ABSTRACT
Fam49 proteins, now referred to as CYRI (CYFIP-related Rac Interactor), are evolutionarily conserved across many phyla. Their closest relative by amino acid sequence is CYFIP, as both proteins contain a domain of unknown function DUF1394. We recently showed that CYRI and the DUF1394 can mediate binding to Rac1 and evidence is building to suggest that CYRI plays important roles in cell migration, chemotaxis and pathogen entry into cells. Here we discuss how CYRI proteins fit into the current framework of the control of actin dynamics by positive and negative feedback loops containing Rac1, the Scar/WAVE Complex, the Arp2/3 Complex and branched actin. We also provide data regarding the interaction between Rac1 and CYRI in an unbiassed mass spectrometry screen for interactors of an active mutant of Rac1.
Regulation of Arp2/3 driven actin assembly at the lamellipodium
Orchestrated directional cell migration is fundamental for multiple aspects of life; from development, wound healing and immune responses to pathogens. At the core of cell migration is the cells ability to rearrange its actin cytoskeleton, which is driven predominantly by the Arp2/3 complex. During this process, extracellular cues trigger intracellular signalling cascades resulting in the activation of the small GTPases Rac1 and Cdc42 at the leading-edge plasma membrane. In the case of Rac1, these signals facilitate the interactions and subsequent activation of the WAVE-regulatory complex (WRC). The WRC is a pentameric complex composed of 5 members; CYFIP1/2 (Sra/PIR121), NckAP1/2 (Hem-1/2), WAVE1-3 (Scar1-3), HSPC300 (Brick1) and Abi1-3 [Citation1]. Interaction with Rac1 causes conformational changes in the complex, releasing the autoinhibited C-terminal Arp2/3 binding domain of WAVE, which in turn activates the Arp2/3 complex [Citation2–Citation4] ().
Figure 1. Negative regulation of actin polymerisation at the lamellipodium.
Actin polymerisation at the lamellipodium is activated through the Rac1-WRC-Arp2/3 cascade. Extracellular stimuli trigger GTP loading of the small GTPase Rac1 and plasma membrane association. Activated Rac1 interacts with WRC through CYFIP1 at either or both the A- and D-sites, releasing and presenting the VCA-domain of WAVE to the Arp2/3 complex. Arp2/3 triggers nucleation of branched F-actin at the leading edge, generating a lamellipodium. Negative regulation of Arp2/3 can occur through the interaction with Arpin, which is activated by Rac1. CYRI proteins compete with the WRC for active Rac1, thereby inhibiting Arp2/3 complex activation. The mechanism of activation of CYRI proteins to constrain protrusions is still unknown. The activation cascade follows grey arrows, while inhibitors are shown in red terminal lines.
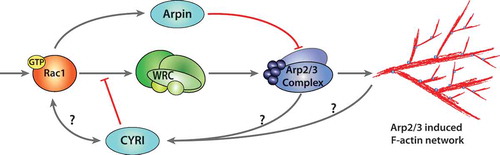
A recent study described two possible binding sites for Rac1 on the WRC. These sites are referred to as A and D- sites, with the D-site being first described by Chen et al., 2017. The D-site has been described by cryo-electron microscopy and is located within the more C-terminal fragile X mental retardation protein (FMRP) region of CYFIP [Citation3,Citation5]. Chen and colleagues found that Rac1 binds to the D-site of CYFIP with a greater affinity than the A-site alone. However, Rac1 binding to both regions on CYFIP is needed to cause full activation of WRC [Citation3]. The A- and D-sites were confirmed to interact with active Rac1 within both mammalian and Dictyostelium cells [Citation5]. Moreover, while the A-site was required for WRC activation, the D-site was surprisingly dispensable for WRC activation but necessary for proper lamellipodial formation and functions, suggesting that both sites differentially contribute to the activation and stability of the WRC for lamellipodium dynamics [Citation5].
