ABSTRACT
Rotavirus (RV) is the leading cause of diarrhea-related death in children worldwide and ninety-five percent of rotavirus deaths occur in Africa and Asia. Rotavirus vaccines (RVV) can dramatically reduce RV deaths, but have low efficacy in low-income settings where they are most needed. The intestinal microbiome may contribute to this decreased RVV efficacy. This pilot study hypothesizes that infants' intestinal microbiota composition correlates with RVV immune responses and that RVV responders have different gut microbiota as compared to non-responders.
We conducted a nested, matched case-control study comparing the pre-vaccination intestinal microbiota composition between 10 6-week old Pakistani RVV-responders, 10 6-week old Pakistani RVV non-responders, and 10 healthy Dutch infants. RVV response was defined as an Immunoglobulin A of ≥20 IU/mL following Rotarix™(RV1) vaccination in an infant with a pre-vaccination IgA<20. Infants were matched in a 1:1 ratio using ranked variables: RV1 dosing schedule (6/10/14; 6/10; or 10/14 weeks), RV season, delivery mode, delivery place, breastfeeding practices, age and gender. Fecal microbiota analysis was performed using a highly reproducible phylogenetic microarray.
RV1 response correlated with a higher relative abundance of bacteria belonging to Clostridium cluster XI and Proteobacteria, including bacteria related to Serratia and Escherichia coli. Remarkably, abundance of these Proteobacteria was also significantly higher in Dutch infants when compared to RV1-non-responders in Pakistan.
This small but carefully matched study showed the intestinal microbiota composition to correlate with RV1 seroconversion in Pakistan infants, identifying signatures shared with healthy Dutch infants.
Introduction
Rotavirus (RV) is one of the most important causes of diarrhea-related death in children under the age of five worldwide, particularly in Africa and Asia, where over 95% of RV deaths occur.Citation1 Since their introduction, rotavirus vaccines (RVV) have significantly reduced the number of cases of serious rotavirus gastroenteritis in developing country settings, however their effectiveness is disappointingly low.Citation2,3 In wealthy countries RVV have an efficacy of as high as 85–98% against severe RV-related gastroenteritis,Citation4-7 however this efficacy drops significantly in low-income countries.Citation8 Well-performed randomized controlled studies in Africa and Asia have shown a combined vaccine efficacy ranging from 48 to 64% for the Rotarix™ and RotaTeq™ vaccines and 53% for the human-bovine (116E) rotavirus vaccine in India.Citation9-12 Rotarix™ is currently being introduced in Pakistan's Expanded Program on Immunization (EPI).
Improving RVV efficacy in low-income settings could save hundreds of thousands of children's lives.Citation13 Several studies have demonstrated that the etiology of RVV's diminished immunogenicity is likely multi-factorial: due to interference from the first dose of co-administered oral polio vaccine (OPV), host FUT2 and FUT3 receptor status and HLA blood group antigen type (HBGA), as well as RVV immune response suppression from maternally derived antibodies.Citation14-17 Yet these explanations fail to completely explain how RVV seroconversion can be even lower than 40% in some settings like an urban slum in Karachi, Pakistan.Citation18,19 Given that the intestinal microbiome differs significantly in different geographic populations,Citation20 evidence is emerging that differences in the intestinal microbiome may help explain this gap in RVV immunogenicity.Citation21,22 This study describes a pilot to explore whether the infant intestinal microbiota may correlate with RVV immunogenicity in a poor urban slum setting. We hypothesized that if the composition of the intestinal microbiota is influencing RVV response, then RVV responders should have a different intestinal microbiome composition as compared to non-responders. To test this, we conducted an exploratory, nested, case-control pilot study in Karachi, Pakistan comparing the differences in pre-vaccination fecal microbiota composition between infants with and without an anti-rotavirus Immunoglobulin (Ig) A seroconversion to RV1.
A second hypothesis was that if there are biologically significant differences in microbiome composition between vaccine responders and non-responders within Pakistan, one would expect similar differences in microbiome composition to exist between developing and developed country infants (with assumed high RV1 seroconversion rate). Specifically, one would assume a positive or negative gradient in microbiome differences across populations – developed country, then Pakistani responders then Pakistani non-responders. To test this hypothesis, we additionally compared the microbiome composition of Pakistani infants to a cohort of healthy Dutch infants.
Results
Study population
A total of 88 infants with pre-vaccination fecal samples were available from the original clinical trials. Of these, 14 children were excluded: 11 children migrated out of the catchment area, 2 children had guardians refuse consent, and 1 died. Seventy-four children were screened for their anti-RV IgA response, and 2 had evidence of previous RV natural exposure and one child had no post-vaccination IgA measurement available. Therefore, 71 children were eligible and enrolled in this study.
