 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Iron deficiency, a common comorbidity of gastrointestinal inflammatory disorders such as inflammatory bowel diseases (IBD), is often treated with oral iron supplementation. However, the safety of oral iron supplementation remains controversial because of its association with exacerbated disease activity in a subset of IBD patients. Because iron modulates bacterial growth and function, one possible mechanism by which iron may exacerbate inflammation in susceptible hosts is by modulating the intestinal microbiota. We, therefore, investigated the impact of dietary iron on the intestinal microbiota, utilizing the conventionalization of germ-free mice as a model of a microbial community in compositional flux to recapitulate the instability of the IBD-associated intestinal microbiota. Our findings demonstrate that altering intestinal iron availability during community assembly modulated the microbiota in non-inflamed wild type (WT) and colitis-susceptible interleukin-10-deficient (Il10−/-) mice. Depletion of luminal iron availability promoted luminal compositional changes associated with dysbiotic states irrespective of host genotype, including an expansion of Enterobacteriaceae such as Escherichia coli. Mechanistic in vitro growth competitions confirmed that high-affinity iron acquisition systems in E. coli enhance its abundance over other bacteria in iron-restricted conditions, thereby enabling pathobiont iron scavenging during dietary iron restriction. In contrast, distinct luminal community assembly was observed with dietary iron supplementation in WT versus Il10−/- mice, suggesting that the effects of increased iron on the microbiota differ with host inflammation status. Taken together, shifts in dietary iron intake during community assembly modulate the ecological structure of the intestinal microbiota and is dependent on host genotype and inflammation status.
Introduction
Iron deficiency anemia (IDA) is the most common micronutrient deficiency, affecting approximately 1 billion individuals worldwideCitation1. Although oral iron supplementation can effectively treat IDA,Citation2,Citation3 the safety and tolerance of oral iron supplementation within specific population subsets such as infants or patients with chronic inflammatory diseases remains controversial. Diarrhea, enhanced burdens of intestinal pathogens and increased fecal markers of inflammation have been reported in iron-deficient infants receiving iron-supplemented foods.Citation4–Citation8 Increased disease activity and functional gastrointestinal (GI) symptoms with oral iron supplementation have also been observed in patients with inflammatory bowel diseases (IBD).Citation9,Citation10 The latter observations have been recapitulated with increased dietary iron administration in rodent models of experimental ileitis and colitis.Citation10–Citation12 Diet-mediated increases in intestinal iron concentrations also enhance luminal production of reactive oxygen species,Citation13 a byproduct of the Fenton reaction, which in turn can increase colonic oxidative stress10 and potentially exacerbate the development of intestinal inflammation in susceptible populations.
The GI tract is home to a complex community of microbes referred to as the intestinal microbiota. Dietary factors, including macronutrient and micronutrient constituents, can compositionally and functionally modulate the intestinal microbiota,Citation11, Citation14–Citation16 which in turn can impact numerous host processes, including metabolism and immune function.Citation17–Citation22 Iron is an essential cofactor for numerous bacterial enzymes and therefore serves as an important micronutrient for most members of the intestinal microbiota.Citation23–Citation26 Iron requirements vary between bacterial taxa, as does the capacity to acquire iron when availability is limited, which includes secretion of small molecules with high affinity for iron known as siderophores. An estimated 5–15% of dietary nonheme iron is absorbed at the duodenum in iron-replete hosts, leaving the remaining unabsorbed iron to pass through the intestines for microbial use. Thus, dietary iron likely impacts the growth and function of resident intestinal bacteria, which in turn may contribute to the development of unfavorable side effects that can occur with oral iron supplementation.
Studies investigating the impact of dietary iron on the intestinal microbiota have primarily focused on rodent models with an established microbial community. However, population subsets that are most vulnerable to IDA, including the infant and IBD populations, frequently harbor an intestinal microbiota that is unstable in composition.Citation27–Citation29 Therefore, wesought to determine how dietary iron restriction and supplementation influences the ecological structure of a microbiota in compositional flux, using the conventionalization of germ-free (GF) mice as a model system. Our findings demonstrate that dietary iron restriction initiated prior to conventionalization of GF mice promoted compositional changes frequently associated with a dysbiotic state, including decreased microbial richness and an increased abundance of Enterobacteriaceae family members such as Escherichia coli. Moreover, decreased intestinal iron availability corresponded with an increased capacity for bacterial iron scavenging. Similar shifts in community composition were observed with dietary iron restriction in colitis-susceptible interleukin-10-deficient (Il10−/-) mice. In contrast, dietary iron supplementation had distinct effects on the microbiotas of WT versus Il10−/- mice. Taken together, luminal iron availability during community assembly influences the resulting structure of the intestinal microbiota and is dependent on host genotype and inflammation status.