The downstream activation of the Arp2/3 complex triggers actin polymerisation at the leading edge and also the formation of branched F-actin [Citation2,Citation6]. This generates broad, flat membrane protrusions termed lamellipodia, which are characteristic of cells migrating on flat rigid substrates. To respond to directional cues rapidly, cells must have a fine-tuned balance between protrusion and retraction to permit steering and chages in direction. Through decades of research, a pathway linking the Rho family GTPase Rac1, the WRC and the Arp2/3 complex has been established for lamellipodia generation and migration [Citation7,Citation8] and reviewed in [Citation9]. While the mechanisms required to activate actin polymerisation at the leading edge are still extensively studied, how negative regulation allows plasticity and retraction is less well understood. Gautreau and Krause have highlighted the known positive and negative feedback loops controlling branched actin assembly [Citation9]. Negative regulators of Arp2/3 complex have been described, such as PICK1, Gadkin and Arpin. However, only Arpin inhibits the Arp2/3 complex at the lamellipodium, while Gadkin and PICK1 play a role at surface endosome and clathrin-coated pits, respectively [Citation10–Citation12]. Interestingly, some activators and inhibitors of the Arp2/3 complex co-localise at membrane sub-compartments of the cell, such as Arpin and WAVE at the lamellipodium, Gadkin and WASH at endosomes and PICK1 and WASP at clathrin-coated pits [Citation13]. Gadkin and PICK1 both contain an acidic motif resembling the tail of WASP-family proteins, but instead of activating Arp2/3, they seem to be antagonistic. Arpin was identified through a bioinformatics search for COOH-terminal acidic A-motifs found in nucleation promoting factors (NPFs). Arpin localises to the lamellipodia and acts downstream of Rac1 to competitively inhibit binding of WAVE to Arp2/3 by exposing the A-motif to Arp2/3 and keeping it in an inactive confirmation [Citation10,Citation13]. This causes a reduction in lamellipodia protrusion lifetime and directional persistence for migration [Citation10], although it is dispensable for chemotaxis [Citation14].
A role for CYRI in membrane protrusions and cell migration
All negative regulators of this pathway identified prior to CYRI are inhibitors of the Arp2/3 complex, raising the question: are there novel regulators at the level of the WRC? Using a tagged version of the Dictyostelium discoideum WRC subunit Nap1 (NckAP1 in human), CYRI was identified by mass spectrometry through a reversible formaldehyde cross-linked, GFP-trap pulldown [Citation15]. CYRI proteins are highly conserved through evolution, where mammals have two isoforms, CYRI-A and CYRI-B. The protein sequence of both CYRI isoforms comprise an N-terminal predicted alpha helix with a concensus myristoylation site, followed by a domain of unknown function 1394 (DUF1394) domain, which has sequence homology to the WRC subunit CYFIP1. The Rac1 binding A-site in CYFIP1 resides in the DUF1394, interacting specifically with active forms of Rac1 but not Cdc42 or RhoA [Citation15]. Depletion of CYRI-B and subsequent optogenetic activation of Rac1 promoted broader and less dynamic lamellipodia, similar to the fried egg phenotype of cells with constitutively active Rac1 as previously described [Citation16]. Conversely, overexpression of CYRI-B resulted in cells shrinking in area and having unproductive protrusions. Localisation of CYRI-B to the leading edge of cells could only be observed in fixed assays with a FLAG-tagged construct, which inhibited many cells from forming lamellipodia. It is possible that larger C- or N-terminal tags such as GFP inhibit the regulation of CYRI-B by interfering with N-terminal myristoylation [Citation15]. CYRI-B functions to restrict protrusion lifetime and promote pseudopod splitting by mimicking the Rac1-CYFIP interaction. It is therefore required to regulate polarity and plasticity of protrusions for effective migration and chemotaxis [Citation15].