Sufficient and good-quality DNA was obtained from 66 of the 71 fecal samples (93%) and these were further characterized using phylogenetic microarray analysis.
Ten (15%) of the 66 infants had an IgA ≥ 20 IU/mL and classified as RV1 responders and these infants were then carefully matched based on six relevant variables to an equal number of RV1 non-responders (see for baseline characteristics). No significant differences between the vaccine responders and non-responders were identified for any of the variables including malnutrition indices.
Table 1. Baseline characteristics RV1 Non-responders and Responders.
Microbiota composition
When evaluating the microbiome composition as a whole, a significantly higher ratio of Gram-negative to Gram-positive bacteria was observed in vaccine responders as compared to non-responders (2.6-fold higher, p = 0.03). Subsequently, when evaluating microbiome composition at the phylum level, RV1 responders had significantly higher levels of Firmicutes, in particular bacteria belonging to the abundant Clostridium cluster XI (p 0.02, FDR 0.36), and Proteobacteria (p 0.04, FDR 0.36) than non-responders. (Supplementary Table 1 and -, and ).
Figure 1. Distribution of the relative abundance of bacteria between Pakistani responders and non-responders. The relative abundance (%) for all phyla (class for Firmicutes) (1A), significantly different phyla (p < 0.05) (1B), and significantly different bacteria at the genus-like level (FDR < 0.5) (1C) are illustrated. Bean plots compare Pakistani responders (1, orange) and non-responders (0, red). The horizontal black line is the median and the height of each bean plot illustrates the distribution of the values for abundance within each group.
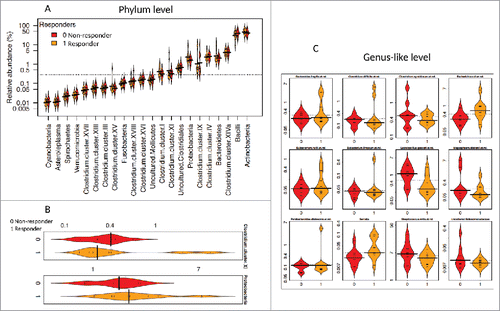
Figure 2. Phylogenetic Heat Tree illustrates the differences in relative bacterial abundance between Pakistani non-responders and responder infants. Colored blue are bacteria where a lower abundance associates with RVV response and colored red are bacterial groups where a higher abundance correlates with RVV response.
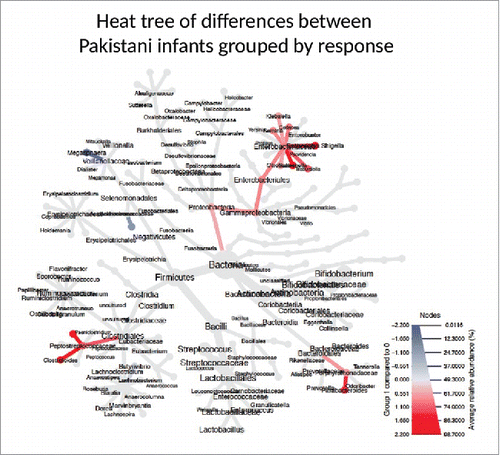
When evaluated at the genus-like level, several bacteria differed between responders and non-responders. The most notable difference observed was in the relative abundance of Gram-negative bacteria related to Serratia and Escherichia coli, which were positively associated with vaccine response and were approximately 2 and 4-fold reduced, respectively in the non-responders (p 0.01, FDR 0.19 for Serratia and p 0.00, FDR 0.05 for E. coli) (Supplementary Table 2, , and ).
