ABSTRACT
Arabinogalactan-proteins (AGPs) are ubiquitous cell wall components present throughout the plant kingdom. They are extensively post translationally modified by conversion of proline to hydroxyproline (Hyp) and by addition of arabinogalactan (AG) polysaccharides to Hyp residues. Two small gene subfamilies within the CAZy GT31 family, referred to as Hyp-galactosyltransferases (Hyp-GALTs and HPGTs), encode enzymes that specifically add galactose to AGP protein backbones as revealed by heterologous expression of the genes coupled with an in vitro enzyme assay and by biochemical characterization of the genetic knock-out mutants. Biochemical analysis of galt2galt5 double and hpgt1hpgt2hpgt3 triple knockout mutants revealed significant reductions in both AGP-specific Hyp-GALT activity and β-Gal-Yariv precipitable AGPs. Further analysis of these mutants demonstrated both overlapping and distinct pleiotropic growth and development phenotypes, indicating the important contributions of the carbohydrate moieties toward AGP function. Current research indicates that all 8 Hyp-GALT/HPGT genes encode enzymes that catalyze the initial step for AGP glycosylation and that AGP glycans play essential roles in plant growth and development.
Introduction
Arabinogalactan-proteins (AGPs) are extensively glycosylated hydroxyproline (Hyp)-rich proteins found in cell walls, plasma membranes, and extracellular secretions of plants. They are involved in various aspects of plant growth and development such as cell proliferation, cell viability, cell division, reproductive development, regeneration of somatic embryos, abiotic stress, and cellular signaling.Citation1-13 AGPs are widely distributed in the plant kingdom, being present in land plants as well as in bryophytes like Physcomitrella patens, basal angiosperms, and even in algae.Citation14-16 AGPs also have several important industrial and biomedical applications including a direct food additive, a food emulsifier, a stabilizer, a sweetener, and an immunomodulatory molecule.Citation17
Structurally, AGPs are diverse and heterogeneous due in part to their various protein backbones as well as the extent and degree of AG polysaccharide addition to the backbones. AGP protein backbones typically consist of repetitive dipeptide motifs such as Ala-Pro, Ser-Pro, Thr-Pro, and Val-Pro in which the Pro residues are converted to Hyp.Citation18 About 40% of the AGPs are tethered to the plasma membrane by a glycosylphosphatidylinositol (GPI) anchor.Citation19,20 AGP glycans or arabinogalactans (AGs) are characterized by the presence of a β−1,3-galactan backbone which is substituted with β−1,6-galactan side chains that are decorated further with arabinose, and less frequently with fucose, rhamnose, and (methyl) glucuronic acid.Citation21 Using NMR analysis of synthetic AGP peptides expressed in tobacco BY2 cells, Tan et al., however, concluded that the AG backbone is composed of repeating β−1,3-galactotriose units with or without side chains, which are connected by β−1,6-linkages, to generate a kinked galactan backbone.Citation22 While debate continues over these 2 models of AG structure, it should be noted that based on linkage and mass spectroscopy analysis, longer β−1,6-galactan side chains have been reported for AGPs from radish root, wheat flour and Arabidopsis leaf.Citation23-25
Several glycosyltransferases (GTs) are involved in assembling the AG chains, and these GTs likely coordinate and regulate the density, length and sequence of the AG chains. Given that the sugar side chains typically account for more than 90% of the mass of AGPs, sugars likely define the interactive molecular surface of AGPs and govern their function. Several of the GTs for AG biosynthesis are known and cloned; however, many await discovery as previously reviewed.Citation21,26 O-glycosylation of AGPs is initiated by a set of 8 Hyp-galactosyltransferases (Hyp-GALTs/HPGTs), which are members of the GT31 family of the CAZy database and designated as GALT2-6 and HPGT1-3.Citation12,27-29 These enzymes add a single galactose residue to Hyp residues in AGP core proteins. Other known and cloned GTs include: one β−1,3-galactosyltransferase in GT31, 2 β−1,6-galactosyltransferases, GALT31A and GALT29A in GT31 and GT29 respectively, 3 β-glucuronosyltransferases, GlcAT14A, GlcAT14B, and GlcAT14C, in GT14,Citation34,35 2 α-fucosyltransferases, FUT4 and FUT6, in GT37, and one β−arabinosyltransferase, Reduced Arabinose Yariv1 or RAY1, in GT77.Citation30-38 Several other GTs for AG biosynthesis await identification and cloning, including α–arabinosyltransferases, α–rhamnosyltransferases, and other β-galactosyltransferases.