Hans Meinhardt proposed that a minimal mathematical model of cell migration could be made using a global inhibitor, an activator and a local inhibitor [Citation17]. The local inhibitor needed to be smaller than the activator (to diffuse more rapidly) and to be recruited by the same signal as the activator. We postulate that WRC and active Rac1 are the activators, while CYRI-B plays the role of the local inhibitor by restricting Rac1’s activity at the cell membrane [Citation15] (). While this model is a good starting point to understand lamellipodial feedback loops, we are still left with further questions surrounding the mechanisms of activation/inactivation of CYRI.
Rac1-mediated pathways in cancer
Migration of tumour and associated cells is a key part of cancer cell metastasis, which relies on the dynamic changes of the actin cytoskeleton. Some studies have correlated expression of components of the actin machinery with cancer progression, but the importance of this is not clear beyond a general correlation with increased actin dynamics when cells are more mesenchymal and thus thought to be more agrressive [Citation13,Citation18–Citation20]. Reduced expression of Cyfip1 or NckAP1 has been linked to increased invasiveness, indicating that they have the potential to act as tumour invasion suppressors [Citation21,Citation22]. In vitro, silencing of either Cyfip1 or NckAP1 in cancer cells migrating in 3D caused cells to become more invasive due to increased reliance on an N-Wasp driven Arp2/3-mediated migration [Citation22]. However, in breast cancer cells, silencing of NckAP1 or disrupting the Arp2/3 complex reduced invasion [Citation23]. Moreover, following subcutaneous implant, there was no difference in primary tumour volume, but a reduction in the number of lung metastases of MDA-MB-231 cells [Citation23], indicating potential involvement of WRC in metastatic dissemination.
Rac1 mediates activation of the WRC and resulting actin polymerisation and can be hyperactivated in cancers. An activating mutant of Rac1 containing a proline to serine change, Rac1P29S, was found in 9% of all sun-exposed melanomas, placing it as one of the top 10 mutated genes (behind BRAF, NRAS, TP53, PTEN and others) [Citation24]. Rac1P29S shows a high GDP/GTP exchange and thus is hyperactive. Similar to the constitutively active Rac1Q61L, the Rac1P29S mutation causes an increase in cell migration, membrane ruffles and proliferation [Citation25,Citation26].
Before it was known to be a Rac1 interacting protein, CYRI-B was highlighted as being highly expressed in pancreatic cancer [Citation27]. CYRI-B knockdown PDAC cells were injected in the tail vein of immunocompromised mice, resulting in increased lung colonisation and enhanced growth compared with control cells [Citation27]. This may be due to enhanced Rac1 activity in CYRI-B knockout cells [Citation15]. Rac1 has previously been implicated in pancreatic cancer progression [Citation28] and is thought to regulate actin localisation and dynamics during dysplastic changes in precancerous regions of pancreas as well [Citation29]. In summary, disrupting the regulation of Rac1 signaling and actin dynamics may drive the invasiveness and spread of various cancers and the role of CYRI in this process warrants further study.
Beyond actin dynamics
Chattaragada et al., found that silencing of CYRI-B in PDAC cells lead to an increase in mitochondrial O2− levels, resulting in stress due to reactive oxidative species (ROS). This rise in ROS levels was linked to enhanced tumour cell proliferation and invasiveness [Citation27]. In neutrophils, activation of Rac1 is well known to enhance superoxide production via direct binding to the phagocyte oxidase complex PHOX [Citation30,Citation31]. However, in other cell types, NOX plays the role of PHOX and its regulation is much less well understood [Citation31,Citation32]. It is possible that higher levels of Rac1 activity in CYRI knockout cells leads to activation of NOX and thus induces greater superoxide production observed by Chattaragada and colleagues [Citation27]. Alternatively, Rac1 has been localised to the mitochondrial membrane and is implicated in maintenance of superoxide production homeostasis through its interaction with BCL-2 [Citation33]. Inhibiting the BCL-2-Rac1 interaction in lymphoma cells, decreased the mitochondrial O2− levels and lead to the triggering of apoptosis [Citation33]. Furthermore, Chattaragada et al. reported mitochondrial localisation of CYRI-B [Citation27]; although we could not confirm this observation either in mammalian cells or Dictyostelium thus far [Citation15]. It remains for the future to discover the mechanism by which CYRI may impact on superoxide production and whether this is related to Rac1 control of NOX or mitochondrial ROS production.