Pakistani infants compared to healthy dutch infants
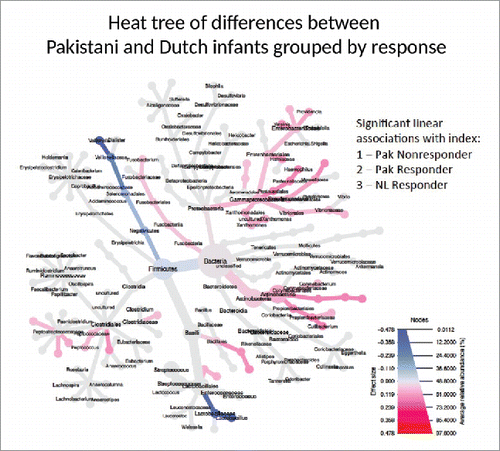
In a second analysis, we hypothesized that if microbial differences identified between RV1 responders and non-responders in Pakistan were actually influencing vaccine response, then similar differences should exist between vaccine non-responders in Pakistan and children in wealthier countries, with high levels of RVV protection. First we compared all Pakistani infants to a set of 154 age-matched Dutch infants. Multivariate analysis demonstrated a significantly different overall microbial composition between all Dutch infants and Pakistani infants (p = 0.014), as expected by geography (See Supplementary Fig. 1). We then made a response index (Pakistani non-responders 1, responders 2, and Dutch infants 3) to evaluate with a heat tree which bacteria correlated with hypothetical RVV response (Illustrated in ). Subsequently, to limit potential confounding, we matched 10 Dutch infants to 10 Pakistani non-responder infants (). Bacterial correlations with RVV response were very similar in the matched and unmatched analyses. There were no differences in diversity, richness, or evenness. Gram-negative Proteobacteria had significantly higher abundance in the Dutch infants (10-fold higher, FDR 9 × 10−12), especially the Gammaproteobacteria (15-fold higher, FDR 9 × 10−12) which includes bacteria such as Serratia, E. coli, Klebsiella, and Enterobacter. (See Supplementary Table 3). In addition, Dutch infants had higher abundance of bacteria related to Staphylococci (4-fold increase, FDR 0.003) and the phylum Verrucomicrobiae (5-fold increase, FDR.0002). (Supplementary Table 3)
Figure 3. Phylogenetic Heat Tree illustrates the differences in relative bacterial abundance between all Pakistani non-responder, Pakistani responder and Dutch infants when indexed by response. Each group's RVV response is indexed (Pakistani non-responder 1, responder 2, and Dutch infants 3) and those bacteria with significant linear associations with index are colored. Colored blue are bacteria where a lower abundance associates with RVV response and colored red are bacterial groups where a higher abundance correlates with RVV response.
Discussion
In a cohort of 66 Pakistani infants, we observed only 10 to be seroconverted after RV1 vaccination, confirming the low response often reported in low-income countries. The pre-vaccination (6 weeks) microbiota of the 10 responding infants was compared with that of a carefully matched group of non-responding Pakistani infants. We observed that the RV1 response was positively correlated with increased ratio of Gram-negative over Gram-positive bacteria, notably reflected in an approximately 3-fold increased abundance of the Proteobacteria related to Serratia and E. coli. Interestingly, when the intestinal microbiota of matched Dutch infants with assumed high RVV immunogenicity was compared with that of the Pakistani RV1 non-responders, again significantly higher abundance of Proteobacteria was observed, particularly Gammaproteobacteria which include bacteria related to Serratia and E. coli.
The results of this pilot study are best regarded as preliminary and hypothesis-generating for an under-developed research field. Gram-negative bacteria, such as Proteobacteria, can stimulate specific innate immune responses, such as through their expression of flagella or toxigenic LPS. In this Pakistani infant population, Proteobacteria or their cell envelope components may therefore be acting as natural immune adjuvants. Murine models have demonstrated that bacterial flagella can cure RV infections through an activation of the innate immune system via TLR5 and NLRC4.Citation23 Accordingly, early exposure to Proteobacteria-expressing flagella may augment innate and subsequent adaptive immune response to RV1. An additional hypothesis is that a higher abundance of LPS derived from E. coli may also be augmenting RV1 immune responses in responders – accumulating data demonstrates that E.coli-derived LPS has stronger immune-stimulatory capacity than LPS in the cell walls of other gram-negative organisms such as those in the Bacteroidetes phylum.Citation24,25 In a parallel study correlating RV1 response with microbiome composition in rural Ghana, we recently observed that a high abundance of Bacteroidetes in the fecal microbiome was negatively correlated with RV1 seroconversion.Citation21 While undeniably speculative, these two studies may complement one another – Proteobacteria and E. coli- derived LPS might boost RV1 responses in some populations whereas Bacteroidetes-derived, and potentially inhibitory, LPS might inhibit RV1 responses in others.
Alternatively, more abundant neonatal colonization with Proteobacteria could have a role in educating the innate immune system. While a high abundance of Proteobacteria is often correlated with an inflammatory gut profile,Citation26 some evidence exists that E. coli may be particularly important in strengthening adaptive B cell immunity. Early colonization with E. coli was associated with higher numbers of memory CD20+ B cells at 4 and 18 months of age in a Swedish cohort.Citation27
Finally, HBGA interactions may be modulating vaccine strain replication. Bacteria might be expressing blood group antigens or glycans needed for RV replication, as has been demonstrated with norovirus.Citation28 Given the known association between HBGAs and microbiome compositionCitation29 as well as HBGA and rotavirus immunogenicity,Citation17,30 host genetics could be interacting with both microbiome composition and viral replication.
