?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
The world is in the midst of a pandemic. We still know little about the disease COVID-19 or about the virus (SARS-CoV-2) that causes it. We do not have a vaccine or a treatment (aside from managing symptoms). We do not know if recovery from COVID-19 produces immunity, and if so for how long, hence we do not know if “herd immunity” will eventually reduce the risk or if a successful vaccine can be developed – and this knowledge may be a long time coming. In the meantime, the COVID-19 pandemic is presenting enormous challenges to medical research, and to clinical trials in particular. This paper identifies some of those challenges and suggests ways in which machine learning can help in response to those challenges. We identify three areas of challenge: ongoing clinical trials for non-COVID-19 drugs; clinical trials for repurposing drugs to treat COVID-19, and clinical trials for new drugs to treat COVID-19. Within each of these areas, we identify aspects for which we believe machine learning can provide invaluable assistance.
1. Introduction
The novel SARS-CoV-2 virus and COVID-19, the disease it causes, have changed the whole world. We are facing a global health crisis – characterized as a pandemic by the World Health Organization – unlike any in recent history. The international scientific community is struggling to understand both the virus and the disease. This requires efforts at an unprecedented level of international focus and cooperation to preserve clinical trial integrity during the pandemic, to develop and to identify treatments, and to find out under what conditions they are safe and effective.
In this paper we discuss challenges in three key areas of clinical research and propose ways in which machine learning (ML) can help to address those challenges. The three areas are: ongoing clinical trials for non-COVID-19 drugs, clinical trials for repurposing drugs to treat COVID-19, and clinical trials for new drugs to treat COVID-19. In each of these three areas we identify opportunities where we believe ML can provide important insights and can help address some of the challenges faced in clinical trials. We are aware that some of what we suggest may not be practicable during the current pandemic – but our discussion, although motivated by the current pandemic also has an eye toward the future. We are also aware that some of what we suggest may not have been used in a regulatory environment previously but the pandemic provides an opportunity to apply novel approaches that can be used in this challenging situation. This may lead to regulatory acceptance of some of these methods and thus lead to changes in future drug development processes. Our discussion is intended as a broad overview; we provide references for deeper reading but in most instances we do not go into detail about ML methods and results. However, in order to give some idea of what has been done and what is possible, we do go into greater detail in two places.
The aim of this paper is to bridge the gap between quantitative research scientists engaged in clinical trials impacted by or related to COVID-19 and the ML community and to help bring these communities together. In what follows we review opportunities for ML applications for clinical trials in the era of COVID-19 to stimulate further research and highlight a few cases where we see particular benefit. More specifically, we review the three distinct areas of challenge mentioned above and discuss selected applications in more detail, which could then serve as a springboard for future research. Table 1 provides a summary and guide to the more detailed discussion that follows. In the various columns we highlight challenges, typical methodologies, opportunities, and the most relevant ML methods. We also include references to the Section(s) in which the challenges are discussed and a more comprehensive list of methods and references can be found.
Table 1. Summary and guide to the more detailed discussion in this paper.
ML has had success in a number of areas. Perhaps the best-known application to medicine is in image recognition, where ML algorithms have proved equal or superior to humans in interpreting X-ray and MRI images and slides. For example, Cruz-Roa et al., (2017) demonstrates that a trained ML algorithm achieves near-perfect detection of breast cancer at a microscopic level. Their algorithm, like other image recognition algorithms, employs supervised learning, in which the algorithm is presented with a training set of instances that provide, for each instance, both the covariates and the ground truth. Less well-known applications employ unsupervised learning, in which the algorithm is presented with a training set of instances that provide only covariates, but is asked only to create clusters of similar instances, and semi-supervised learning, in which the training set provides a few instances with both the covariates and the ground truth and many instances with only the covariates. Both unsupervised and semi-supervised learning are frequently employed in cluster analysis. Segar et al., (2020) provide a recent application of unsupervised learning to cluster analysis of heart failure and Filipovych et al., (2011) provide a recent example of semi-supervised learning to image recognition. In addition to supervised learning, the work that we discuss here employs reinforcement learning (RL), in which the algorithm learns from previous experience and adjusts its behaviour in response to what it has learned, and causal inference. Both RL and causal inference have their roots in statistics. Indeed, the application of RL to clinical trials, using the framework of multi-armed bandits, derives from the seminal work of Thompson (1933), Gittins (1979) and Lai and Robbins (1985). Causal inference has its roots in the work of Neyman (1923) and Rubin (1978) and ML work in causal inference most frequently employs what has now become the ``standard’’ Neyman-Rubin potential outcomes framework.
2. Clinical trials for non-COVID drugs
The societal response to COVID-19 pandemic has included travel restrictions, social distancing and even confinements all over the world. All of these will significantly reduce the ability and/or willingness of trial subjects and staff to access clinical sites and affect data collection: some data will be missing and some data may be gathered in a different way (e.g. remotely vs. on-site). Moreover, the possibility of transmission of COVID-19 from trial subjects to medical personnel and vice-versa presents a substantial risk, especially because it appears that the disease is transmissible before the onset of symptoms and some infected individuals never display symptoms. These concerns have led the US Food and Drug Administration to issue special guidelines for the conduct of clinical trials during the pandemic (FDA 2020b). All these concerns will undoubtedly result in complications that lead to compromised trial data and challenges in the interpretation of clinical trial results (Akacha et al. 2020; Meyer et al., 2020).
The extent of these challenges will depend on, for example, the duration of the current COVID-19 pandemic, the number of impacted subjects, the disease condition being studied and various trial design elements, and may result in the halting of many ongoing clinical trials (EMA 2020a). The halting of a clinical trial and the resulting absence of data may make it difficult to gather and document the knowledge that was expected in the trial design, especially if the trial was halted in its early stages.
Another problem is that data collected before the pandemic may be of different quality than data collected after the pandemic for many reasons, some identifiable and some non-identifiable. For example, there will likely be significant impact on the day-to-day operations of clinical sites, leading to missed visits, increased protocol deviations, late data entry, data collected using different modalities (e.g. collected via a remote visit) and slow follow-up to queries. The times at which COVID-19 cases first occurred and were first observed, the time of “lock-down” and the time of “reopening” will vary dramatically across countries, states, counties, cities, towns, sites or even specific units, but trialists may have limited access to this information. For those subjects who are infected with SARS-CoV-2 the variation in observed symptoms may be enormous – some subjects may be asymptomatic while others may die. This variation may make it difficult to assign causation to the drug under study and hence to identify safety violations. It may also affect changes in laboratory markers and affect the course of treatment for all but the most severe diseases (e.g., advanced cancers).
These and many other impacts will be felt during the pandemic and for an extended time as global healthcare systems deal with the aftermath. Addressing these issues when the trial is resumed will require effective and reliable methods for extracting knowledge from data of different quality and for establishing confidence in that knowledge.