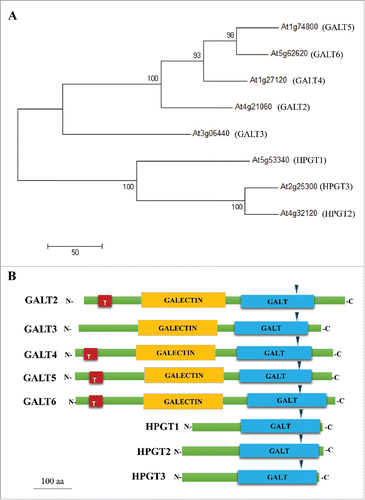
With regard to the 2 small gene families within GT31 that exhibit Hyp-GALT enzyme activity, which are the focus of this addendum, some additional information is warranted. First, the functional identity of these genes was verified by heterologous expression of the genes coupled with an in vitro enzyme assay and by biochemical characterization of genetic knock-out mutants.Citation12,27-29 Second, one gene family consists of 5 genes/enzymes (GALT2-At4g21060, GALT3-At3g06440, GALT4-At1g27120, GALT5-At1g74800, and GALT6-At5g62620) each of which contains both a GALT domain as well as a GALECTIN domain, while the other family consists of 3 genes/enzymes (HPGT1-At5g53340, HPGT2-At4g32120, and HPGT3-At2g25300), each of which contains only a GALT domain.Citation12,27 Third, this paper presents the first direct comparison between members of these 2 related gene families in terms of their biochemical activity and mutant phenotypes.
Results and discussion
The importance of AGPs is reflected in their wide range of physiological functions related to vegetative processes, sexual reproduction, development, and signaling throughout the plant kingdom. Given the complexity and heterogeneity of the numerous AGPs, little is known about the contribution of AG polysaccharides to AGP function. Identification of the genes responsible for AGP glycosylation and their genetic mutants, however, is beginning to reveal such contributions.
In vitro GALT assays and functional analysis led to the identification of 8 Hyp-GALTs within the GT31 family.Citation12,27,28 It is interesting that one subclade consists of 5 proteins (named GALT2-6) each with 2 protein domains, a catalytic GALT domain and a GALECTIN domain, whereas the other subclade consists of 3 proteins (named HPGT1-3), each of which only contains a GALT domain ().
Figure 1. In silico analysis of Hyp-GALTs for AGPs. (A) Phylogenetic tree of the Arabidopsis Hyp-GALT members which act on AGPs. The glycosyltransferase sequences were retrieved from the Carbohydrate-Active enZYmes (CAZY) database (http://www.cazy.org/GT1_eukaryota.html) and NCBI database. The full-length amino acid sequences of the Arabidopsis Hyp-GALTs were aligned with Clustal Omega and the maximum parsimony tree was generated using MEGA 6.Citation39 Numbers at the nodes indicate bootstrap values calculated for 1000 replicates (>50%). The scale bar represents 50 amino acid substitutions. (B) Protein structure of the 8 Hyp-GALT genes found in Arabidopsis. Blue arrowheads indicate the position of the DXD motif, which is predicted for UDP-galactose binding. TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) was used to predict the transmembrane domain (T); Pfam domain predictions: Pf01762 identified the Galactosyltransferase (GALT) domain (http://www.sanger.ac.uk/Software/Pfam/) and Pf00337 identified the Galactose-binding lectin (GALECTIN) domain.
Based on genotyping and phenotypic analysis of single, double, and triple T-DNA insertion mutants for the GALT and HPGT genes, these lines have a number of phenotypic alterations compared to wild-type.Citation12,27 In particular, here we directly examined and compared the galt2galt5 double mutant with the hpgt1hpgt2hpgt3 triple mutant. The galt double and hpgt triple knockout mutants displayed 43% and 70% reductions in β-Gal-Yariv precipitable AGPs compared to wild type, respectively. In addition, the double and triple mutants showed a 31% and 18% decrease in Hyp-galactosylation activity compared to wild type using our in vitro enzyme assay, respectively (). These results indicate that both subclades affect AGPs and that a greater reduction is observed when 3 genes were knocked out as opposed to two. However, it is not known whether this greater reduction is due to the greater effect of the HPGT genes or to the greater number of genes that were knocked out (i.e., 3 versus 2). It is also interesting to note that while both the double and triple mutants show a decrease in Hyp-GALT activity using our in vitro assay, the decrease is not proportional to the reductions in β-Gal-Yariv precipitable AGPs observed in these mutants. One likely explanation for this is that HPGTs may have a different AGP peptide substrate preference compared to the GALTs.
