ABSTRACT
From yeast to man, an evolutionary distance of 1.3 billion years, the F-actin filament structure has been conserved largely in line with the 94% sequence identity. The situation is entirely different in bacteria. In comparison to eukaryotic actins, the bacterial actin-like proteins (ALPs) show medium to low levels of sequence identity. This is extreme in the case of the ParM family of proteins, which often display less than 20% identity. ParMs are plasmid segregation proteins that form the polymerizing motors that propel pairs of plasmids to the extremities of a cell prior to cell division, ensuring faithful inheritance of the plasmid. Recently, exotic ParM filament structures have been elucidated that show ParM filament geometries are not limited to the standard polar pair of strands typified by actin. Four-stranded non-polar ParM filaments existing as open or closed nanotubules are found in Clostridium tetani and Bacillus thuringiensis, respectively. These diverse architectures indicate that the actin fold is capable of forming a large variety of filament morphologies, and that the conception of the “actin” filament has been heavily influenced by its conservation in eukaryotes. Here, we review the history of the structure determination of the eukaryotic actin filament to give a sense of context for the discovery of the new ParM filament structures. We describe the novel ParM geometries and predict that even more complex actin-like filaments may exist in bacteria. Finally, we compare the architectures of filaments arising from the actin and tubulin folds and conclude that the basic units possess similar properties that can each form a range of structures. Thus, the use of the actin fold in microfilaments and the tubulin fold for microtubules likely arose from a wider range of filament possibilities, but became entrenched as those architectures in early eukaryotes.
The history of the eukaryotic actin filament structure
Actin has fascinated scientists for 75 years. We begin by charting the history of the elucidation of the eukaryotic actin filament structure. Actin, in its filamentous form (F-actin), was first discovered by Straub in the early 1940s as an integral component of muscle.Citation1 Twenty years later, Hanson & Lowy used the novel structural tool of the early 1960s - electron microscopy – to show that F-actin, when polymerized in vitro, forms straight right-handed helices composed of 2 tightly intertwining strands.Citation2 Using myosin, which binds F-actin strongly in the absence of nucleotide (corresponding to the rigor state of muscle), Hugh Huxley, who invented negative stain and spearheaded the early stages of structural electron microscopy, observed a unique ‘arrow head’ pattern under the electron microscope. He concluded that the 2 strands forming the F-actin helical filament were in the same orientation and the filament therefore is polar.Citation3 Huxley's myosin labeling technique for F-actin was exploited several years later to show that not only muscle cells but eukaryotic cells contain substantial amounts of F-actin,Citation4 and Pollard and colleagues in 1975 determined that the “barbed” and “pointed” ends of the filament, indicated by the arrow heads, related to the fast and slow growing ends of the filament.Citation5
Revolutionary work by Toshio Yanagida in the mid-1980s showed that actin filaments could be visualized with fluorescence microscopy by labeling with rhodamine-phalloidin, which binds tightly at the interface between 3 actin protomers.Citation6 This methodology aided the characterization of a myriad of cellular proteins associated with actin, many of which regulate the ability of actin to polymerize or depolymerize.Citation7,8 In 1990, back-to-back publications of the actin monomer X-ray structureCitation9 with the determination of its orientation within the filament, based on fitting of the monomer structure into X-ray fiber diffraction data, produced the “Holmes” model of the filament.Citation10 Despite some initial controversy,Citation11-13 this model stood the test of time, with the near-atomic resolution details being slowly revealed as cryo-electron microscopy techniques improved. The current model was determined at 3.6 Å by the Raunser group in 2016.Citation14 At the time of the Holmes model, there was no reason to think that there would be more than a single filament architecture produced by actin-like sequences. The absolute conservation of eukaryotic actin filament structure likely arises from its role as an exceptionally highly connected hubCitation15 where the “universal-actin-pool” is harnessed by several filament nucleating machineriesCitation8,16 to provide force and architecture to a wide variety of biological processes. Essentially, once the eukaryotic actin filament was integrated into more than one biological process, a high degree of negative selection pressure restricted the actin sequence and structure to become frozen in time, since genetic driftCitation17 favoring one biological process would have a negative impact on biological processes competing for actin.