Analysis of data from ongoing clinical trials
During the pandemic, on-site assessment of patients may be less frequent, which will lead to missing data. Moreover, potential differences in visit frequency before, during and after the pandemic may mean that patient data is not sampled at the usual intervals. Both of these issues could be addressed using existing ML methods. Missing data might be imputed using ML methods specifically designed to impute missing data in temporal data streams (Yoon et al. 2017, 2018; Yin and Cheung 2019). These methods make it possible to infer the patient state during the period in which on-site monitoring was less frequent. On this task, ML methods, using Multi-Dimensional Recurrent Neural Networks (Yoon et al. 2018c) and Generative Adversarial Imputation Nets (Yoon et al., 2018b), substantially out-perform previous methods, including Multiple Imputation by Chained Equations, Matrix Completion and Expectation Maximization on a variety of datasets (from the online UCI repository). All of these methods rely on the assumption that data is missing at random, i.e., that the reason data is missing is recorded in the data and unrelated to the patients’ unobserved state. This may or may not be a reasonable assumption in the context of the pandemic; for instance, variables that influence whether visits will be cancelled (such as local conditions and a patient’s risk-status) are likely to be recorded, but others (such as illness of a family member and difficulty in traveling) may not be. Moreover, this assumption may or may not conform to current regulatory guidelines. In either case, estimation will remain a problem if not enough information is available for imputation or prediction models (Akacha et al., 2020; Meyer et al., 2020) to be applicable Accessing these records may require integrating site-level operational data with patient data from the study. Other ML models are specifically designed for the analysis of irregularly sampled temporal data (Neil et al. 2016; Shukla and Marlin 2019; Alaa and van der Schaar 2017b).
As clinical trials continue throughout the pandemic, the validity of trial results may be compromised by the many differences between the periods before, during and after the pandemic (EMA 2020a). It is very likely that measures taken during the pandemic will alter the daily lives of trial subjects, the general standard of care they receive, the application of the treatment being tested and even the control. Such alterations might affect both the clinical outcomes and the effectiveness of treatments, and the effects of these alterations must be untangled from the treatment effects. This will require including variables that capture a subject’s individual history during the pandemic (especially changes in medical treatment, but also changes in diet, exercise, etc., if relevant) and using recent machine learning methods to estimate treatment effects. Because the true shape of the relationship between a subject’s pandemic history and treatment effects is completely unknown at this point, the inherent flexibility and data-driven nature of ML methods provide them an advantage over standard statistical approaches in this scenario.
If the trial continues during the pandemic, it is likely that the participation and recruitment of subjects will be altered (if not stopped entirely). If these alterations affect particular subgroups disproportionately, they will change the composition of the patient population, and may bias the estimated population-level treatment effect, unless such confounding effects are adjusted for in the analysis. It will therefore be crucial to identify the extent of these alterations and determine the implications for both the estimates of treatment effects for the various subgroups and for the confidence that can be placed in these estimates (see below).
Extracting knowledge from data of suspended trials
To extract knowledge from the data of trials that have been terminated before their intended endpoint, we first need to learn both what we do know and what we do not know on the basis of the available data. ML methods for estimating heterogeneous treatment responses (Athey and Imbens 2016; Tran and Zheleva 2019) may be well suited to these tasks. These and other methods for estimating heterogeneous treatment responses begin with a method for estimating individualized treatment effects (ITE) and construct subgroups and estimates of heterogeneous treatment responses using the chosen method of estimating ITE. Hill (2011), Athey and Imbens (2016), Alaa and van der Schaar (2017, 2018) and Yoon et al. (2018a) provide an array of different methods for estimating ITE. We refer to the section ‘Exploiting observational data in the design of new trials’ below and Bica et al. (2020) for more discussions.
These models can identify subgroups that have similar covariates and treatment responses and estimate the treatment responses in each subgroup. Because the trial will have been terminated before its intended endpoint, data will necessarily be incomplete. Moreover, the burden of the pandemic on the health care system and the concomitant limitation of resources may have interfered with the collection and recording of subject covariates and outcomes. Thus there may be concerns about the reliability of these treatment response estimates. To manage these concerns and to separate reliable estimates from unreliable estimates requires assessing the confidence in these estimates. Recent ML methods for systematically quantifying the uncertainty of estimation (Lei et al. 2018) are designed for such tasks. A particular issue that may arise in suspended clinical trials is that the population distribution of subjects in the trial (by age, gender, income, geographic location, etc.) may be distorted relative to the intended population and/or the real-world population. Such distortion would affect both the reliability of, and the confidence in, the extracted knowledge, and need to be taken into account using appropriate methods, see e.g. Akacha et al. (2020) and the references therein. ML methods for quantifying uncertainty under covariate shift (Tibshirani et al. 2019) may be suited to address this issue as well.
Adjusting re-started clinical trials for efficient resource utilization
When the situation is normalized, it is likely that many halted clinical trials will be re-started. When re-starting a blinded trial with a fixed format, little or nothing can be changed. However, if only a small fraction of the trial had been conducted prior to halting, one might consider stopping the trial, unblinding the data, and using the knowledge extracted from that data as prior information in the design of a new trial. Conversely, if the trial had been almost complete prior to halting, one might again consider stopping the trial and unblinding the data. In early stages of drug development the knowledge extracted from that data could be used to decide whether a new trial is warranted, and, if so, how to design that new trial; in late stages of drug development, the knowledge extracted could be used to decide whether the drug is ready to be submitted for regulatory approval. When permitted – as in trials with an adaptive design – the knowledge extracted from pre-pandemic data may be valuable in adjusting design elements such as recruitment plans, sample sizes and treatment allocations. More broadly, the knowledge learned from halted trials (e.g. identified subgroups, estimates of treatment effects and confidence levels for those estimates) can be used as prior information for re-started trials that leverage adaptive (Kunz et al., 2020) or Bayesian clinical trial designs (Lee and Chu 2012).
3. Drug repurposing trials
Covid-19 is currently not a well understood disease, with multiple biological and clinical manifestations – e.g. respiratory, immune-related, coagulation, gastro-intestinal. ML can play an important role in finding patterns and signatures in the underlying molecular biology of COVID-19 mechanisms, and linking those to the clinical characteristics of the disease. This in turn can facilitate the identification of both existing medicines that could potentially be repurposed, as well as validating in-silico, whether novel medicines may be effective. Thus which clinical trials to run could potentially be driven by this biomedical insight from machine learning conducted on existing data. An existing approach for doing this is knowledge graph inference, where a vast network of existing, interrelated data is formed, and ML is used to reason over this network, extracting new insights which would not be possible from looking at individual data sets on their own (Alaimo and Pulvirenti, 2018). As more data is accumulated the graph can continually be built out. Enabling samples to be taken from patients participating in COVID-19 trials, would add even more richness to the picture, adding potential temporal changes to be inferred as well.
At the moment, no drugs have been approved for the treatment of COVID-19 except on an emergency basis. However, 40 or more existing drugs have been identified as having promise, some of which have been approved for clinical trials (EMA 2020c). The most prominent example is Hydroxychloroquine, which is approved for treatment of malaria; other possibilities include Lopinavir/Ritonavir, which is approved for treatment of HIV, Acalabrutinib, which is approved for treatment of chronic lymphocytic leukemia and Remdesivir, which was not previously approved for treatment of any condition but which appears promising in current trials and has been authorized for emergency use (FDA 2020a). Some of these same drugs have been approved for emergency and compassionate use in various other countries (EMA 2020c). To complement anecdotal evidence, promising in-vitro evidence and evidence from small experiments and small-scale clinical trials, the efficacy of these drugs for treatment of COVID-19 in humans will need to be established in large-scale clinical trials. Repurposing existing drugs for treatment of COVID-19 presents a potentially much faster route to finding an effective treatment, both because new drugs do not have to be developed and because, for many of the existing drugs, safety and efficacy for some conditions in humans has already been established on the basis of previous clinical trials (although perhaps only for some particular population(s) and in some dose(s) that might be different than needed to treat COVID-19). Such knowledge may speed the process of determining safety and efficacy in treating COVID-19.