Figure 2. Biochemical phenotype analysis of the galt2galt5 double mutant and the hpgt1hpgt2hpgt3 triple mutant. AGPs were extracted from leaves (5 g) of 3 week old WT, galt2galt5 and hpgt1hpgt2hpgt3 plants as described in Schultz et al.Citation40 AGPs were quantified following the method described by Gao et al.Citation41 The GALT assay contained 100 μg of detergent (1% Triton X-100) permealized Golgi enriched microsomes as the enzyme source, [AO]7 as the acceptor substrate, 3 µM UDP-[14C]Gal as the sugar donor, 0.1 M HEPES-KOH, pH 7, 0.4 M Sucrose, 0.1% BSA, 1 mM dithiothreitol, 5 mM MgCl2, 5 mM MnCl2, 1 mM phenylmethylsulfonyl fluoride, and one tablet of Roche EDTA-free complete protease inhibitor cocktail. The assays were incubated at room temperature for 2 h and analyzed by reverse phase HPLC. The assay reactions were terminated by mixing with 400 μL anion-exchange resin (DOWEX 1×8–100), and incorporation of UDP-[14C]Gal was determined using a liquid scintillation counter. Data represent the mean and SD of n = 2 independent experiments. Different letters indicate statistically significant differences (one way ANOVA, P < 0.05); blue letters denote differences in AGP content whereas gray letters denote differences in GALT activity.
![Figure 2. Biochemical phenotype analysis of the galt2galt5 double mutant and the hpgt1hpgt2hpgt3 triple mutant. AGPs were extracted from leaves (5 g) of 3 week old WT, galt2galt5 and hpgt1hpgt2hpgt3 plants as described in Schultz et al.Citation40 AGPs were quantified following the method described by Gao et al.Citation41 The GALT assay contained 100 μg of detergent (1% Triton X-100) permealized Golgi enriched microsomes as the enzyme source, [AO]7 as the acceptor substrate, 3 µM UDP-[14C]Gal as the sugar donor, 0.1 M HEPES-KOH, pH 7, 0.4 M Sucrose, 0.1% BSA, 1 mM dithiothreitol, 5 mM MgCl2, 5 mM MnCl2, 1 mM phenylmethylsulfonyl fluoride, and one tablet of Roche EDTA-free complete protease inhibitor cocktail. The assays were incubated at room temperature for 2 h and analyzed by reverse phase HPLC. The assay reactions were terminated by mixing with 400 μL anion-exchange resin (DOWEX 1×8–100), and incorporation of UDP-[14C]Gal was determined using a liquid scintillation counter. Data represent the mean and SD of n = 2 independent experiments. Different letters indicate statistically significant differences (one way ANOVA, P < 0.05); blue letters denote differences in AGP content whereas gray letters denote differences in GALT activity.](/cms/asset/0b73970c-a1c8-40f7-bd39-2a8a41f725da/kcib_a_1177687_f0002_oc.gif)
The reduced AGP glycosylation and reduced Hyp-GALT activity observed in the galt and hpgt single and higher order mutants were also reflected in their physiological phenotypes.Citation12,27 Both the galt and the hpgt mutants demonstrated pleiotropic effects, which were generally subtle or non-existent in the single mutants but were more severe in the galt double and hpgt triple mutants. The galt mutants demonstrated reduced root hair growth, reduced seed coat mucilage, reduced seed set, and accelerated leaf senescence. The galt mutants also displayed several conditional phenotypes including impaired root growth and root tip swelling in response to salt and sucrose; these mutants were also less sensitive to the growth inhibitory effects of Yariv reagent in root and pollen tubes. In contrast, while the hpgt single mutants showed no obvious phenotypes, the hpgt triple mutants exhibited the pleotropic phenotypes of longer lateral roots, longer and denser root hairs, swollen root tips, smaller leaves, shorter inflorescence stems, reduced fertility, and shorter siliques. Thus, while the galt and hpgt mutant phenotypes share some similarities, there are also some differences, which may reflect the action of the GALTs and HPGTs on different sets of AGPs. Regardless of these differences, these results provide clear evidence that Hyp-linked AG polysaccharide chains of AGPs, which are initiated by the action of the GALTs and HPGTs, are essential for AGP function.