This exploratory study has significant limitations restricting the generalizability of our findings despite having high internal validity. The study is primarily limited by power – the low vaccine response (15%) among infants with an available fecal sample meant that we were only able to evaluate 10 RV1 responders in depth. Additionally, in the original dosing study, RV IgA seroconversion was 36% to 39%, depending on study arm, so the cohort of RV1 responder infants that we were able to examine might be a skewed patient population. Anti-RV IgA sero-conversion is also only a correlate of vaccine protection, and clinical vaccine efficacy would require larger sample sizes and follow-up. Additionally, the infant microbiome is characterized by a low diversity and high inter-individual variability with large microbiome fluctuations over time.Citation20 We examined one time point directly prior to RV1 vaccination and our study is unable to account for variation over time. Nevertheless, we believe that the time point directly prior to RV1 vaccination is the most important window into the potential interaction between the rotavirus vaccine and the infant's microbiome. A potential technical limitation is that the HITChip phylogenetic microarray used in this study, while validated in 1000s of subjects, has never been tested in a developing country setting like Pakistan and may not identify novel bacterial phylotypes. Since most if not all intestinal genera are covered on this microarray, we expect that the cross-hybridization that occurs would permit detection of the most abundant microbial populations, even if some phylotypes are not covered by the used probes. However, a similar problem could also occur with sequencing techniques: sequences are mapped to a reference database. If the reference database does not contain a phylotype because it is novel the phylotype will be identified as the most similar, known phylotype, which is the same as what occurs with HITChip cross-hybridization.
Confirming these intriguing study results in other low-income settings could help substantiate a role for the intestinal microbiome in influencing rotavirus vaccine efficacy. Identification of key bacterial phylotypes that correlate with RVV response could also be the first step designing an intervention to improve RVV efficacy. Such a future intervention could have several theoretical forms – modulation of the infant microbiota prior to vaccination with targeted antibiotics (not an ideal strategy in infant populations), evidence-based probiotic or modulation via targeted bacteriophage therapy. Alternatively, understanding of which bacteria elicit rotavirus vaccine immunogenicity could support testing for novel adjuvantation of this oral vaccine. Finally, microbiota profiles might be used to predict which infants are at risk for RVV failure and in need of additional vaccine doses or targeted education.
Methods
Study design and participants
All participating Pakistani infants were healthy, 4–6 weeks of age at enrolment, with a birth-weight > 1500g (exact gestational age unknown), and had been previously enrolled in a phase IV, open-label randomized clinical trial of RV1 immunogenicity (NCT01199874, clinicaltrials.gov) examining the effect of different RV1 dosing schedules (6/10/14, 6/10 and 10/14 weeks) and withholding breastfeeding around the time of vaccine administration on RV1 immunogenicity.Citation18,19
The trial was conducted in a peri-urban, low-income slum along the seacoast in Karachi, Pakistan. Serum samples were collected prior to the first dose of vaccine and 28 days following the last dose of vaccine (14 or 18 weeks depending on study arm) for anti-rotavirus IgA antibody measurements. Fecal samples were collected directly (0–2 days) prior to and in the week following vaccination for the first 88 infants enrolled in these trials, further fecal sample collection was halted due to logistical issues.
Infants were included in the current study if guardians had provided informed consent in the original phase IV study and additionally consented to collection of a new fecal sample collection (results not described here). Further inclusion criteria specified the availability of a baseline, pre-vaccination (0–2 days preceding vaccination) fecal sample, and that there was no evidence of natural RV infection prior to vaccination (a pre-vaccination IgA ≥20 IU/mL). IgA seroconversion to greater than 20 IU/mL following vaccination was considered a surrogate marker for RV1 protection against severe rotavirus gastroenteritis.Citation31 Participating infants were grouped as either RV1 responders (post-vaccination 14 and/or 18 week anti-RV IgA antibody ≥ 20 IU/mL in infants with a pre-vaccination IgA < 20 IU/mL) or non-responders and matched in a 1:1 ratio using the ranked variables: RV1 study arm (dose and breastfeeding arms), RV season in Karachi (defined as the date of the 28 days post-vaccination serum IgA falling between December 1st, 2011 and February 1st, 2012), mode of delivery (vaginal or caesarean section), breastfeeding practices (exclusive breastfeeding, breastfeeding and formula, or formula only), place of delivery (home or health care facility), age of fecal sample collection (in weeks), and sex.
The nested trial was approved by the institutional review board of Aga Khan University and was conducted in accordance with good clinical practice guidelines.