‘Virtual’ clinical trials
The central problem in assessing the effectiveness of a new drug is the comparison against existing drugs or a placebo. Clinical trials address this problem by creating a control group that is either untreated or treated with existing drugs but drawn randomly from the same population as the treated group. However, there are situations when the use of such a control group may not be possible, or its size might be limited, for either practical or ethical reasons, including during an outbreak. The rapid spread of the covid-19 pandemic and the lack of knowledge about effective drugs (or treatments) has led hospitals around the world to experiment with drugs that have not undergone proper clinical trials and are not likely to undergo such trials in the immediate future. By integrating data across hospitals, data-driven methods can be used to identify patients who have received standard treatments but are otherwise similar to patients who have received experimental treatments (Zhu et al. 2016; Suo et al. 2018). Machine learning methods in particular can be utilised to create, ex post, a ‘virtual’ control group, especially in situations where highly complex or non-linear interactions between covariates and outcomes need to be captured. Data from such a ‘virtual’ clinical trial may not be entirely comparable to data obtained from a standard clinical trial, but knowledge learned from such ‘virtual’ trials can identify those drugs that should undergo the first formal clinical trials, inform hospitals and physicians about the most promising candidates for compassionate use, and inform researchers about the most promising experimental drugs.
Exploiting observational data in the design of new trials
In addition to identifying particularly promising drugs, the experimental and compassionate use of drugs to treat COVID-19 is yielding a large body of data, which can be exploited to produce prior information for the design of future (Bayesian) controlled trials (Schmidli et al. 2020). Machine learning methods for causal inference from observational data are especially well-suited to this task.
To illustrate, consider the problem of estimating the effect of a new drug for the treatment of COVID-19. For each patient , the observation will provide an array
of patient features, a treatment indicator
and an observed outcome
. Here we assume that only one treatment and a control are under consideration. In a different setting, there might be several possible treatments and a control; in such a setting, the treatment indicator might take on more than two possible values. However the same framework and methods can still be applied.
Using the potential outcomes framework of Neyman (1923) and Rubin (1978), we define the patient outcome without treatment () to be
and the patient outcome with treatment (
) to be
. In the observed data, we have information only about the factual patient outcome; because one of
,
is 1 and the other is 0, we can write the factual outcome as
. The counterfactual outcome – i.e. the patient outcome that would have occurred under the option that was not applied – is not observed, but must be estimated from the observed data. The individualized treatment effect is the difference in potential outcomes:
Machine learning has developed a number of methods for estimating individualized treatment effects from observed data. As illustrated in Figure 1, the observed outcomes for the treated patients (red) and the observed outcomes for the control patients (blue) can be used to estimate the response surfaces and
. By modeling the response surfaces using one shared function
and simply including the treatment indicator
as a feature, such that
, many machine learning models could be used to flexibly estimate treatment effects. A popular example of this approach is Hill (2011), using Bayesian Additive Regression Trees (BART). More sophisticated methods for causal inference such as Alaa and van der Schaar (2017), Shalit et al. (2017), Alaa and van der Schaar (2018), and Yoon et al. (2018a) approximate
through multi-task learning, which involves using a shared structure between the two response functions, while at the same time fitting separate outcome models for the control and treated populations. These ML methods are flexible and capable of learning non-linear interactions among the patient features, treatments and potential outcomes. The method of Alaa and van der Schaar (2017) also uncovers, for each patient, the features that are most important for estimating patient”s potential outcomes.
Figure 1. The observed data contains information about patient characteristics , assigned treatments and observed (factual outcomes) outcomes. The observed outcomes for the control (blue) and treated (red) patients can be used to train machine learning methods to estimate the response surfaces
for each treatment option. Using these response functions we can estimate individualized treatment effects and thus identify patients who would benefit most and patients who would benefit least most from receiving the treatment. This would not be possible if we only estimated the average treatment effect.
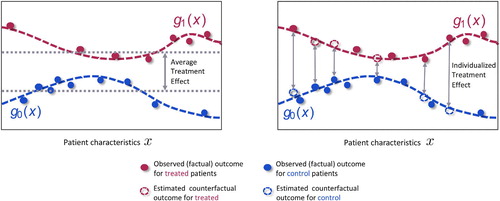
In some circumstances, selection bias may be present in the observed data; i.e. the treatment assignment depended on the patient characteristics. This bias must be accounted for in estimating the response surfaces. To this end, Alaa et al. (2017) use the propensity score, and Shalit et al.(2017) build treatment invariant representations of patient characteristics.
By obtaining unbiased estimates of the response functions we can compute both potential outcomes
conditioned on the patient’s characteristic and thus obtain the individualized treatment effect
. Using the estimated individualized treatment effect for each person, we can then identify patients for whom the treatments are more or less effective. This information can be subsequently leveraged to find the patient subgroups that would benefit most from the treatments.
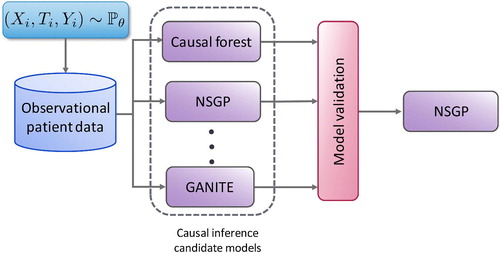
The arsenal of causal inference methods has grown substantially in the past few years. This creates opportunities for more reliable inference, but also complicates the choices that researchers have to make and defend when selecting a causal inference method for the available observational data. Because counterfactual data is not available, we cannot use cross-validation to decide which model to use for an observational dataset nor to tune the hyperparameters of any such model. Validation of causal inference models is crucial for translating recent advances in machine learning-based causal inference into practice. To address this challenge, Alaa and van der Schaar (2019) propose the use of influence functions, a technique from robust statistics, to approximate the loss of causal inference methods without requiring access to counterfactual data. Their method achieves promising results on causal inference model evaluation and selection and, as illustrated in Figure 2, can be used to identify the most appropriate causal inference model for each observational dataset of interest.
Figure 2. Given an observational dataset with patient features , assigned treatments
and factual outcomes
jointly sampled from the distribution
, validation is needed (e.g. Alaa and van der Schaar 2019) to select the causal inference method, out of the large number available (e.g. Causal Forests (Athey and Imbens 2016), NSGP (Alaa and van der Schaar 2019), and GANITE (Yoon et al. 2018)) that will achieve the best estimate of the individualized treatment effects.
When we have information about patient outcomes conditional on time-dependent treatments and patient covariates, causal inference methods that can estimate treatment effects over time can be used (Lim et al. 2018; Bica et al. 2020). These methods can estimate counterfactual patient outcomes under sequences of possible treatment assignments and thus help us understand what treatments should be given to patients and in what order.