Finally, identification of these 8 Hyp-GALTs raises the following questions that should be addressed in future studies. 1. Do these 8 enzymes act on all AGPs or only on a subset of AGPs? 2. Do these enzymes act in a complex as is apparently the case for biosynthesis of the β−1,6-galactan side chains by GALT31A-GALT29A?Citation32 3. Do these enzymes have different preferences for AGP peptide sequences? 4. Are there any other AGP-specific Hyp-GALTs/HPGTs that remain to be identified and characterized? Nonetheless, based upon the progress to date, we are now beginning to understand how the Hyp-AG polysaccharides of AGPs are made and their important contributions to biological function.
Abbreviations
| AG | = | arabinogalactan |
| AGPs | = | arabinogalactan-proteins |
| HPGT | = | hydroxyproline galactosyltransferase |
| Hyp | = | hydroxyproline |
| GALT | = | galactosyltransferase |
| GTs | = | glycosyltransferases |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- van Hengel AJ, Roberts K. Fucosylated arabinogalactan-proteins are required for full root cell elongation in Arabidopsis. Plant J 2002; 32:105-13; PMID:12366804; http://dx.doi.org/10.1046/j.1365-313X.2002.01406.x
- Marzec M, Szarejko I, Melzer M. Arabinogalactan proteins are involved in root hair development in barley. J Exp Bot 2015; 66:1245-57; PMID:25465033; http://dx.doi.org/10.1093/jxb/eru475
- Gao M, Showalter AM. Yariv reagent treatment induces programmed cell death in Arabidopsis cell cultures and implicates arabinogalactan-protein involvement. Plant J 1999; 19:321-31; PMID:10476079; http://dx.doi.org/10.1046/j.1365-313X.1999.00544.x
- Chaves I, Regalado AP, Chen M, Ricardo CP, Showalter AM. Programmed cell death induced by (β-D-galactosyl)3 Yariv reagent in Nicotiana tabacum BY-2 suspension-cultured cells. Physiol Plant 2002; 116:548-53; http://dx.doi.org/10.1034/j.1399-3054.2002.1160414.x
- Levitin B, Richter D, Markovich I, Zik M. Arabinogalactan proteins 6 and 11 are required for stamen and pollen function in Arabidopsis. Plant J 2008; 56:351-63; PMID:18644001; http://dx.doi.org/10.1111/j.1365-313X.2008.03607.x
- Demesa-Arevalo E, Vielle-Calzada JP. The classical arabinogalactan protein AGP18 mediates megaspore selection in Arabidopsis. Plant Cell 2013; 25:1274-87; PMID:23572547; http://dx.doi.org/10.1105/tpc.112.106237
- Coimbra S, Costa M, Mendes MA, Pereira AM, Pinto J, Pereira LG. Early germination of Arabidopsis pollen in a double null mutant for the arabinogalactan protein genes AGP6 and AGP11. Sex Plant Reprod 2010; 23:199-205; PMID:20162305; http://dx.doi.org/10.1007/s00497-010-0136-x
- Qin Y, Zhao J. Localization of arabinogalactan-proteins in different stages of embryos and their role in cotyledon formation of Nicotiana tabacum L. Sex Plant Reprod 2007; 20:213-24; http://dx.doi.org/10.1007/s00497-007-0058-4
- Shi H, Kim Y, Guo Y, Stevenson B, Zhu JK. The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 2003; 15:19-32; PMID:12509519; http://dx.doi.org/10.1105/tpc.007872
- Motose H, Sugiyama M, Fukuda H. A proteoglycan mediates inductive interaction during plant vascular development. Nature 2004; 429:873-8; PMID:15215864; http://dx.doi.org/10.1038/nature02613
- Gens JS, Fujiki M, Pickard BG. Arabinogalactan proteins and wall associated kinases in a plasmalemmal reticulum with specialized vesicles. Protoplasma 2000; 212:115-34; PMID:11543565; http://dx.doi.org/10.1007/BF01279353
