41591-021-01318-5.
- Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, Wang M, Yu J, Zhang B, Kwong PD, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–35. doi:10.1038/s41586-021-03398-2.
- Yang L, Liu W, Yu X, Wu M, Reichert JM, Ho M. COVID-19 antibody therapeutics tracker: a global online database of antibody therapeutics for the prevention and treatment of COVID-19. Antib Ther. 2020;3:205–12.
- Tu YF, Chien CS, Yarmishyn AA, Lin YY, Luo YH, Lin YT, Lai WY, Yang DM, Chou SJ, Yang YP, et al. A Review of SARS-CoV-2 and the Ongoing Clinical Trials. Int J Mol Sci. 2020;21:2657.
- Yuan M, Wu NC, Zhu X, Lee CD, So RTY, Lv H, Mok CKP, Wilson IA. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368(6491):630–33. doi:10.1126/science.abb7269.
- Pinto D, Park Y-J, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583(7815):290–95. doi:10.1038/s41586-020-2349-y.
- Liu H, Wu NC, Yuan M, Bangaru S, Torres JL, Caniels TG, van Schooten J, Zhu X, Lee CCD, Brouwer PJM, et al. Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity. Immunity. 2020;53(6):1272–80 e5. doi:10.1016/j.immuni.2020.10.023.
- Wan Y, Shang J, Sun S, Tai W, Chen J, Geng Q, He L, Chen Y, Wu J, Shi Z, et al. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J Virol. 2020;94(5):e02015-19.
- Yip MS, Leung HL, Li PH, Cheung CY, Dutry I, Li D, Daeron M, Bruzzone R, Peiris JS, Jaume M. Antibody-dependent enhancement of SARS coronavirus infection and its role in the pathogenesis of SARS. Hong Kong Med J. 2016;22:25–31.
- Arvin AM, Fink K, Schmid MA, Cathcart A, Spreafico R, Havenar-Daughton C, Lanzavecchia A, Corti D, Virgin HW. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584(7821):353–63. doi:10.1038/s41586-020-2538-8.
- Wang TT, Sewatanon J, Memoli MJ, Wrammert J, Bournazos S, Bhaumik SK, Pinsky BA, Chokephaibulkit K, Onlamoon N, Pattanapanyasat K, et al. IgG antibodies to dengue enhanced for FcγRIIIA binding determine disease severity. Science. 2017;355(6323):395–98. doi:10.1126/science.aai8128.
- Mulangu S, Dodd LE, Davey RT Jr., Tshiani Mbaya O, Proschan M, Mukadi D, Lusakibanza Manzo M, Nzolo D, Tshomba Oloma A, Ibanda A, et al. A Randomized, Controlled Trial of Ebola Virus Disease Therapeutics. N Engl J Med. 2019;381(24):2293–303. doi:10.1056/NEJMoa1910993.
- Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, Musser BJ, Soo Y, Rofail D, Im J, et al. REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N Engl J Med. 2021;384(3):238–51.
- Starr TN, Greaney AJ, Addetia A, Hannon WW, Choudhary MC, Dingens AS, Li JZ, Bloom JD. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371(6531):850–54. doi:10.1126/science.abf9302.
- Zhang W-Z, Tang J-C, Wang -S-S, Wang Z-J, Qin W-M, He J-H. The protein complex crystallography beamline (BL19U1) at the Shanghai Synchrotron Radiation Facility. Nucl Sci Tech. 2019;30(11):170.
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–26.
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40(4):658–74. doi:10.1107/S0021889807021206.
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(4):235–42. doi:10.1107/S0907444910045749.
- Emsley P, Cowtan K. Coot : model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(12):2126–32. doi:10.1107/S0907444904019158.
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX : a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(2):213–21. doi:10.1107/S0907444909052925.