The optimal dosages of drugs (or combinations of drugs) re-purposed to treat COVID-19 patients may be very different from the optimal dosages for their originally-intended applications, as they may be influenced both by effectiveness in treating the disease and the likelihood of adverse interactions with the disease itself. ML models for individualized dose-response estimation can be applied to this problem (Bica et al. 2020). In each of these applications, the estimation uncertainty can be quantified to establish confidence in the estimates produced (Lei et al. 2018; Tibshirani et al. 2019), which will enable more reliable exploitation of the observed data.
Execution and evaluation of actual clinical trials
More than 300 trials to investigate the efficacy of medical treatments against COVID-19 are already registered with the Word Health Organization (IDDO 2020). However, bodies that exercise oversight, such as the European Committee for Medicinal Products for Human Use (CHMP) have expressed concern that small studies will not be able to generate convincing evidence; instead of many small studies, they have called for large multi-arm, multi-site trials to evaluate a multitude of therapeutic options (CHMP 2020). As a result, a number of large adaptive clinical trials for evaluating repurposed medications for COVID-19, such as Solidarity (WHO 2020) and RECOVERY (Oxford 2020), are currently underway and are recruiting patients at a multitude of sites to be randomly assigned across available treatment arms. Machine learning methods have the potential to improve the design, execution and evaluation of such trials, as we will illustrate below.
Because of their inherent flexibility and efficiency, adaptive trials (Bretz et al. 2017; Pallmann et al. 2018) are especially suited to the current volatile situation. Instead of randomizing patients to fixed treatment arms in fixed proportions throughout the trial, adaptive designs use interim analyses to reconfigure patient recruitment criteria, assignment rules and treatment options (Park et al. 2018). In recent years, there has been a growing trend to leverage ML approaches, especially tools from reinforcement learning (RL) such as Markov decision processes and multi-armed bandits, to improve and expedite adaptive clinical trial designs (Villar et al. 2015; Varatharajah et al. 2018; Atan et al. 2019). The framework of multi-armed bandits is particularly useful in the context of clinical trials because it fits easily and well and because there is an enormous literature on multi-armed bandits, going back to Gittins (1983). All of this work is designed to address the exploration-exploitation trade-off, which can be interpreted as a trade-off between clinical research (to discover knowledge about treatments) and clinical practice (to benefit the participants) in clinical trials (Berry 2004), by assigning new patients to treatment arms on the basis of information from previous patients. These methods have been shown to speed up learning and identify subgroups for which different treatments might be employed and different treatment responses might be expected (Lee et al. 2020). Because these methods are automatic, they are easy to implement (when trial logistics permit). Moreover, the Bayesian nature of many of these algorithms permits smooth incorporation of previously discussed observational evidence as prior information.
Another important potential application of ML methods in this setting is to perform post-hoc analyses of existing trial data in order to identify heterogeneous treatment response across different patient subgroups (e.g. Athey and Imbens (2016)). ML methods can establish the validity of these analyses by producing systematic confidence guarantees (Tibshirani et al. 2019). This is especially important for COVID-19 because of the broad range of characteristics and comorbidities of patients, the wide variation in the disease trajectory of infected patients and our current limited understanding of the disease mechanism.
Robust recursive partitioning for subgroup analysis
The understanding of treatment effects plays an important role in shaping interventions and treatments. When – as is often the case – treatment effects are different for different segments of the patient population, it is important to identify those segments for which the treatment is effective and those for which it is ineffective, and those for which it has unacceptable side effects and those for which it does not. COVID-19 provides a striking example of the possibilities, because it appears to manifest in different ways – including as a respiratory disorder and as a hematological disorder – and over different time horizons, and to manifest differently for patients of different ages and with different underlying conditions. A treatment that is effective against early manifestation as a respiratory disorder for a patient with asthma might not be effective against late manifestation as a hematological disorder for a patient with diabetes, etc. Such differences have been observed in the initial clinical trial of remdesivir in adults with severe COVID-19 (Wang et al. 2020). In such situations, heterogeneous treatment effect (HTE) analysis (also known as subgroup analysis) can be extremely useful in finding subgroups consisting of patients who have similar covariates and display similar treatment responses. In the context of a clinical trial, HTE analysis can increase the likelihood of identifying subgroups of the population for whom a particular treatment is effective, even when it is found to be ineffective for the population as a whole
Identifying subjects who have similar covariates and display similar treatment responses, requires reliable estimates of the treatment responses of individual subjects; i.e. of ITEs. As we have mentioned earlier, there are a number of methods for estimating ITE; some may be more appropriate in a particular circumstance than others. Several recent approaches for estimating HTE proceed by simultaneously estimating ITE and recursively partitioning the subject population; see especially Athey and Imbens (2016), Su et al. (2009) and Tran and Zheleva (2019). These methods chose partitions to maximize the heterogeneity of treatment effects across subgroups (using a sample mean estimator) under the assumption that treatment effects are homogeneous within subgroups. However, this assumption is not true in practice; as a result, these methods often identify subgroups for which the heterogeneity within the subgroups is comparable to the heterogeneity across subgroups. This leads to wide confidence intervals and often to false discovery – even identifying groups for which the treatment effect is estimated to be positive even though it is simply noise. Obviously, decisions based on such false discovery are useless, if not worse.
We describe a novel robust recursive partitioning (R2P) method for subgroup analysis that overcomes this critical challenge (Lee et al., 2020b). R2P has three distinctive features that separate it from previous methods: it makes a deliberate effort to minimize heterogeneity of treatment effects within each of the subgroups while maximizing heterogeneity across subgroups, it produces confidence guarantees with narrow confidence intervals and it can take make use of any ITE estimator, including estimators that are yet to be proposed.
We formalize the robust partitioning problem in the following way. Combining the given ITE estimator with the method of split conformal regression (Lei et al. 2018) provides ITE estimates together with valid confidence intervals (given any pre-assigned coverage rate). On the basis of these estimates, we define, for any subset of the covariate space (a potential subgroup), the expected absolute deviation
, which is a measure of the heterogeneity of ITE within
, and the expected width of confidence intervals
, which is a measure of how confident we are about the estimates. Ideally, we would like to create a partition
of the covariate space that minimizes both
and
for each element of the partition; because this is impossible, we choose a hyperparameter
and minimize the sum of a convex combination of
and
. That is we formalize the problem as finding a partition
of the covariate space to minimize
It might seem that the solution to this problem would be to choose a very fine partition into small subsets, but this is not so: although the expected deviation may shrink when
does, the expected width of confidence intervals
grows. This problem formulation indirectly balances heterogeneity within subgroups and heterogeneity across subgroups.
To provide empirical evidence that this method works well, we must resort to simulated data. This is unavoidable because in real data we never know both potential outcomes: the subject either received the treatment or did not; in the former case we know the treated outcome and in the latter case we know the untreated outcome. Of course all other empirical studies also use simulated data, for precisely the same reason. Figure 3 presents box plot comparisons of R2P against four benchmark methods for subgroup analysis: standard regression trees for causal effects (CT-A) (Breiman et al. 1984) conformal regression trees for causal effects (CCT) (Johansson et al. 2018), causal trees with honest criterion (CT-H) (Athey and Imbens, 2016) and causal trees with generalization costs (CT-L) (Tran and Zheleva, 2019).