Additionally, Yuki et al. [Citation34] report that CYRI-B restricts Salmonella infection in mice, as they identified CYRI in a screen for novel genes that when mutated affected susceptibility to infection. This suggests a possible immune function for CYRI-B, possibly at the level of bacterial uptake, which requires engagement of the actin cytoskeletal machinery and Rac1 activation [Citation34]. They further show in this study that CYRI-B can be destroyed by ubiquitination downstream of Rac1 activation by the Salmonella effector SopE, suggesting interesting possibilities for regulation of CYRI-B in cells [Citation34].
Proteins that bind specifically to active Rac1
CYRI-B interacts with active forms of Rac1, such as Rac1P29S, Rac1Q61L [Citation15,Citation34] and interestingly appears to show increased affinity to double mutant Rac1P29S/Q61L [Citation15]. Rac1P29S/Q61L was first described by Chen and colleagues [Citation3] but is not a naturally occurring mutation (to our knowledge). It is unclear how this double mutation affects Rac1 structurally, including GTP binding or hydrolysis. We speculated that this mutant might have higher affinity for DUF1394 domains, since we and Chen both found enhanced binding to CYRI and CYFIP. To test whether any other motifs or proteins were preferred by Rac1P29S/Q61L over the more conventional single mutation Rac1Q61L, we transfected cells with either GFP, GFP-Rac1Q61L or GFP-Rac1P29S/Q61L () and used GFP-trap and mass spectrometry to compare binding partners. We retrieved many known Rac1-interacting proteins that were grouped into Gene Ontology (GO)-terms (). Additionally many of the target genes were in clusters around membrane-bound organelles such as the nucleus and mitochondria ( and see also Supplementary Table 1). Details of proteins interacting with either mutant active Rac1 protein can be found in Supplementary Table 1 and the data are available via ProteomeXchange with identifier PXD013941.
Figure 2. Screen for active Rac1 interacting proteins.
A) Cos7 cells transfected with GFP, GFP-Rac1Q61L or GFP-Rac1P29S/Q61L for immunoprecipitation with GFP-pulldown, Scale bar 25 μm. B) GO-term analysis (Homo sapiens) to sort significantly enriched proteins that bound to both forms of active Rac1 compared to GFP into categories based on number of hits. Adjusted p-values using the Benjamini False discovery rate (FDR) scoring shows significance of those hits using a rainbow scale with p-value 0.05 highlighted. C) Volcano plot illustrating results from t-test applied on protein intensity differences between the two Rac-1 mutants (GFP-Rac1Q61L and GFP-Rac1P29S/Q61L) measured in liquid chromatography–tandem mass spectrometry experiments. The colour coding is based on density of the data points, the scale is indicated on the right, and the curved line shows the 5% FDR. D) Schematic representation of a cell and the important regions of Rac1 activity based on the GO-term locations.
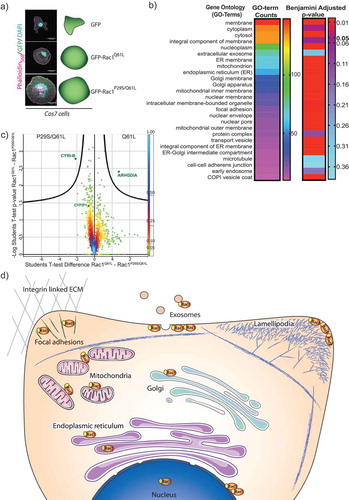
To our surprise, CYRI-B was the only protein retrieved with a statistically significant ratio that interacted preferentially with Rac1P29S/Q61L compared to Rac1Q61L (). On the other hand, ARHGDIA (also known as RhoGDI1) showed significantly less binding to the double mutant Rac1P29S/Q61L than Rac1Q61L. RhoGDI is the major regulator of GDP-bound inactive Rho GTPases Rac1Q61L [Citation35] (). This could be a clue as to why the double mutant Rac is more available for binding to CYRI-B, but does not explain why we did not retrieve CYFIP1 in this experiment nor why CYRI-B and presumably the DUF1394 uniquely prefers this mutant form of Rac1. Notably, the NonO proteins, found to control nuclear actin and transcription [Citation36], also bound preferentially to Rac1Q61L. Finally, we highlight proteins from our screen that interact with Rac1 Q61L and Rac1 P29S/Q61L at specific regions within the cell (). This follows recent literature showing diverse functions for Rac1 activity throughout the cell [Citation37].