Prokaryotic actin-like filaments discovered
The concept of there being a single actin began to change in the early 1990s, when the homologous structure of the non-polymerizing 70-kDa heat shock cognate protein was determined.Citation18 Subsequent bioinformatics analysis predicted that eukaryotic actin is related to a larger family of proteins including hexokinase, heat shock proteins and the bacterial proteins FtsA, MreB and StbA/ParM.Citation19 Similarity in these sequences was characterized by the conservation in amino acids participating in the ATP-binding pocket, later known as the actin fold.Citation19,20 Yet it was not until 2001 that experimental evidence emerged that functional filament-forming actins exist in prokaryotes. These bacterial actins were subsequently termed actin-like proteins (ALPs).Citation21
Three common classes of ALPs have been identified in bacteria, with a forth being the rare MamKCitation22 forming the scaffold of the magnetosome in magnetobacteria. MreBs form single stranded protofilaments, which can be arranged as antiparallel pairs, are present in most rod-shaped prokaryotes and are involved establishing cell shape.Citation23 FtsAs contain a membrane inserting C-terminal amphipathic helix and recruit the cell division protein and tubulin homolog FtsZ to the mid cell surface via FtsZ's C-terminal peptide. In concert both proteins self-organize into protofilament systems.Citation24 MreBs and FtsAs are the most commonly found ALPs in prokaryotes. Lastly the plasmid segregating actins, the first characterized being ParM from the R1 plasmid in E. coli (EcParM).Citation25 Together these ALPs all contain the conserved actin fold, but often with very low overall sequence identities of below 20%.Citation26 Despite these large variations, all monomer structures solved to date by X-ray crystallography proved to be very similar.Citation27
Variety in prokaryotic actin-like filaments
Despite the structural similarity of the monomers, this did not prove true for the filament structures of ParM's, which turned out to have huge variations. In the initial paper describing EcParM,Citation25 this filament was thought to be just a small variation of F-actin in its helical parameters. Only after more extensive EM reconstructions did it become apparent that the structure differed substantially, in being a left-handed helical filament as opposed to the right-handed F-actinCitation28 (). The evolutionary pressure that determined the handedness of the various biological filaments is not known at present. Other ParM's investigated in the following years obtained by electron microscopy (AlfA from Bacillus subtilis plasmid pBET131 (BsParM), ParM from Staphylococcus aureus plasmid pSK41 (SaParM)) showed that the helical parameters could differ even more substantially from F-actin. Yet all these filaments (EcParM, BsParM and SaParM) were still polar double stranded straight helices, like F-actin.Citation29,30
Figure 1. Two views of the structures of filaments formed from the actin (blue/orange) and tubulin (red/yellow) folds. The actins are: the twisted single-stranded crenactin from the archaeon Pyrobaculum calidifontis,Citation33,39 Caulobacter crescentus MreB filament formed from an antiparallel non-twisted pair of strands (CcMreB),Citation23 right-handed eukaryotic F-actin,Citation10 left-handed Escherichia coli ParM from the R1 plasmid (EcParM),Citation35 Clostridium tetani open nanotubules from the pE88 plasmid (CtParM) which are 2 antiparallel related copies of a parallel pair of strandsCitation31 and Bacillus thuringiensis supercoiled antiparallel filaments and nanotubules from the pBMB67 plasmid (BtParM).Citation34 The tubulins are: the eukaryotic microtubule,Citation40 Bacillus thuringiensis TubZ from the pBtoxis plasmid (BtTubZ)Citation41 and Methanococcus jannaschii FtsZ (MjFtsZ).Citation42 In addition the tubulin fold of BtubA/B from Prosthecobacter vanneervenii can form tubules comprised of 5 strands.Citation43