Figure 3. Distribution of treatment effects for subgroups identified by R2P and four benchmark methods using simulated data. The vertical axis is the estimated treatment effect; the horizontal axis indexes the subgroups identified by each method. R2P, CCT and CT-A each identify 5 subgroups, CT-H identifies 4 subgroups and CT-L identifies 3. (See the text for the description of the four benchmark methods.) Each box represents the range between the 25th and 75th percentiles of the treatment effects of the test samples; each whisker represents the range between the 5th and 95th percentiles.

As Figure 3 shows, R2P identifies subgroups reliably: different subgroups display very different average treatment effects and the distributions of the different groups are well-discriminated (non-overlapping). The benchmark methods are unreliable: the distributions of treatment effects are not well-discriminated and false discovery occurs for all four other methods, and occurs frequently for three of the four. The numerical results tell the same story. The objective is to create a partition into subgroups with the property that treatment effects are very heterogeneous across subgroups but very homogeneous within subgroups. The extent to which a partition achieves this objective can be measured by the ratio of the variance in the average treatment effect across subgroups to the variance of the average treatment effect within subgroups; we would like this ratio to be as big as possible. For R2P, this ratio is greater than 20; the ratio for CT-A is less than 8, the ratios for CCT and CT-H are less than 4, and the ratio for CT-L is less than 1 (Lee et al., 2020b).
4. Trials for new drugs designed for treating COVID-19
Existing clinical validation processes for new drugs, such as the processes adopted by regulatory agencies like the U.S. Food and Drug Administration or the European Medicines Agency are well designed but coarse-grained and static: they are conducted sequentially in phases with each phase conducted largely independently of other phases. This process emphasizes confidence in the safety and efficacy of drugs at the cost of long delay; a fully phased sequence of trials can take years – and many drugs fail their trials. During an active outbreak of a pandemic disease for which no treatment is currently known – such as the current COVID-19 pandemic – but during which anecdotal evidence for certain drugs or drug combinations can emerge quickly, a more dynamic and fine-grained validation process should be considered. In the early stages of such a process, confidence in the safety and efficacy of a drug (or combination of drugs) will be lower, but during an ongoing active and dangerous outbreak, continued aggressive testing may be warranted even with a lower confidence level. During a pandemic, a rapid feedback loop of testing and validation cannot be completely separated from treatment. Implementing such a rapid feedback loop will require persuading regulatory agencies to grant approval for investigations that are based on sound models and provide commitment to continued real-time monitoring and the development and use of a global control database.
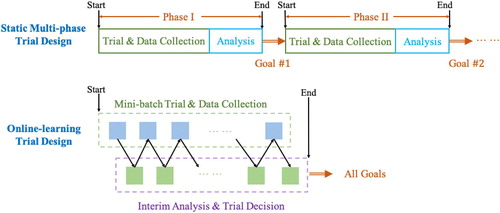
Online learning for design of dynamic clinical trials
A key to expediting clinical trials without sacrificing confidence will be to break the multi-phase paradigm and convert the process into a continuous and adaptive trial-collection-retrial loop, where the data collected previously is used to determine the continuation trial strategy. This online learning paradigm is illustrated in Figure 4. As noted above, in the static clinical trial design, information is not fully utilized: each phase is intended to serve only one purpose, and the data that is collected in each phase is usually not used for design or inference in later stages. In a pandemic setting it would be justifiable and more efficient for the design and execution of the entire sequence of trials to be adjusted "on the fly" in a fine-grained manner on the basis of observed outcomes, rather than determined in advance. This fine-grained dynamic paradigm is particularly well-suited for Bayesian designs because it enabling posterior updating for multiple objectives simultaneously.
Applying a dynamic online-learning-based framework offers the possibility to learn simultaneously about toxicity and efficacy of a new drug. Because this methodology is more efficient, and reduces learning time, it can be particularly useful for time-sensitive clinical trials of COVID-19 treatments. Moreover, given the dire situation of the COVID-19 pandemic, any trial of potential treatments must also take efficacy into account because ethical considerations would require expected therapeutic benefit for participants. Both factors call for the trial design to include efficacy as a co-primary endpoint from the beginning – and not just in the second and third phases.
To highlight the aforementioned challenges and demonstrate the benefits of online learning, we describe an early-phase clinical trial design with a (possibly restricted) class of patients for whom the potential for therapeutic benefit is required. Standard multi-phased designs are not optimal for this purpose because they are not sample-efficient and they do not necessarily maximize the treatment effect for trial participants. The framework we describe addresses these issues by considering toxicity and efficacy jointly in a single trial, rather than sequentially in separate trials. Previous work has focused on jointly modeling efficacy and toxicity and on designing the trial methodology on the basis of statistical algorithms that exploit a multivariate model (Bekele and Shen 2005; Zhang et al. 2006; Yin et al. 2006; Dette et al. 2019). Instead, Shen et al. (2020) formulates this as a proper online learning problem and proposes a method to solve this problem.
Assume a total of dose levels of a new drug are to be tested on a maximum of n patients. Each trial event on a particular patient results in a toxicity event and an efficacy event. For simplicity we assume here that both of these events can be classified as 0 or 1; i.e. that dose-limiting toxicity (DLT) occurred
or did not
, and that the patient’s condition improved
or did not
. (More generally, we might assign degrees of severity of side-effects and degrees of improvement or degradation in condition.) In order to guide the study to improve the condition of as many participating patients as possible, we define the objective of the study to be the maximization of cumulative (expected) efficacy over all patients. In order to insure that as few patients as possible are exposed to unsafe doses, we take as given to us (by regulators, for instance) a toxicity threshold
and a failure threshold
; we then impose the constraint that the probability that the average observed toxicity exceeds
should be no greater than
. This allows us to pursue high efficacy while assuring that toxicity events are unlikely.
We formalize the online learning problem as:
Note that the objective is exactly the average efficacy of treatment and that the constraint is the required degree of safety. This online-learning formulation requires safe exploration for the most effective dose level. For a trial design to meet this requirement it must, in deciding the dose for every new patient, consider both the expected toxicity and the expected efficacy.
To solve this online learning problem Shen et al. (2020) develop a novel method: Safe Efficacy Exploration Dose Allocation (SEEDA). SEEDA employs a new multi-armed bandit algorithm to maximize the cumulative reward function subject to the constraint that the current choice of arm has a low probability of violating the given safety threshold. Shen et al. (2020) demonstrate (both theoretically and empirically) that SEEDA outperforms previously used designs when both efficacy and toxicity are considered and the patient budget is limited.