In summary, CYRI proteins are novel regulators of lamellipodia dynamics through their interaction with Rac1 at the DUF1394 domain. CYRI proteins negatively regulate the activity of the WRC and suppress lamellipodia spread and dynamics of protrusions. Cells need CYRI proteins to maintain plasticity and to allow leading edge actin dynamics to be able to rapidly respond to changing environments. While CYRI’s role in cells is starting to become clear, much remains to be learned about the roles of CYRI proteins in healthy tissues, organisms and in cancer.
Methods
Transfection and protein isolation
Cos-7 (chlorocebus sabeus fibroblast) cells grown at 37°C and 5% CO2, were transfected with either GFP, GFP-Rac1Q61L (human) or GFP-Rac1P29S/Q61L using Lipofectamine2000 (Invitrogen) following the manufacturer’s protocol. The transfected cells were left overnight at 37°C, 5% CO2 and replated the next day on laminin-coated dishes and incubated overnight.
The plates were washed twice in ice-cold PBS and lysed in GFP-trap buffer (25mM Tris HCl, pH7.5, 100mM NaCl, 5mM MgCl2, 0.5% NP-40, containing protease and phosphatase inhibitors). Around 1.5 mg of lysates were incubated with GFP-Trap_A beads (ChromoTek) for 2 hours at 4°C following the manufacturer’s protocol. Beads were washed three times with Wash buffer (25mM Tris HCl, pH7.5, 100mM NaCl, 5mM MgCl2, containing protease and phosphatase inhibitors). The beads were stored at −20°C in a solution of 100 mM ammonium bicarbonate + 2 M UREA Prior to digestion.
On-beads proteolytic digestion
Immunoprecipitations were performed in triplicate, and purified proteins were digested on beads using the method described in [Citation38].
Liquid chromatography–mass spectrometry
Tryptic digests were obtained and separated by nanoscale C18 reverse-phase liquid chromatography using an EASY-nLC 1200 (Thermo Fisher Scientific) coupled to an Orbitrap Q-Exactive HF mass spectrometer (Thermo Fisher Scientific). The eluting peptide solutions were introduced into the mass spectrometer via a nanoelectrospray ion source (Sonation). The mass spectrometer was operated in positive ion mode and used in data-dependent acquisition.
Data analysis
The MS Raw data were processed with MaxQuant software [Citation39] and searched with Andromeda search engine [Citation40], querying both UniProt [Citation41] Chlorocebus sabaeus (13/03/2017; 19,228 entries) and Homo sapiens (09/07/2016; 92,939 entries) plus the sequence of GFP-Rac1 (Q61→L) and GFP-Rac1 (P29→S and Q61→L) construct used in the experiment. Combined databases were searched assuming the digestion enzyme trypsin allowing for two miscleavages. Methionine oxidation and N-terminal acetylation were specified as variable modifications, and Cysteine carbamidomethylation was specified as fixed modifications. The peptide, protein and site false discovery rate (FDR) was set to 1%. The common reverse and contaminant hits (as defined in MaxQuant output) were removed. Only protein groups identified with at least one uniquely assigned peptide were used for the analysis. For label-free quantification, proteins were quantified according to the label-free quantification algorithm available in MaxQuant [Citation42]. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Three independent replicates were generated per condition (GFP control and two RAC-1 mutants used in the immunoprecipitations), and significantly enriched proteins were selected using a t-test based analysis (5% False Discovery Rate).