In parallel, we anonymously compared the microbiota of healthy, full-term Dutch infants to our Pakistani RV1 non-responders and responders. These healthy Dutch infants had not received RVV as RVV is not included in the Dutch national vaccination program, but it was assumed that if they had, their RVV seroconversion rate would be well over 90% based on extensive efficacy and effectiveness data for RVV in Northern Europe.Citation4,7 Only Dutch infants without colic, co-morbidities, or maternal stress were eligible for inclusion. The Pakistani infants were compared to an age-matched only cohort of 154 infants and then compared to Dutch infants matched for gender, age at fecal sample collection (4–5 weeks of age), place of delivery, mode of delivery, breast-feeding practices, and birth weight. Fecal samples from Dutch infants were collected as part of a previously published study (Bibo study) in which infants and mothers were followed from the third trimester to 5–6 years of age. All infants' fecal samples were stored, processed for DNA extraction and further analysed in an identical protocol using the Human Intestinal Tract Chip (HITChip) phylogenetic microarray (see below).Citation32,33
Laboratory evaluations
IgA assay
Serum obtained 0–2 days prior to vaccination and 28 days post last RV1 dose was evaluated for anti-RV IgA antibody. Anti-RV IgA antibody was measured in serum using an enzyme-linked immunosorbent assay (ELISA) described elsewhere, expressed as international units per milliliter (IU/mL).Citation34
Fecal microbiota analysis
Fecal samples obtained 0–2 days prior to vaccination, were kept at 0–4C after collection and were frozen to −80C within 24–48 hours of collection. Total DNA was extracted from the fecal material by a repeated bead beating procedure using a modified protocol for the QiaAmp DNA MiniStool Kit (Qiagen, Hilden, Germany) as previously described.Citation35
Analysis of the microbiota composition was performed using the HITChip phylogenetic microarray, which contains oligonucleotide probes for hypervariable regions on the 16S rRNA gene.Citation36 The HITChip is a comprehensive and highly reproducible phylogenetic microarray that enables the parallel profiling and the semi-quantitative analysis of over 1100 phylotypes representing all major intestinal phyla grouped in 130 genus-like groups described for the human intestinal microbiota. Hybridization of extracted DNA from sample to the oligonucleotide probes on the HITChip yields a signal intensity per probe and can thereby provide a quantitative and qualitative phylogenetic profiling of the microbiome composition. The HITChip has been used in over a dozen studies and validated in analyses of over 1000 subjects.Citation32,37
All samples were analyzed on two independent microarray experiments and the data only passed the quality control if the inter-experiment Pearson correlation was >0.97. The signal intensities were normalized using the fRPA methodCitation38 and summarized at different levels of phylogenetic resolution: species, genus, and phylum, except for the Firmicutes, which was further divided to Clostridium clusters and Bacilli. This high-throughput technique has been bench marked with ultra-deep sequencing of 16S rRNA and next-generation parallel sequencing of intestinal metagenomes.Citation39,40
Statistical analysis
Cases were defined as RV1 responders – infants with a serum RV IgA < 20 IU/mL pre-vaccination and IgA ≥ 20 IU/mL post-vaccination. Controls were defined as RV1 non-responders – infants with a serum RV IgA < 20 IU/mL pre-vaccination and post-vaccination.
A Chi-square test was used to determine statistically significant differences in baseline characteristics between RV1 responders (case) and non-responders (controls).
Comprehensive multivariate statistical analyses were performed using Canoco 5.0 software for Windows.Citation41 Principal coordinate (PCA) and redundancy analyses (RDA) were used to evaluate differences in the overall microbial composition between the study groups. The 130 genus-like bacterial groups targeted by the HITChip were used as biological variables and environmental variables were the matching variables named above. Monte Carlo permutation testing (MCPT) assessed the significance of the effect of these variables in the data set.
The relative abundance of specific bacterial groups in the fecal microbiota was determined at the genus-like level and at the phylum level (class for the Firmicutes). Generalized linear model with negative binomial distribution or generalized least squares model were used to determine significant differences in composition and p-values were corrected for false discovery rate (FDR) by the Benjamini-Hochberg method.
Graphical representations and modelling of the relative abundance and correlations with RVV response were performed with the package mareCitation42 within R,Citation43 utilizing the packages nlme,Citation44 MASS,Citation45 bean plot,Citation46 metacoder,Citation47 and vegan.Citation48
Disclosure of potential conflicts of interest
The authors report no disclosures of interest.
KGMI_A_1376162_Supplemental.docx
Download MS Word (421 KB)Acknowledgments
This study is dedicated to Professor Joseph (Joep) Lange, who died prior to its completion. He was a global health activist and scientist and this work would not have been possible without his initiative, support and encouragement. The authors acknowledge and thank all the Pakistani and Dutch families who participated in this study and the staff members of the trial team in Pakistan for their work in conducting this study as well as members of the BIBO study team in the Netherlands. The authors would like to thank Monica McNeal and her staff at the Laboratory of Specialized Clinical Studies at the Cincinnati Children's Hospital, who performed all of the immunoglobulin testing for the original dosing study. The authors would also like to thank Duncan Steele and Jessica Fleming for their contribution to the original dosing study. This study is registered at Clinicaltrials.gov: NCT02220439.