Sequential patient recruitment
Randomized controlled trials (RCTs) are the gold standard for comparing the effectiveness of a new treatment to the current one. But most RCTs are slow and many RCTs fail (Printz 2015). Most RCTs allocate the patients to the treatment group and the control group by uniform randomization. If patients can be recruited in cohorts (rather than all at once) and the effects on each cohort can be observed before recruiting the next cohort, then ML-based methods developed in recent years (Atan et al. 2019; Harrer et al. 2019) have shown that learning can be dramatically improved – especially if the effects are heterogeneous across identifiable subgroups of patients. In such situations, the patient allocation problem can be formulated as a finite stage Markov decision process (a standard RL model), but with a carefully selected clinical trial design objective (e.g., minimizing a weighted combination of Type I and Type II errors as in the RCT-KG algorithm of Atan et al. (2019)). In particular, the RL based method preserves the randomization feature of RCTs, and enables more efficient adaptive designs by using what has been learned from previous cohorts to adaptively recruit patients to subgroups and allocate patients (to treatment/control). ML methods achieve significant reduction in error and require many fewer patients to achieve a prescribed level of confidence (Atan et al. 2019; Lee et al. 2020a). Furthermore, these new methods can provide significant benefits that outweigh potential inflation of confidence under the circumstances of an active outbreak, see also Dodd et al. (2016); Proschan et al. (2016); Mulangu et al. (2019), see Stallard et al. (2020) for recent overview of such methods in the context of COVID-19 therapies.
5. Conclusion
The current SARS-CoV-2/COVID-19 pandemic represents the greatest global healthcare challenge of our lifetime. Now, and in the immediate future, the need is to identify, approve and distribute treatments and vaccines for COVID-19 – but what we learn in this effort will yield benefits that affect the entire future course of drug development and change the lives of patients across the world.
Many of the technical issues discussed above are particularly acute in the context of a pandemic – but they are by no means uniquely relevant to the current context. The needs to assess temporal shifts in treatment effect, to distinguish the characteristics of a sample of patients recruited to a clinical trial against the real-world disease population and to speed up the process of investigation and approval – to cite only a few examples – have always existed. The challenge today is uniquely acute because of the scale of the pandemic and the variety of unknowns. In the face of this task, the traditional biostatistician might be tempted to follow a familiar path: to rely on the unique skills and methods that have served well in the design and execution of traditional clinical trials and drug-development programs; to approach each individual trial as a separate problem; to find a (locally) optimal way to handle data-integrity issues for a given study; to generate a small dataset for each novel agent; to reach conclusions from that dataset that are at odds with similar datasets generated and conclusions reached by hundreds or thousands of others following a similar path. We have attempted here to suggest a different path: to reach out across disciplines to leverage insights, knowledge and methods from many areas. We believe this will be essential to harness the necessary expertise to address the kind of challenges we now face. We have focused here on machine learning and clinical trials because those are the areas of our expertise, but areas such as epidemiology, natural language processing, operations research, statistics and systems biology – and even advertising and finance – may provide important and necessary contributions.
The scale of the pandemic means that an enormous volume of data is being generated on modes of infection, risk factors, symptoms, treatments, outcomes and on the nature of the virus itself. Because these data come from many sources, they will arrive as fragments, and these fragments must be first be integrated before they can be understood. This will require making these data widely available and easily accessible – while still preserving patient privacy; this is a challenge in itself – but it has been done in various contexts and is becoming easier with the widespread adoption of electronic health records, at least in the developed countries. Indeed, it may be useful to have and integrate not just medical and biological data, but demographic data, geographic data, etc. This is certainly not easy but in can be done – and has been done. (See for instance Alaa and van der Schaar (2020), which provides details of the work of the Cambridge Adjutorium, integrating hospital-level data from across the UK to predict the demand for COVID-19-related resources. The trained algorithm is currently under testing for adoption across the UK.) It has been said that data science is a team sport – and never has a team been more necessary to bringing the required tools to the hands of clinicians, researchers, patients, regulators, payers and many others.
Companies are launching new trials for COVID-19 at unprecedented speed. Adaptations to impacted trials and global trial platforms provide a glimpse of what will be possible post-pandemic, from changing operational aspects (accelerated transitions to virtual visits, digital endpoints from wearables, home-based labs and pharmacokinetics, networks of distributed sites, etc.) to fundamental shifts in the design paradigm (increased platform studies, real-world studies, cross-company collaborations etc.). An important challenge will be to decide which of the different approaches used during the pandemic – because traditional approaches were too slow or not possible – should become standard after the pandemic. Diverse quantitative communities are coming together to address the challenges of this pandemic; our hope is that they will stay together – not just for this pandemic but in the long run, which will greatly improve the conduct of clinical trials in the future.
The authors would like to finish with a huge thank you to all those whose work on the front line of this pandemic is making it possible for us to continue our part of this effort.
References
- Akacha, M., Branson, J., Bretz, F., Dharan, B., Gallo, P., Gathmann, I., Hemmings, R., Jones, J., Xi, D., and Zuber, E. (2020), “Challenges in assessing the impact of the COVID-19 pandemic on the integrity and interpretability of clinical trials”, Statistics in Biopharmaceutical Research, DOI: 10.1080/19466315.2020.1788984
- Alaa, A. M., and van der Schaar, M. (2017), “Bayesian inference of individualized treatment effects using multi-task Gaussian processes,” in Advances in Neural Information Processing Systems 30, 3424–3432.
- Alaa, A. M., and van der Schaar, M. (2017), “Learning from Clinical Judgments: Semi-Markov-Modulated Marked Hawkes Processes for Risk Prognosis,” in Proceedings of the 34th International Conference on Machine Learning, PMLR, 70, 60-69.
- Alaa, A. M., and van der Schaar, M. (2018), “Limits of estimating heterogeneous treatment effects: Guidelines for practical algorithm design,” in Proceedings of the 35th International Conference on Machine Learning, PMLR, 80, 129–138.
- Alaa, A. M. and van der Schaar, M. (2019), “Validating causal inference models via influence functions,” in Proceedings of the 36th International Conference on Machine Learning, PMLR, 97, 191–201.
- Alaa, A. M., and van der Schaar, M. (2019), “Adjutorium COVID-19: Technical documentation,” available on line at http://www.vanderschaar-lab.com/NewWebsite/covid-19/200414_Adjutorium_Documentation.pdf
- Alaa, A. M., Weisz, M., and van der Schaar, M. (2017), “Deep counterfactual networks with propensity-dropout,” International Conference on Machine Learning – Deep learning workshop, 1-6
- Alaimo, S. and Pulvirenti, A. (2018), “Network-based drug repositioning: Approaches, resources and research directions,” in Computational Methods for Drug Repurposing, Humana Press, New York, NY, Springer, 97-113.
- Atan, O., Zame, W. R., and van der Schaar, M. (2019), “Sequential patient recruitment and allocation for adaptive clinical trials,” in Proceedings of the 22nd International Conference on Artificial Intelligence and Statistics (AISTATS), PMLR, 89, 1891–1900.
- Athey, S., and Imbens, G. (2016), “Recursive partitioning for heterogeneous causal effects,” Proceedings of the National Academy of Sciences, 113, 7353–7360.
- Bekele, B. N., and Shen, Y. (2005), “A Bayesian approach to jointly modeling toxicity and biomarker expression in a phase I/II dose-finding trial,” Biometrics, 61, 343–354.
- Berry, D. A. (2004), “Bayesian statistics and the efficiency and ethics of clinical trials,” Statistical Science, 19, 175–187.