Go-term analysis
Genes derived from mass spectrometry results were scored for significance for both active Rac1 screens and absent from GFP controls were then uploaded to DAVID (Database for Annotation, Visualisation and Integrated Discovery) v6.8 for GO-term themed searches following the protocol described in [Citation43]. We used Homo Sapiens as the species as this closely relates to Chlorocebus sabaeus but has more GO-terms available.
Supplemental Material
Download MS Excel (866.8 KB)Acknowledgments
We would like to thank the Core Services and Advanced Technologies at the Cancer Research UK Beatson Institute (C596/A17196).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Chen Z, Borek D, Padrick SB, et al. Structure and control of the actin regulatory WAVE complex. Nature. 2010 Nov 25;468(7323):533–538.
- Machesky L, Mullins RD, Higgs HN, et al. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. PNAS. 1999;96(1):3739–3744.
- Chen B, Chou HT, Brautigam CA, et al. Rac1 GTPase activates the WAVE regulatory complex through two distinct binding sites. Elife. 2017 Sep 26;6.
- Davidson AJ, Insall RH. Actin-based motility: WAVE regulatory complex structure reopens old SCARs. Curr Biol. 2011 Jan 25;21(2):R66–8.
- Schaks M, Singh SP, Kage F, et al. Distinct Interaction Sites of Rac GTPase with WAVE Regulatory Complex Have Non-redundant Functions in Vivo. Curr Biol. 2018 Nov 19;28(22):3674–3684 e6.
- Svitkina TM, Borisy GG. Arp2/3 Complex and Actin Depolymerizing Factor/Cofilin in Dendritic Organization and Treadmilling of Actin Filament Array in Lamellipodia. J Cell Biol. 1999;145(5):1009–1026.
- Machesky L, Insall RH. Scar1 and the related Wiskott–aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8(1):1347–1356.
- Ismail AM, Padrick SB, Chen B, et al. The WAVE regulatory complex is inhibited. Nat Struct Mol Biol. 2009 May;16(5):561–563.
- Krause M, Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol. 2014 Sep;15(9):577–590.
- Dang I, Gorelik R, Sousa-Blin C, et al. Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature. 2013 Nov 14;503(7475):281–284.
- Maritzen T, Zech T, Schmidt MR, et al. Gadkin negatively regulates cell spreading and motility via sequestration of the actin-nucleating ARP2/3 complex. PNAS. 2012;109(26):10382–10387.
- Rocca DL, Martin S, Jenkins EL, et al. Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat Cell Biol. 2008 Mar;10(3):259–271.
- Molinie N, Gautreau A. The Arp2/3 Regulatory System and Its Deregulation in Cancer. Physiol Rev. 2018 Jan 1;98(1):215–238.
- Dang I, Linkner J, Yan J, et al. The Arp2/3 inhibitory protein Arpin is dispensable for chemotaxis. Biol Cell. 2017 Apr;109(4):162–166.
- Fort L, Batista JM, Thomason PA, et al. Fam49/CYRI interacts with Rac1 and locally suppresses protrusions. Nat Cell Biol. 2018 Oct;20(10):1159–1171.
- Welch H, Coadwell WJ, Ellson CD, et al. P-Rex1, a PtdIns(3,4,5)P3-andGBy-Regulated Guanine-Nucleotide Exchange Factor for Rac. Cell. 2002;108(1):809–821.
- Meinhardt H. Orientation of chemotactic cells and growth cones: models and mechanisms. J Cell Sci. 1999;112(1):2867–2874.
- Otsubo T, Iwaya K, Mukai Y, et al. Involvement of Arp2/3 complex in the process of colorectal carcinogenesis. Mod Pathol. 2004 Apr;17(4):461–467.