- Basu D, Tian L, Wang W, Bobbs S, Herock H, Travers A, Showalter AM. A small multigene hydroxyproline-O-galactosyltransferase family functions in arabinogalactan-protein glycosylation, growth and development in Arabidopsis. BMC Plant Biology 2015; 15:295; PMID:26690932; http://dx.doi.org/10.1186/s12870-015-0670-7
- Lamport DTA, Várnai P. Periplasmic arabinogalactan glycoproteins act as a calcium capacitor that regulates plant growth and development. New Phytologist 2013; 197:58-64; http://dx.doi.org/10.1111/nph.12005
- Fu H, Yadav MP, Nothnagel EA. Physcomitrella patens arabinogalactan proteins contain abundant terminal 3-O-methyl- L -rhamnosyl residues not found in angiosperms. Planta 2007; 226:1511-24; PMID:17653569; http://dx.doi.org/10.1007/s00425-007-0587-y
- Costa M, Pereira AM, Rudall PJ, Coimbra S. Immunolocalization of arabinogalactan proteins (AGPs) in reproductive structures of an early-divergent angiosperm, Trithuria (Hydatellaceae). Ann Bot 2013; 111:183-90; PMID:23186834; http://dx.doi.org/10.1093/aob/mcs256
- Popper ZA, Michel G, Herve C, Domozych D S, Willats W G T, Tuohy M G, Kloareg B, Stengel D B. Evolution and diversity of plant cell walls: from algae to flowering plants. Annu Rev Plant Biol 2011; 62:567-90; PMID:21351878; http://dx.doi.org/10.1146/annurev-arplant-042110-103809
- Classen B, Thude S, Blaschek W, Wack M, Bodinet C. Immunomodulatory effects of arabinogalactan–proteins from Baptisia and Echinacea. Phytomedicine 2006; 13:688-94; PMID:17085292; http://dx.doi.org/10.1016/j.phymed.2005.10.004
- Kieliszewski MJ. The latest hype on Hyp-O-glycosylation codes. Phytochemistry 2001; 57:319-23; PMID:11393510; http://dx.doi.org/10.1016/S0031-9422(01)00029-2
- Schultz CJ, Ferguson KL, Lahnstein J, Bacic A. Post-translational modifications of arabinogalactan-peptides of Arabidopsis thaliana. J Biol Chem. 2004; 279:45503-11; PMID:15322080; http://dx.doi.org/10.1074/jbc.M407594200
- Showalter AM, Keppler B, Lichtenberg J, Gu D, Welch LR. A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol 2010; 153:485-513; PMID:20395450; http://dx.doi.org/10.1104/pp.110.156554
- Tan L, Showalter AM, Egelund J, Hernandez-Sanchez A, Doblin MS, Bacic A. Arabinogalactan-proteins and the research challenges for these enigmatic plant cell surface proteoglycans. Front Plant Sci 2012; 3:1-10; PMID:22645563; http://dx.doi.org/10.3389/fpls.2012.00140
- Tan L, Varnai P, Lamport DTA, Yuan C, Xu J, Qiu F, Kieliszewski MJ. Plant O-hydroxyproline arabinogalactans are composed of repeating trigalactosyl subunits with short bifurcated side chains. J Biol Chem. 2010; 285:24575-83; PMID:20489210; http://dx.doi.org/10.1074/jbc.M109.100149
- Haque MA, Kotake T, Tsumuraya Y. Mode of action of beta-glucuronidase from Aspergillus niger on the sugar chains of arabinogalactan-protein. Biosci Biotechnol Biochem 2005; 69:2170-7; PMID:16306699; http://dx.doi.org/10.1271/bbb.69.2170
- Tryfona T, Liang HC, Kotake T, Kaneko S, Marsh J, Ichinose H, Lovegrove A, Tsumuraya Y, Shewry PR, Stephens E, et al. Carbohydrate structural analysis of wheat flour arabinogalactan protein. Carbohydr Res 2010; 345:2648-56; PMID:20971454; http://dx.doi.org/10.1016/j.carres.2010.09.018
- Tryfona T, Liang HC, Kotake T, Tsumuraya Y, Stephens E, Dupree P. Structural characterization of Arabidopsis leaf arabinogalactan polysaccharides. Plant Physiol. 2012; 160:653-66; PMID:22891237; http://dx.doi.org/10.1104/pp.112.202309
- Knoch E, Dilokpimol A, Geshi N. Arabinogalactan proteins: focus on carbohydrate active enzymes. Front Plant Sci. 2014; 11:198; PMID:24966860.