- Bica, I., Alaa, A. M., Jordon, J., and van der Schaar, M. (2020), “Estimating counterfactual treatment outcomes over time through adversarially balanced representations,” in Proceedings of the 8th International Conference on Learning Representations, 1-28
- Bica, I., Alaa, A.M., Lambert, C. and van der Schaar, M. (2020), From real-world patient data to individualized treatment effects using machine learning: Current and future methods to address underlying challenges. Clin. Pharmacol. Ther. Accepted Author Manuscript. doi:10.1002/cpt.1907.
- Bica, I., Jordon, J., and van der Schaar, M. (2020), “Estimating the effects of continuous-valued interventions using generative adversarial networks,” arXiv preprint arXiv:2002.12326.
- Breiman, L., Friedman, J., Stone, C.J. and Olshen, R.A. Classification and Regression Trees. CRC Press (1984)
- Bretz, F., Gallo, P., and Maurer, W. (2017), “Adaptive designs: The Swiss Army knife among clinical trial designs?” Clinical Trials, 14, 417–424.
- Committee for Medicinal Products for Human Use [CHMP] (2020), “A call to pool EU research resources into large-scale, multi-centre, multi-arm clinical trials against COVID-19,” [online]. Available at https://www.ema.europa.eu/en/documents/other/call-pool-eu-research-resources-large-scale-multi-centre-multi-arm-clinical-trials-against-COVID-19_en.pdf.
- Cruz-Roa, A., Gilmore, H., Basavanhally, A., Feldman, M., Ganesan, S., Shih, N.N.C., Tomaszewski,J.,Gonzalez, F.A. and Madabhushi, A. (2017), “Accurate and reproducible invasive breast cancer detection in whole-slide images: A Deep Learning approach for quantifying tumor extent,” Scientific Reports 7 (Article number 46450).
- Dette, H., Möllenhoff, K., and Bretz, F. (2019), “Equivalence tests for binary efficacy-toxicity responses,” arXiv preprint arXiv:1910.08769.
- Dodd, L. E., Proschan, M. A., Neuhaus, J., Koopmeiners, J. S., Neaton, J., Beigel, J. D., Barrett, K., Lane, H. C., and Davey Jr, R. T. (2016), “Design of a randomized controlled trial for Ebola virus disease medical countermeasures: PREVAIL II, the Ebola MCM Study,” The Journal of Infectious Diseases, 213(12), 1906–1913.
- European Medicines Agency [EMA] (2020a), “Points to consider on implications of Coronavirus disease (COVID-19) on methodological aspects of ongoing clinical trials,” [online]. Available at https://www.ema.europa.eu/en/documents/scientific-guideline/points-consider-implications-coronavirus-disease-COVID-19-methodological-aspects-ongoing-clinical_en.pdf.
- European Medicines Agency [EMA] (2020b), “Guidance on the management of clinical trials during the COVID-19 (Coronavirus) pandemic,” [online]. Available at https://ec.europa.eu/health/sites/health/files/files/eudralex/vol-10/guidanceclinicaltrials_covid19_en.pdf.
- European Medicines Agency [EMA] (2020c), “Coronavirus disease (COVID-19),” [online]. Available at https://www.ema.europa.eu/en/human-regulatory/overview/public-health-threats/coronavirus-disease-COVID-19.
- Filipovych, R., Resnick, S.M., and Davatzikos, C. (2011), “Semi-supervised cluster analysis of imaging data,” NeuroImage, 54, 2185-2197
- Food and Drug Administration [FDA] (2020a), “Letter of Emergency Use Authorization (EUA) for emergency use of remdesivir for the treatment of hospitalized 2019 coronavirus disease (COVID-19) patients”. Available at https://www.fda.gov/media/137564/download.
- Food and Drug Administration [FDA] (2020b), “US Food and Drug Administration. Guidance for Industry, Investigators, and Institutional Review Boards. FDA Guidance on Conduct of Clinical Trials of Medical Products during COVID-19 Pandemic.” Available at https://www.fda.gov/media/136238/download.
- Gittins, J. C. (1979). Bandit processes and dynamic allocation indices. Journal of the Royal Statistical Society, Series B (Methodological), 41, 148-164.
- Harrer, S., Shah, P., Antony, B., and Hu, J. (2019), “Artificial intelligence for clinical trial design,” Trends in Pharmacological Sciences, 40, 577-591.
- Hill, J. L. (2011). Bayesian nonparametric modeling for causal inference. Journal of Computational and Graphical Statistics, 20, 217-240.
- Hobbs, B. P., Carlin, B. P., Mandrekar, S. J., and Sargent, D. J. (2011), “Hierarchical commensurate and power prior models for adaptive incorporation of historical information in clinical trials,” Biometrics, 67, 1047–1056.
- Infectious Diseases Data Observatory [IDDO] (2020), “Living systematic review COVID-19. Coronavirus disease 2019 registered clinical trials,” [online]. Available at https://www.iddo.org/research-themes/COVID-19/live-systematic-clinical-trial-review.
- Johansson, U., Linusson, H., Löfström, T. and Boström, H., (2018) “Interpretable regression trees using conformal prediction”, Expert Systems with Applications, 97:394–404, 2018.
- Kunz, C. U., Joergens, S., Bretz, F., Stallard, N., Van Lancker, K., Xi, D., Zohar, S., Gerlinger, C., and Friede, T. (2020) “Clinical trials impacted by the COVID-19 pandemic: Adaptive designs to the rescue?” Statistics in Biopharmaceutical Research (under review)
- Lai, T. L., and Robbins, H. (1985). Asymptotically efficient adaptive allocation rules. Advances in Applied Mathematics, 6, 4-22.
- Lee, H.-S., Shen, C., Jordon, J., and van der Schaar, M. (2020a), “Contextual constrained learning for dose-finding clinical trials”, in Proceedings of the 23rd International Conference on Artificial Intelligence and Statistics (AISTATS), PMLR, 108, 2645-2654.
- Lee, H.-S., Zhang, Y., Zame, W., Shen, C., Lee, J.-W. and van der Schaar, M. (2020b),”Robust Recursive Partitioning for Heterogeneous Treatment Effects with Uncertainty Quantification,” arXiv preprint arXiv:2006.07917.
- Lee, J. J., and Chu, C. T. (2012), “Bayesian clinical trials in action,” Statistics in Medicine, 31, 2955–2972.
- Lei, J., G’Sell, M., Rinaldo, A., Tibshirani, R. J., and Wasserman, L. (2018), “Distribution-free predictive inference for regression,” Journal of the American Statistical Association, 113, 1094–1111.
- Lim, B., Alaa, A. M., and van der Schaar, M. (2018), “Forecasting treatment responses over time using recurrent marginal structural networks,” in Advances in Neural Information Processing Systems 31, 7483–7493.
- Meyer, R. D., Ratitch, B., Wolbers, M., Marchenko, O., Quan, H., Li, D., Fletcher, C., Li, X., Wright, D., Shentu, Y., Englert, S., Shen, W., Dey, J., Liu, T., Zhou, M., Bohidar, N., Zhao, P.-L., and Hale, M. (2020) “Statistical issues and recommendations for clinical trials conducted during the COVID-19 pandemic,” Statistics in Biopharmaceutical Research, doi: 10.1080/19466315.2020.1779122.