- Rauhala HE, Teppo S, Niemela S, et al. Silencing of the ARP2/3 Complex Disturbs Pancreatic Cancer Cell Migration. Anticancer Res. 2013;33:45–52.
- Kazazian K, Go C, Wu H, et al. Plk4 Promotes Cancer Invasion and Metastasis through Arp2/3 Complex Regulation of the Actin Cytoskeleton. Cancer Res. 2017 Jan 15;77(2):434–447.
- Silva JM, Ezhkova E, Silva J, et al. Cyfip1 is a putative invasion suppressor in epithelial cancers. Cell. 2009 Jun 12;137(6):1047–1061.
- Tang H, Li A, Bi J, et al. Loss of Scar/WAVE complex promotes N-WASP- and FAK-dependent invasion. Curr Biol. 2013 Jan 21;23(2):107–117.
- Teng Y, Qin H, Bahassan A, et al. The WASF3-NCKAP1-CYFIP1 Complex Is Essential for Breast Cancer Metastasis. Cancer Res. 2016 Sep 01;76(17):5133–5142.
- Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell. 2012 Jul 20;150(2):251–263.
- Davis MJ, Ha BH, Holman EC, et al. RAC1P29S is a spontaneously activating cancer-associated GTPase. Proc Natl Acad Sci U S A. 2013 Jan 15;110(3):912–917.
- Krauthammer M, Kong Y, Ha BH, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012 Sep;44(9):1006–1014.
- Chattaragada MS, Riganti C, Sassoe M, et al. FAM49B, a novel regulator of mitochondrial function and integrity that suppresses tumor metastasis. Oncogene. 2018 Feb 8;37(6):697–709.
- Heid I, Lubeseder-Martellato C, Sipos B, et al. Early requirement of Rac1 in a mouse model of pancreatic cancer. Gastroenterology. 2011 Aug;141(2):719–30, 730 e1-7.
- Morris HT, Machesky LM. Actin cytoskeletal control during epithelial to mesenchymal transition: focus on the pancreas and intestinal tract. Br J Cancer. 2015 Feb 17;112(4):613–620.
- Pick E. Role of the Rho GTPase Rac in the activation of the phagocyte NADPH oxidase: outsourcing a key task. Small GTPases. 2014;5:e27952.
- Panday A, Sahoo MK, Osorio D, et al. NADPH oxidases: an overview from structure to innate immunity-associated pathologies. Cell Mol Immunol. 2015 Jan;12(1):5–23.
- Hordijk PL. Regulation of NADPH oxidases: the role of Rac proteins. Circ Res. 2006 Mar 3;98(4):453–462.
- Velaithan R, Kang J, Hirpara JL, et al. The small GTPase Rac1 is a novel binding partner of Bcl-2 and stabilizes its antiapoptotic activity. Blood. 2011 Jun 9;117(23):6214–6226.
- Yuki KE, Marei H, Fiskin E, et al. CYRI1-mediated inhibition of RAC1 signalling restricts Salmonella Typhimurium infection. bioRxiv. 2017.
- Garcia-Mata R, Boulter E, Burridge K. The ‘invisible hand’: regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol. 2011 Jul 22;12(8):493–504.
- Rajakyla EK, Vartiainen MK. Rho, nuclear actin, and actin-binding proteins in the regulation of transcription and gene expression. Small GTPases. 2014;5:e27539.
- Payapilly A, Malliri A. Compartmentalisation of RAC1 signalling. Curr Opin Cell Biol. 2018 Oct;54:50–56.
- Hubner NC, Mann M. Extracting gene function from protein-protein interactions using Quantitative BAC InteraCtomics (QUBIC). Methods. 2010;53(4):453–459.
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008 Dec;26(12):1367–1372.
- Cox J, Neuhauser N, Michalski A, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011 Apr 1;10(4):1794–1805.
- UniProt C. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010 Jan;38(Database issue):D142–8.
- Cox J, Hein MY, Luber CA, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014 Sep;13(9):2513–2526.
- Huang Da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57.