Funding
The original study Pakistani study was supported by PATH through funding from the Bill and Melinda Gates Foundation. The authors also thank GSK for their support in funding part of this work. GSK was provided the opportunity to review a preliminary version of this manuscript for factual accuracy, but the authors are solely responsible for final content and interpretation. The BIBO study was funded by the Netherlands Organization for Scientific Research (NWO) and the Behavioral Science Institute (BSI) of Radboud University. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of GSK, PATH, the Bill and Melinda Gates Foundation, Centers for Disease Control and Prevention, NWO, or BSI.
References
- Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD, WHO-coordinated Global Rotavirus Surveillance Network. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12:136-41. doi:10.1016/S1473-3099(11)70253-5.
- Bar-Zeev N, Kapanda L, Tate JE, Khuzwayo CJ, Iturriza-Gomez M, Nakagomi O, Mwansambo C, Costello A, Parashar UD, Heyderman RS a, et al. Effectiveness of a monovalent rotavirus vaccine in infants in Malawi after programmatic roll-out: an observational and case-control study. Lancet Infect Dis. 2015;15:422-8. doi:10.1016/S1473-3099(14)71060-6.
- Abeid KA, Jani B, Cortese MM, Kamugisha C, Mwenda JM, Pandu AS, Msaada KA, Mohamed AS, Khamis AU, Parashar UD, et al. Monovalent Rotavirus Vaccine effectiveness and impact on rotavirus hospitalizations in Zanzibar, Tanzania: data from the first 3 years after introduction. J Infect Dis. 2016;jiw524. doi:10.1093/infdis/jiw524.
- Ruiz-Palacios GM, Pérez-Schael I, Velázquez FR, Abate H, Breuer T, Clemens SC, Cheuvart B, Espinoza F, Gillard P, Innis BL, et al. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N Engl J Med. 2006;354:11-22. doi:10.1056/NEJMoa052434.
- Linhares AC, Velázquez FR, Pérez-Schael I, Sáez-Llorens X, Abate H, Espinoza F, López P, Macías-Parra M, Ortega-Barría E, Rivera-Medina DM, et al. Efficacy and safety of an oral live attenuated human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in Latin American infants: a randomised, double-blind, placebo-controlled phase III study. Lancet. 2008;371:1181-9. .doi:10.1016/S0140-6736(08)60524-3.
- Vesikari T, Karvonen A, Prymula R, Schuster V, Tejedor JC, Cohen R, Meurice F, Han HH, Damaso S, Bouckenooghe A. Efficacy of human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in European infants: randomised, double-blind controlled study. Lancet. 2007;370:1757-63. doi:10.1016/S0140-6736(07)61744-9.
- Vesikari T, Matson DO, Dennehy P, Van Damme P, Santosham M, Rodriguez Z, Dallas MJ, Heyse JF, Goveia MG, Black SB, et al. Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N Engl J Med. 2006;354:23-33. doi:10.1056/NEJMoa052664.
- Fischer Walker CL, Black RE. Rotavirus vaccine and diarrhea mortality: quantifying regional variation in effect size. BMC Public Health. 2011;11 Suppl 3:S16. doi:10.1186/1471-2458-11-S3-S16.
- Armah GE, Sow SO, Breiman RF, Dallas MJ, Tapia MD, Feikin DR, Binka FN, Steele AD, Laserson KF, Ansah NA, et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-Saharan Africa: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;376:606-14. doi:10.1016/S0140-6736(10)60889-6.
- Madhi SA, Cunliffe NA, Steele D, Witte D, Kirsten M, Louw C, Ngwira B, Victor JC, Gillard PH, Cheuvart BB, et al. Effect of human rotavirus vaccine on severe diarrhea in African infants. N Engl J Med. 2010;362:289-98. doi:10.1056/NEJMoa0904797.
- Zaman K, Dang DA, Victor JC, Shin S, Yunus M, Dallas MJ, Podder G, Vu DT, Le TPM, Luby SP, et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in Asia: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;376:615-23. doi:10.1016/S0140-6736(10)60755-6.
- Bhandari N, Rongsen-Chandola T, Bavdekar A, John J, Antony K, Taneja S, Goyal N, Kawade A, Kang G, Rathore SS, et al. Efficacy of a monovalent human-bovine (116E) rotavirus vaccine in Indian infants: a randomised, double-blind, placebo-controlled trial. Lancet. 2014;383:2136-43. doi:10.1016/S0140-6736(13)62630-6.
- Atherly D, Dreibelbis R, Parashar UD, Levin C, Wecker J, Rheingans RD. Rotavirus vaccination: cost-effectiveness and impact on child mortality in developing countries. J Infect Dis. 2009;200 Suppl 1:S28-38. doi:10.1086/605033.