- Mulangu, S., Dodd, L. E., Davey Jr, R. T., Tshiani Mbaya, O., Proschan, M., Mukadi, D., Lusakibanza Manzo, M., Nzolo, D., Tshomba Oloma, A., Ibanda, A., et al. (2019), “A randomized, controlled trial of Ebola virus disease therapeutics,” New England Journal of Medicine, 381(24), 2293–2303.
- Neil, D., Pfeiffer, M., and Liu, S.-C. (2016), “Phased LSTM: Accelerating recurrent network training for long or event-based sequences,” in Advances in neural information processing systems 29, 3882–3890.
- Neyman, J. (1923), “Sur les applications de la théorie des probabilités aux experiences agricoles: Essai des principes,” Roczniki Nauk Rolniczych, 10, 1–51.
- Oxford (2020), “University of Oxford “RECOVERY” trial for COVID-19 treatments,” [online]. Available at https://www.recoverytrial.net/.
- Pallmann, P., Bedding, A. W., Choodari-Oskooei, B., Dimairo, M., Flight, L., Hampson, L. V., Holmes, J., Mander, A. P., Sydes, M. R., Villar, S. S., Wason, J. M. S., Weir, C. J., Wheeler, G. M., Yap, C., and Jaki, T. (2018), “Adaptive designs in clinical trials: why use them, and how to run and report them,” BMC medicine, 16(1), 29.
- Park, J. J., Thorlund, K., and Mills, E. J. (2018), “Critical concepts in adaptive clinical trials,” Clinical epidemiology, 10, 343-351.
- Printz, C. (2015), “Failure rate: Why many cancer drugs don’t receive FDA approval, and what can be done about it,” Cancer, 121(10), 1529–1530.
- Proschan, M. A., Dodd, L. E., and Price, D. (2016), “Statistical considerations for a trial of Ebola virus disease therapeutics,” Clinical Trials, 13(1), 39–48.
- Rubin, D. B. (1978), “Bayesian inference for causal effects: The role of randomization,” The Annals of statistics, 6(1) 34–58.
- Schmidli, H., Häring, D. A., Thomas, M., Cassidy, A., Weber, S., and Bretz, F. (2019), “Beyond randomized clinical trials: Use of external controls,” Clinical Pharmacology & Therapeutics, 107(4), 806-816.
- Segar.,M., Patel, K.V., Ayers, C., Basit, M., Tang, W.H.W., Willett, D., Berry, J., Grodin, J.L. and Pandey. A. (2020), “Phenomapping of patients with heart failure with preserved ejection fraction using machine learning based unsupervised cluster analysis,” European Journal of Heart Failure 22, 148-158.
- Shalit, U., Johansson, F. D., and Sontag, D. (2017), “Estimating individual treatment effect: generalization bounds and algorithms,” in Proceedings of the 34th International Conference on Machine Learning, PMLR, 70, 3076–3085.
- Shen, C., Wang, Z., Villa, S., and van der Schaar, M. (2020). Learning for Dose Allocation in Adaptive Clinical Trials with Safety Constraints. in Proceedings of the 37th International Conference on Machine Learning.
- Shukla, S. N., and Marlin, B. (2019), “Interpolation-prediction networks for irregularly sampled time series,” in Proceedings of the 7th International Conference on Learning Representations.
- Stallard, N., Hampson, L., Benda, N., Brannath, W., Burnett, T., Friede, T., Kimani, P.K., Koenig, F., Krisam, J., Mozgunov, P., Posch, M., Wason, J., Wassmer, G., Whitehead, J., Williamson, S.F., Zohar, S., and Jaki, T. (2020) "Efficient adaptive designs for clinical trials of interventions for COVID-19," Statistics in Biopharmaceutical Research, DOI: 10.1080/19466315.2020.1790415
- Su, Q., Ma, F., Yuan, Y., Huai, M., Zhong, W., Gao, J., and Zhang, A. (2018), “Deep patient similarity learning for personalized healthcare,” IEEE Transactions on Nanobioscience, 17(3), 219–227.
- Thompson, W. R. (1933). On the likelihood that one unknown probability exceeds another in view of the evidence of two samples. Biometrika, 25(3/4), 285-294.
- Tibshirani, R. J., Barber, R. F., Candes, E., and Ramdas, A. (2019), “Conformal prediction under covariate shift,” in Advances in Neural Information Processing Systems 32, 2526–2536.
- Tran, C., and Zheleva, E. (2019), “Learning triggers for heterogeneous treatment effects,” in Proceedings of the AAAI Conference on Artificial Intelligence, 33, 5183–5190.
- Varatharajah, Y., Berry, B., Koyejo, S., and Iyer, R. (2018), “A Contextual-bandit-based approach for informed decision-making in clinical trials,” arXiv preprint arXiv:1809.00258.
- Villar, S. S., Bowden, J., and Wason, J. (2015), “Multi-armed bandit models for the optimal design of clinical trials: benefits and challenges,” Statistical Science, 30(2), 199-215.
- Wang, Y., Zhang, D., Du, G., Du, R., Zhao, J., Jin, Y., Fu, S., Gao, L., Cheng, Z., Lu, Q. and Hu, Y. (2020). Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. The Lancet 2020, 395, 1569-78.
- World Health Organization [WHO] (2020), “Solidarity” clinical trial for COVID-19 treatments,” [online]. Available at https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-clinical-trial-for-COVID-19-treatments.
- Yin, G., Li, Y., and Ji, Y. (2006), “Bayesian dose-finding in phase I/II clinical trials using toxicity and efficacy odds ratios,” Biometrics, 62, 777–787.
- Yin, K., and Cheung, W. K. (2019), “Context-aware imputation for clinical time series,” in Proceedings of 2019 IEEE International Conference on Healthcare Informatics (ICHI), 1-3.
- Yoon, J., Jordon, J., and van der Schaar, M. (2018a), “GANITE: Estimation of individualized treatment effects using generative adversarial nets,” in Proceedings of the 6th International Conference on Learning Representations, 1-22.
- Yoon, J., Jordon, J., and van der Schaar, M. (2018b), “GAIN: Missing data imputation using generative adversarial nets,” Proceedings of the 35th International Conference on Machine Learning, PMLR 80, 5689-5698.
- Yoon, J., Zame, W. R., and van der Schaar, M. (2017), “Multi-directional recurrent neural networks: A novel method for estimating missing data,” in Time Series Workshop at the 34th International Conference on Machine, 1-5.
- Yoon, J., Zame, W. R., and van der Schaar, M. (2018c), “Estimating missing data in temporal data streams using multi-directional recurrent neural networks,” IEEE Transactions on Biomedical Engineering, 66, 1477–1490.
- Zhang, W., Sargent, D. J., and Mandrekar, S. (2006), “An adaptive dose-finding design incorporating both toxicity and efficacy,” Statistics in Medicine, 25, 2365–2383.
- Zhu, Z., Yin, C., Qian, B., Cheng, Y., Wei, J., and Wang, F. (2016), “Measuring patient similarities via a deep architecture with medical concept embedding,” in Proceedings of 2016 IEEE 16th International Conference on Data Mining (ICDM), 749–758.