- Ogawa-Ohnishi M, Matsubayashi Y. Identification of three potent hydroxyproline O-galactosyltransferases in Arabidopsis. Plant J 2015; 81:736-46; PMID:25600942; http://dx.doi.org/10.1111/tpj.12764
- Basu D, Liang Y, Liu X, Himmeldirk K, Faik A, Kieliszewski M, Held M, Showalter AM. Functional identification of a hydroxyproline-O-galactosyltransferase specific for arabinogalactan protein biosynthesis in Arabidopsis. J Biol Chem 2013; 288:10132-143; PMID:23430255; http://dx.doi.org/10.1074/jbc.M112.432609
- Basu D, Wang W, Ma S, DeBrosse T, Poirier E, Emch K, Soukup E, Tian L, Showalter AM. Two hydroxyproline galactosyltransferases, GALT5 and GALT2, function in arabinogalactan-protein glycosylation, growth and development in Arabidopsis. PLoS One 2015; 10:e0125624; PMID:25974423; http://dx.doi.org/10.1371/journal.pone.0125624
- Qu Y, Egelund J, Gilson PR, Houghton F, Gleeson PA, Schultz CJ, Bacic A. Identification of a novel group of putative Arabidopsis thaliana β-(1,3)-galactosyl transferases. Plant Mol Biol 2008; 68:43-59; PMID:18548197; http://dx.doi.org/10.1007/s11103-008-9351-3
- Geshi N, Johansen JN, Dilokpimol A, Rolland A, Belcram K, Verger S, Kotake T, Tsumuraya Y, Kaneko S, Tryfona T. A galactosyltransferase acting on arabinogalactan protein glycans is essential for embryo development in Arabidopsis. Plant J 2013; 76:128-37; PMID:23837821.
- Dilokpimol A, Poulsen CP, Vereb G, Kaneko S, Schulz A, Geshi N. Galactosyltransferases from Arabidopsis thaliana in the biosynthesis of type II arabinogalactan: molecular interaction enhances enzyme activity. BMC Plant Biol. 2014; 14:90; PMID:24693939; http://dx.doi.org/10.1186/1471-2229-14-90
- Knoch E, Dilokpimol A, Tryfona T, Poulsen C P, Xiong G, Harholt J, Petersen BL, Ulvskov P, Hadi MZ, Kotake T. A β-glucuronosyltransferase from Arabidopsis thaliana involved in biosynthesis of type II arabinogalactan has a role in cell elongation during seedling growth. Plant J 2013; 76:1016-29; PMID:24128328; http://dx.doi.org/10.1111/tpj.12353
- Dilokpimol A, Geshi N. Arabidopsis thaliana glucuronosyltransferase in family GT14. Plant Signal Behav. 2014; 9:e28891; PMID:24739253; http://dx.doi.org/10.4161/psb.28891
- Wu Y, Williams M, Bernard S, Driouich A, Showalter AM, Faik A. Functional identification of two nonredundant Arabidopsis alpha (1,2) fucosyltransferases specific to arabinogalactan proteins. J Biol Chem. 2010; 285:13638-45; PMID:20194500; http://dx.doi.org/10.1074/jbc.M110.102715
- Liang Y, Basu D, Pattathil S, Xu W-L, Venetos A, Martin SL, Faik A, Hahn MG, Showalter AM. Biochemical and physiological characterization of fut4 and fut6 mutants defective in arabinogalactan-protein fucosylation in Arabidopsis. J Exp Bot. 2013; 64:5537-51; PMID:24127514; http://dx.doi.org/10.1093/jxb/ert321
- Tryfona T, Theys TE, Wagner T, Stott K, Keegstra K, Dupree P. Characterisation of FUT4 and FUT6 α-(1®2)-Fucosyltransferases reveals that absence of root arabinogalactan fucosylation increases Arabidopsis root growth salt sensitivity. PLoS One 2014; 9:e93291; PMID:24667545.
- Gille S, Sharma V, Baidoo EEK, Keasling JD, Scheller HV, Pauly M. Arabinosylation of a Yariv-precipitable cell wall polymer impacts plant growth as exemplified by the Arabidopsis glycosyltransferase mutant ray1. Mol Plant 2013; 6:1369-72; PMID:23396039; http://dx.doi.org/10.1093/mp/sst029
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol 2013; 30:2725-9; PMID:24132122; http://dx.doi.org/10.1093/molbev/mst197
- Schultz CJ, Johnson KL, Currie G, Bacic A. The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell 2000; 12:1751-67; PMID:11006345; http://dx.doi.org/10.1105/tpc.12.9.1751
- Gao M, Kieliszewski MJ, Lamport DT, Showalter AM. Isolation, characterization, and immunolocalization of a novel, modular tomato arabinogalactan protein corresponding to the LeAGP-1 gene. Plant J 1999; 18:43-55; PMID:10341442; http://dx.doi.org/10.1046/j.1365-313X.1999.00428.x