- Emperador DM, Velasquez DE, Estivariz CF, Ben Lopman, Jiang B, Parashar U, Anand A, Zaman K. Interference of Monovalent, Bivalent, and Trivalent Oral Poliovirus Vaccines on Monovalent Rotavirus Vaccine Immunogenicity in Rural Bangladesh. Clin Infect Dis. 2016;62:150-6. doi:10.1093/cid/civ807.
- Moon S-S, Groome MJ, Velasquez DE, Parashar UD, Jones S, Koen A, van Niekerk N, Jiang B, Madhi SA. Prevaccination Rotavirus Serum IgG and IgA Are Associated With Lower Immunogenicity of Live, Oral Human Rotavirus Vaccine in South African Infants. Clin Infect Dis. 2016;62:157-65. doi:10.1093/cid/civ828.
- Chilengi R, Simuyandi M, Beach L, Mwila K, Becker-Dreps S, Emperador DM, Velasquez DE, Bosomprah S, Jiang B. Association of Maternal Immunity with Rotavirus Vaccine Immunogenicity in Zambian Infants. PLoS ONE. 2016;11(3) 11:e0150100. doi:10.1371/journal.pone.0150100.
- Nordgren J, Sharma S, Bucardo F, Nasir W, Gunayd n G, Ouermi D, Nitiema LW, Becker-Dreps S, Simpore J, Hammarstrom L, et al. Both Lewis and Secretor Status Mediate Susceptibility to Rotavirus Infections in a Rotavirus Genotype-Dependent Manner. Clin Infect Dis. 2014;59(11):1567-73. doi:10.1093/cid/ciu633.
- Ali A, Kazi AM, Cortese MM, Fleming JA, Moon S, Parashar UD, Jiang B, Mcneal MM, Steele D, Bhutta Z, et al. Impact of Withholding Breastfeeding at the Time of Vaccination on the Immunogenicity of Oral Rotavirus Vaccine—A Randomized Trial. PLoS ONE. 2015;10: e0127622. doi:10.1371/journal.pone.0127622.
- Ali SA, Kazi AM, Cortese MM, Fleming JA, Parashar UD, Jiang B, McNeal MM, Steele D, Bhutta Z, Zaidi A. Impact of Different Dosing Schedules on the Immunogenicity of the Human Rotavirus Vaccine in Infants in Pakistan: A Randomized Trial. J Infect Dis. 2014 Dec 1;210(11):1772-9. doi:10.1093/infdis/jiu335.
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222-7. doi:10.1038/nature11053.
- Harris VC, Armah G, Fuentes S, Korpela KE, Parashar U, Victor JC, Tate J, de Weerth C, Giaquinto C, Wiersinga WJ, et al. Significant Correlation Between the Infant Gut Microbiome and Rotavirus Vaccine Response in Rural Ghana. J Infect Dis. 2017;2017 Jan 1;215(1):34-41. doi:10.1093/infdis/jiw518.
- Isolauri E, Joensuu J, Suomalainen H, Luomala M, Vesikari T. Improved immunogenicity of oral D x RRV reassortant rotavirus vaccine by Lactobacillus casei GG. Vaccine. 2003;13:310-2. doi:10.1016/0264-410X(95)93319-5.
- Zhang B, Chassaing B, Shi Z, Uchiyama R, Zhang Z, Denning TL, Crawford SE, Pruijssers AJ, Iskarpatyoti JA, Estes MK, et al. Prevention and cure of rotavirus infection via TLR5/NLRC4-mediated production of IL-22 and IL-18. Science. 2014;346:861-5. doi:10.1126/science.1256999.
- Vatanen T, Kostic AD, d'Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur TD, Hämäläinen A-M, et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell. 2016;165:842-53. doi:10.1016/j.cell.2016.04.007.
- Alexander C, Rietschel ET. Bacterial lipopolysaccharides and innate immunity. J Endotoxin Res. 2001;7:167-202.
- Shin N-R, Whon TW, Bae J-W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015 Sep;33(9):496-503. .doi:10.1016/j.tibtech.2015.06.011.
- Rudin A, Lundell A-C. Infant B cell memory and gut bacterial colonization. Gut Microbes. 2014;3:474-5. doi:10.4161/gmic.21419.
- J Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinje J, et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science. 2014;346:755-9. doi:10.1126/science.1257147.
- Wacklin P, Tuimala J, Nikkilä J, Sebastian Tims, Mäkivuokko H, Alakulppi N, Laine P, Rajilić-Stojanović M, Paulin L, De Vos WM, et al. Faecal Microbiota Composition in Adults Is Associated with the FUT2 Gene Determining the Secretor Status. PLoS ONE. 2014;2014 Apr 14;9(4):e94863. doi:10.1371/journal.pone.0094863.