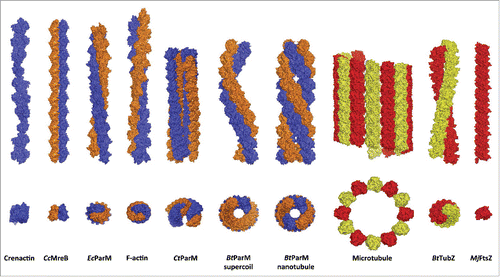
The first departure from the dogma that all ParM's formed only double stranded polar filaments came with the structure of Alp12 (CtParM) from Clostridium tetani, which segregates the pE88 plasmid encoding the lethal tetanus toxin.Citation31 CtParM formed 4-stranded filaments (2 double strands) arranged in an open cylinder separated by a wide cleft (). Subunits within a single-stranded protofilament associated through subdomain interactions that have parallels to all other known actin structures.Citation32 At the macroscopic level, subdomain 3 to subdomain 4 inter-subunit connections are the constant feature that lead to relatively unidirectional protofilaments,Citation32 despite the lack of conservation of inter-subunit interacting residues or their positions within the protein sequences.Citation33 Yet the 2 protofilaments formed a polar double-stranded filament through a completely different interface than observed for other actins. Although each double strand is polar, the 2 double stranded filaments are paired in anti-parallel fashion, by 2 β sheets of subdomain 3 to construct this novel open cylindrical architecture.Citation31
More recently, ParM from the pBMB67 plasmid in Bacillus thuringiensis (BtParM) was shown to form supercoiled, rather than straight filaments, which although double stranded, are anti-parallel rather than polar (), a far departure from the construction of F-actin or any other ParM.Citation34 ParM's are plasmid segregation polymerizing actins that are linked to the parC regions of the plasmid DNA via adaptor proteins (ParRs), which are specific to each individual ParM/parC combination. Within the ParCMR system from E. coli (EcParCMR), the ParR/parC complex was shown to pair 2 or more filaments into randomly oriented bundles.Citation35 In this system other forces, such as cellular crowding, also paired EcParM filaments in the absence of the associated molecules. The roles of the 2 competing effects (direct binding of ParR/parC versus osmotic pressure) are difficult to separate, and hence their relative contribution is ambiguous.Citation28 The ParCMR system from Bacillus thuringiensis (BtParCMR) proved to be entirely different and more amenable to teasing apart each contribution. Polymerization of BtParM in the presence of BtParR stimulated ATP hydrolysis by BtParM and formed a cylinder, comprised of 4 antiparallel strands, with inner and outer diameters of 57 Å and 145 Å, respectively, which is also formed in the presence of the BtParR/parC complex (). The structure of the BtParM cylinder is composed of 2 interwoven supercoiled antiparallel filaments, which is the geometry of the BtParM filament formed in the absence of ParR and parC (). Here osmotic pressure originating from molecular crowding also paired filaments, but in this case they form rafts of individual supercoiled filaments arranged in parallel ().Citation34 Thus in the case of BtParCMR, the unique BtParM nanotubule geometry requires a second component of the BtParCMR system, ParR or ParR/parC.
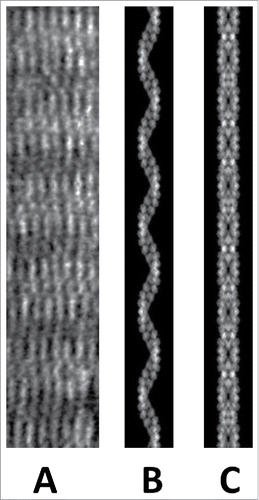
Figure 2. In the presence of molecular crowding BtParM formed rafts of filaments lying in parallel as observed by electron microscopy (A). The filaments within the rafts were not paired into a cylinder, but were supercoiled single BtParM filaments. (B) A projection image calculated from the supercoiled BtParM filament model for comparison. (C) A projection image calculated from the nanotubule model for comparison. Pairing of filaments only occurs in the presence of ParR or the ParR/parC complex.
Eukaryotic actin filaments function as in a universal-pool-of-actin in which variety in actin-binding proteins is able to harness the same molecular polymerizing motor for many cellular processes.Citation16 Prokaryotic cells have developed different mechanisms for force generation. We have previously speculated that since bacteria have one-filament-one-function systems to create force through polymerization, they evolved distinct actin filaments to power specific functional processes within a single cell.Citation16 This leads to several questions, firstly: Are there more novel actin filament architectures out there? Given the vast numbers of uncharacterized ParM's that are known to exist from phylogenetic analyses, the answer can only be “yes.” A more pertinent question is: How much further will these filaments differ from F-actin? The answer to this question will likely be quickly revealed by cryo-electron microscopy, which in just a few years has become the structural biology tool of choice to study filament systems at near-atomic resolution, due to the improvements in electron microscopes, and in particular, direct electron detectors. Finally, why did Nature create so many different designs of actin-like filaments for plasmid segregation?
The answer is probably for at least 2 reasons. Firstly, when 2 different plasmids reside in the same cell they will require different segregation systems to be faithfully inherited, hence, there may have been some positive evolutionary pressure for the ParMs to diversify. Secondly, if evolution is truly deterministic, the filament structures of plasmid segregating actin-like filaments are likely related to the sizes of the individual plasmids to be segregated. That is a ParM filament consisting of 4 strands (like CtParM) should be substantially stronger than a double stranded filament (like EcParM), thus being able to push a bigger load. This hypothesis is currently under investigation.
Comparison of actin-like and tubulin-like filament structures
Actin-like and tubulin-like filaments have transposable roles in biology. In prokaryotes, the tubulin-like protein FtsZ assembles to form the cytokinetic ring, whereas this function is performed by F-actin in eukaryotes. In contrast, plasmid DNA segregation is often orchestrated by ParM actin-like proteins in bacteria, while chromosome DNA segregation is choreographed by microtubules in eukaryotes. Comparison of the known structures formed by the actin and tubulin folds, indicates that these basic building blocks are capable of forming extensive ranges of structures. Both folds can form linear single stand filaments, twisted parallel pairs of filaments and tubules of various dimensions (). The ability to alter a tubule's dimensions, though changing the number of strands in the tubule, has been demonstrated through protein engineering to require relatively minor surface amino acid modifications in the case of barrels formed from α-helices.Citation36 Thus, various diameters of tubules formed from the actin and tubulin folds are likely to have been sampled during evolution. Whether a superior property is intrinsic to the actin fold for forming microfilament-like architectures, and to the tubulin fold for forming microtubule-like structures, or whether the use of these protein scaffolds were stochastic events that became entrenched in early eukaryotes remains open to debate. Nevertheless, it is clear that the highly specialized filament geometries of microfilaments and microtubules are just 2 of many possibilities that are available in nature.
Conclusions
The structures of F-actin and microtubules have been conserved over a billion years in eukaryotic cells. These structures are maintained through evolution by their interactions with large numbers of binding proteins, which have likely restricted their genetic drift and have allowed for the filament properties to be exploited by many cellular processes. In contrast, the one-filament-one-function design observed in many prokaryotic filaments has allowed for the adoption of a large variety of different filament structures. Despite the many variances in filament structures, 2 features are preserved between all actins. Firstly, the individual strands forming actin-like filaments (protofilaments) share grossly similar contacts, subdomains 1 and 3 from one monomer interact with subdomains 2 and subdomain 4, respectively in the neighboring monomer in the protofilaments. Secondly the nucleotide-binding site, which accepts GTP in some ALPs,Citation37 acts as a conformational switch activated by polymerization controling ATP/GTP hydrolysis and phosphate release. The ATP/GTP switch (converting the initially bound ATP/GTP to ADP-Pi/GDP-Pi and subsequently to ADP/GDP) acts as a timing mechanism, which coordinates the depolymerization of actin filaments via the conserved contacts in the protofilaments.Citation23
The determinants as to whether filaments treadmill or are dynamically unstable largely remain unexplored for the ALPs. These activities will be impacted by: 1) off and on rates for each nucleotide-bound state of the monomers at both ends of the filament in its different nucleotide-bound forms; 2) nucleotide-exchange rates in monomers and at filament ends; 3) hydrolysis and phosphate-release rates on polymerization; 4) concentrations of ALPs and nucleotides; 5) higher order mechanisms for monomer association/dissociation with the filament; and 6) binding partners. A simple case for an in vitro dynamically unstable filament occurs when the nucleotide-exchange rate for monomers is slow relative to the off rate for ADP-bound protomers at the ends off the filament. On approaching steady state the filaments will depolymerize until the dissociated monomer pool has regenerated sufficient ATP-bound monomers to support repolymerization, as is suggested for EcParM.Citation37 In contrast, in vitro treadmilling for actin results from the 2 ends of the filament having different on and off rates for monomers.Citation38 However, these are just 2 of the multitude of possible scenarios.