- Payne DC, Currier RL, Staat MA, Sahni LC, Selvarangan R, Halasa NB, Englund JA, Weinberg GA, Boom JA, Szilagyi PG, et al. Epidemiologic Association Between FUT2 Secretor Status and Severe Rotavirus Gastroenteritis in Children in the United States. JAMA Pediatr. 2015;69(11):1040-5. doi:10.1001/jamapediatrics.2015.2002.
- Patel M, Glass RI, Jiang B, Santosham M, Lopman B, Parashar U. A Systematic Review of Anti-Rotavirus Serum IgA Antibody Titer as a Potential Correlate of Rotavirus Vaccine Efficacy. J Infect Dis. 2013;208:284-94. doi:10.1093/infdis/jit166.
- de Weerth C, Fuentes S, Puylaert P, De Vos WM. Intestinal microbiota of infants with colic: development and specific signatures. Pediatrics. 2013;131:e550-8. doi:10.1542/peds.2012-1449.
- Zijlmans MAC, Korpela K, Riksen-Walraven JM, De Vos WM, de Weerth C. Maternal prenatal stress is associated with the infant intestinal microbiota. Psychoneuroendocrinology. 2015;53:233-45. doi:10.1016/j.psyneuen.2015.01.006.
- Bernstein DI, Smith VE, Sherwood JR, Schiff GM, Sander DS, DeFeudis D, Spriggs DR, Ward RL. Safety and immunogenicity of live, attenuated human rotavirus vaccine 89–12. Vaccine. 1997;16:381-7. doi:10.1016/S0264-410X(97)00210-7.
- Salonen A, Nikkilä J, Jalanka-Tuovinen J, Immonen O, Rajilić-Stojanović M, Kekkonen RA, Palva A, De Vos WM. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: Effective recovery of bacterial and archaeal DNA using mechanical cell lysis, J Microbiol Methods. 2010;81(2):127-34. doi: 10.1016/j.mimet.2010.02.007.
- Rajilić-Stojanović M, Heilig HGHJ, Molenaar D, Kajander K, Surakka A, Smidt H, De Vos WM. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol. 2009;11:1736-51. doi:10.1111/j.1462-2920.2009.01900.x.
- Lahti L, Salojärvi J, Salonen A, Scheffer M, De Vos WM. Tipping elements in the human intestinal ecosystem. Nat Commun. 2014;5:59. doi:10.1038/ncomms5344.
- Lahti L, Torrente A, Elo LL, Brazma A, Rung J. A fully scalable online pre-processing algorithm for short oligonucleotide microarray atlases. Nucleic Acids Research. 2013;41:e110-0. doi:10.1093/nar/gkt229.
- Claesson MJ, O'Sullivan O, Wang Q, Nikkilä J, Marchesi JR, Smidt H, De Vos WM, Ross RP, O'toole PW. Comparative Analysis of Pyrosequencing and a Phylogenetic Microarray for Exploring Microbial Community Structures in the Human Distal Intestine. PLoS ONE. 2009;2009 Aug 20;4(8):e6669. doi:10.1371/journal.pone.0006669.
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto J-M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174-80. doi:10.1038/nature09944.
- Leps J, Smilauer P. Multivariate Analysis of Ecological Data Using CANOCO [Internet]. Cambridge:Cambridge University Press;2003. Available from: http://www.cambridge.org/9780521814096.
- Korpela K. Mare: Microbiota analysis in R Easily. R Package version 1.0. 2016. Available from https://github.com/katrikorpela/mare
- R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. 2011. Available from http://www.R-project.org/
- Pinheiro JC, Bates D, DebRoy S, Sarkar D, and R Core Team. nlme: Linear and Nonlinear Mixed Effects Models. R package version. 2016;3:1-128.
- Venables WN, Ripley BD. Modern Applied Statistics with S. Fourth Edition. New York:Springer; 2002. ISBN 0-387-95456-0.
- Kampstra P. Beanplot: A Boxplot Alternative for Visual Comparison of Distributions. Journal of Statistical Software. 2008; Code Snippets 28 (1):1-9. URL http://www.jstatsoftsoft.org/v28/c01/.
- Foster ZSL, Sharpton TJ, Grünwald NJ. Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput Biol. 2017;13(2):e1005404. doi:10.1371/journal.pcbi.1005404
- Oksanen J, Guillaume Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, OHara RB, Gavin L, Simpson P, Solymos P. vegan: Community Ecology Package. Ordination methods, diversity analysis and other functions for community and vegetation ecologists. Version 2.4-0. Available from: https://CRAN.R-project.org/package=vegan.