Filament architectures reflect their function. MreB and FtsA filaments form non-helical protofilaments allowing them to present a consistent binding interface to the membrane, which would not be possible if they were twisted. ParM's, in contrast, form 2 to 4 (and perhaps even more) stranded helical filaments. As motor proteins, the helical design brings greater rigidity to the filaments and by increasing the number of strands expands the possibilities to segregate larger payloads. The functional requirement of any ParM filament is to polymerize and interact with the specific plasmid through the complementary adaptor protein. This has allowed for far more latitude in exploring diverse filament architectures during evolution in comparison to filament systems that have more extensive interactions. Thus, we speculate that the currently known actin filament structures are likely to be a fraction of the total diversity.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Straub FB. Actin. Stud Inst Med Chem Univ Szeged 1942; II:3-15
- Hanson J, Lowy J. The structure of F-actin and of actin filaments isolated from muscle. J Mol Biol 1963; 6:46-60; http://dx.doi.org/10.1016/S0022-2836(63)80081-9
- Huxley HE. Electron microscope studies on the structure of natural and synthetic protein filaments from striated muscle. J Mol Biol 1963; 7:281-308; PMID:14064165; http://dx.doi.org/10.1016/S0022-2836(63)80008-X
- Ishikawa H, Bischoff R, Holtzer H. Formation of arrowhead complexes with heavy meromyosin in a variety of cell types. J Cell Biol 1969; 43:312-28; PMID:5344150; http://dx.doi.org/10.1083/jcb.43.2.312
- Woodrum DT, Rich SA, Pollard TD. Evidence for biased bidirectional polymerization of actin filaments using heavy meromyosin prepared by an improved method. J Cell Biol 1975; 67:231-7; PMID:240859; http://dx.doi.org/10.1083/jcb.67.1.231
- Yanagida T, Nakase M, Nishiyama K, Oosawa F. Direct observation of motion of single F-actin filaments in the presence of myosin. Nature 1984; 307:58-60; PMID:6537825; http://dx.doi.org/10.1038/307058a0
- dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol Rev 2003; 83:433-73; PMID:12663865; http://dx.doi.org/10.1152/physrev.00026.2002
- Xue B, Robinson RC. Guardians of the actin monomer. Eur J Cell Biol 2013; 92:316-32; PMID:24268205; http://dx.doi.org/10.1016/j.ejcb.2013.10.012
- Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature 1990; 347:37-44; PMID:2395459; http://dx.doi.org/10.1038/347037a0
- Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature 1990; 347:44-9; PMID:2395461; http://dx.doi.org/10.1038/347044a0
- Schutt CE, Lindberg U, Myslik J, Strauss N. Molecular packing in profilin: actin crystals and its implications. J Mol Biol 1989; 209:735-46; PMID:2585507; http://dx.doi.org/10.1016/0022-2836(89)90603-7
- Schutt CE, Rozycki MD, Lindberg U. What's the matter with the ribbon? Curr Biol 1994; 4:185-6; PMID:7953529; http://dx.doi.org/10.1016/S0960-9822(94)00046-1
- Egelman EH. Actin filament structure. The ghost of ribbons past. Curr Biol 1994; 4:79-81; PMID:7922321; http://dx.doi.org/10.1016/S0960-9822(00)00020-8
- von der Ecken J, Heissler SM, Pathan-Chhatbar S, Manstein DJ, Raunser S. Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature 2016; 534:724-8; PMID:27324845; http://dx.doi.org/10.1038/nature18295
- Carlson MR, Zhang B, Fang Z, Mischel PS, Horvath S, Nelson SF. Gene connectivity, function, and sequence conservation: predictions from modular yeast co-expression networks. BMC Genomics 2006; 7:40; PMID:16515682; http://dx.doi.org/10.1186/1471-2164-7-40
- Gunning PW, Ghoshdastider U, Whitaker S, Popp D, Robinson RC. The evolution of compositionally and functionally distinct actin filaments. J Cell Sci 2015; 128:2009-19; PMID:25788699; http://dx.doi.org/10.1242/jcs.165563
- Kimura M. Evolutionary rate at the molecular level. Nature 1968; 217:624-6; PMID:5637732; http://dx.doi.org/10.1038/217624a0
- Flaherty KM, McKay DB, Kabsch W, Holmes KC. Similarity of the three-dimensional structures of actin and the ATPase fragment of a 70-kDa heat shock cognate protein. Proc Natl Acad Sci U S A 1991; 88:5041-45; PMID:1828889; http://dx.doi.org/10.1073/pnas.88.11.5041
- Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci U S A 1992; 89:7290-4; PMID:1323828; http://dx.doi.org/10.1073/pnas.89.16.7290
- Kabsch W, Holmes KC. The actin fold. FASEB J 1995; 9:167-74; PMID:7781919
- van den Ent F, Amos LA, Lowe J. Prokaryotic origin of the actin cytoskeleton. Nature 2001; 413:39-44; PMID:11544518; http://dx.doi.org/10.1038/35092500
- Ozyamak E, Kollman J, Agard DA, Komeili A. The bacterial actin MamK: in vitro assembly behavior and filament architecture. J Biol Chem 2013; 288:4265-77; PMID:23204522; http://dx.doi.org/10.1074/jbc.M112.417030
- van den Ent F, Izore T, Bharat TA, Johnson CM, Lowe J. Bacterial actin MreB forms antiparallel double filaments. Elife 2014; 3:e02634; PMID:24843005
- Loose M, Mitchison TJ. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat Cell Biol 2014; 16:38-46; PMID:24316672; http://dx.doi.org/10.1038/ncb2885
- van den Ent F, Moller-Jensen J, Amos LA, Gerdes K, Lowe J. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J 2002; 21:6935-43; PMID:12486014; http://dx.doi.org/10.1093/emboj/cdf672
- Derman AI, Becker EC, Truong BD, Fujioka A, Tucey TM, Erb ML, Patterson PC, Pogliano J. Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol Microbiol 2009; 73:534-52; PMID:19602153; http://dx.doi.org/10.1111/j.1365-2958.2009.06771.x
- Dominguez R, Holmes KC. Actin structure and function. Annu Rev Biophys 2011; 40:169-86; PMID:21314430; http://dx.doi.org/10.1146/annurev-biophys-042910-155359
- Popp D, Narita A, Iwasa M, Maeda Y, Robinson RC. Molecular mechanism of bundle formation by the bacterial actin ParM. Biochem Biophys Res Commun 2010; 391:1598-603; PMID:20026051; http://dx.doi.org/10.1016/j.bbrc.2009.12.078
- Popp D, Narita A, Ghoshdastider U, Maeda K, Maéda Y, Oda T, Fujisawa T, Onishi H, Ito K, Robinson RC. Polymeric structures and dynamic properties of the bacterial actin AlfA. J Mol Biol 2010; 397:1031-41; PMID:20156449; http://dx.doi.org/10.1016/j.jmb.2010.02.010
- Popp D, Xu W, Narita A, Brzoska AJ, Skurray RA, Firth N, Ghoshdastider U, Maéda Y, Robinson RC, Schumacher MA. Structure and filament dynamics of the pSK41 actin-like ParM protein: implications for plasmid DNA segregation. J Biol Chem 2010; 285:10130-40; PMID:20106979; http://dx.doi.org/10.1074/jbc.M109.071613
- Popp D, Narita A, Lee LJ, Ghoshdastider U, Xue B, Srinivasan R, Balasubramanian MK, Tanaka T, Robinson RC. Novel actin-like filament structure from Clostridium tetani. J Biol Chem 2012; 287:21121-9; PMID:22514279; http://dx.doi.org/10.1074/jbc.M112.341016
- Ghoshdastider U, Jiang S, Popp D, Robinson RC. In search of the primordial actin filament. Proc Natl Acad Sci U S A 2015; 112:9150-1; PMID:26178194; http://dx.doi.org/10.1073/pnas.1511568112
- Braun T, Orlova A, Valegård K, Lindås AC, Schröder GF, Egelman EH. Archaeal actin from a hyperthermophile forms a single-stranded filament. Proc Natl Acad Sci U S A 2015; 112:9340-5; PMID:26124094; http://dx.doi.org/10.1073/pnas.1509069112
- Jiang S, Narita A, Popp D, Ghoshdastider U, Lee LJ, Srinivasan R, Balasubramanian MK, Oda T, Koh F, Larsson M, et al. Novel actin filaments from Bacillus thuringiensis form nanotubules for plasmid DNA segregation. Proc Natl Acad Sci U S A 2016; 113:E1200-1205; PMID:26873105; http://dx.doi.org/10.1073/pnas.1600129113
- Gayathri P, Fujii T, Møller-Jensen J, van den Ent F, Namba K, Löwe J. A bipolar spindle of antiparallel ParM filaments drives bacterial plasmid segregation. Science 2012; 338:1334-7; PMID:23112295; http://dx.doi.org/10.1126/science.1229091
- Woolfson DN, Bartlett GJ, Burton AJ, Heal JW, Niitsu A, Thomson AR, Wood CW. De novo protein design: how do we expand into the universe of possible protein structures? Curr Opin Struct Biol 2015; 33:16-26; PMID:26093060; http://dx.doi.org/10.1016/j.sbi.2015.05.009
- Popp D, Narita A, Lee LJ, Larsson M, Robinson RC. Microtubule-like properties of the bacterial actin homolog ParM-R1. J Biol Chem 2012; 287:37078-88; PMID:22908230; http://dx.doi.org/10.1074/jbc.M111.319491
- Neuhaus JM, Wanger M, Keiser T, Wegner A. Treadmilling of actin. J Muscle Res Cell Motil 1983; 4:507-27; PMID:6358256; http://dx.doi.org/10.1007/BF00712112
- Lindås AC, Chruszcz M, Bernander R, Valegård K. Structure of crenactin, an archaeal actin homologue active at 90°C. Acta Crystallogr D Biol Crystallogr 2014; 70:492-500; http://dx.doi.org/10.1107/S1399004714000935
- Sui H, Downing KH. Structural basis of interprotofilament interaction and lateral deformation of microtubules. Structure 2010; 18:1022-31; PMID:20696402; http://dx.doi.org/10.1016/j.str.2010.05.010
- Aylett CH, Wang Q, Michie KA, Amos LA, Lowe J. Filament structure of bacterial tubulin homologue TubZ. Proc Natl Acad Sci U S A 2010; 107:19766-71; PMID:20974911; http://dx.doi.org/10.1073/pnas.1010176107
- Oliva MA, Cordell SC, Lowe J. Structural insights into FtsZ protofilament formation. Nat Struct Mol Biol 2004; 11:1243-50; PMID:15558053; http://dx.doi.org/10.1038/nsmb855
- Pilhofer M, Ladinsky MS, McDowall AW, Petroni G, Jensen GJ. Microtubules in bacteria: Ancient tubulins build a five-protofilament homolog of the eukaryotic cytoskeleton. PLoS Biol 2011; 9:e1001213; PMID:22162949; http://dx.doi.org/10.1371/journal.pbio.1001213