Results
Intestinal iron availability during community assembly impacts the resulting structure of the intestinal microbiota
Dietary iron interventions are frequently administered during infancy when the intestinal microbiota is undergoing ecological successionCitation28,Citation29 or during disease states when the gut microbiota exhibits transient instability.Citation27,Citation30 We, therefore, investigated the impact of dietary iron restriction or supplementation on the developing intestinal microbiota as a model of a bacterial community in compositional flux. GF WT mice were placed on an iron-deficient (“low”), control or iron-supplemented (“high”) diet, and then were colonized with a fecal microbiota from a WT donor. . Fecal and cecal tissue samples were collected for 16S rRNA sequencing at 4 weeks post-colonization, when the microbiota exhibits initial compositional stability following conventionalization.Citation31,Citation32 Because host systemic iron levels have been correlated with compositional changes to the intestinal microbiotaCitation7,Citation33,Citation34 and iron deficient diets can induce systemic iron deficiency as early as 4 weeks,Citation35,Citation36 mice on the low iron diet received weekly i.p. iron injections to prevent systemic iron depletion. Following the 4-week dietary intervention and i.p. injections, liver iron stores did not differ between dietary groups (Figure S1). Moreover, as described,Citation13,Citation37 dietary iron restriction and supplementation significantly altered fecal iron concentrations (Figure S2), confirming that our diets modulated luminal iron concentrations.
Principal Coordinate Analysis (PCoA) using Bray–Curtis dissimilarity at the operational taxonomic unit (OTU) level revealed compositional differences in the fecal microbiota between WT mice administered the different iron diets at 4 weeks post-conventionalization ()). Fecal communities in the high iron diet mice significantly differed from those in the low iron diet mice (PCoA axis 1; FDR-corrected p-value = 4.4x10−9). The composition of the fecal microbiota in mice fed the control diet exhibited the most interindividual variability as assessed by Bray Curtis distances between samples within each diet group (Figure S3), suggesting that dietary iron supplementation or depletion exerts strong selective pressure in shaping the fecal microbiota. Dietary iron restriction also significantly reduced microbial richness relative to iron supplementation (–)). Thus, manipulation of intestinal iron availability through dietary interventions impacts the ecological structure of the fecal microbiota in WT mice.
Figure 1. Dietary iron alters the structure of the fecal and mucosal microbiota. A + D) Principal coordinate analysis based on Bray–Curtis metrics for the (a) fecal and (d) mucosal microbiota in ex-GF WT mice maintained on an iron-deficient (low), control or iron supplemented (high) diet 4 weeks after fecal bacterial colonization. Microbial richness as measured by (b, e) observed OTUs and (c, f) Shannon diversity index for the (b, c) fecal and (e, f) mucosal microbiota. Each symbol represents an individual mouse, n = 7–9 mice per group. Box and whisker plots show the minimum, first quartile, median, third quartile, and maximum values. Comparisons and FDR-corrected p-values were determined using a mixed linear model with the cage as a random effect. * p< 0.05, *** p < 0.001.
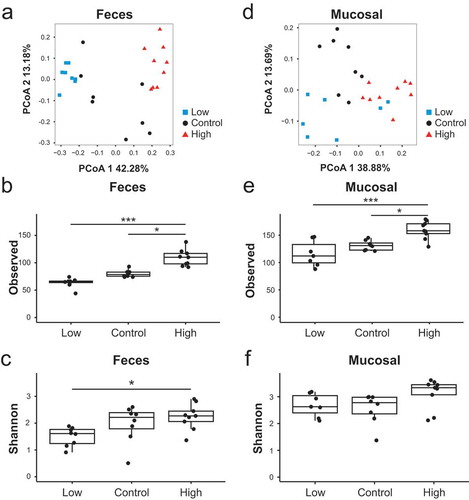
We next examined the impact of dietary iron supplementation and restriction on the structure of the mucosal microbiota. PCoA analysis revealed partial clustering of the mucosal microbiota by dietary treatment, including significant differences between the low and high iron diet groups (PCoA axis 1; FDR-corrected p-value = 0.02) ()) and increased microbial richness with dietary iron supplementation (–)). Collectively, these results suggest that dietary iron influences biodiversity and community assembly of the fecal and mucosal microbiotas.
Dietary iron supplementation alters fecal abundances of distinct bacterial taxa
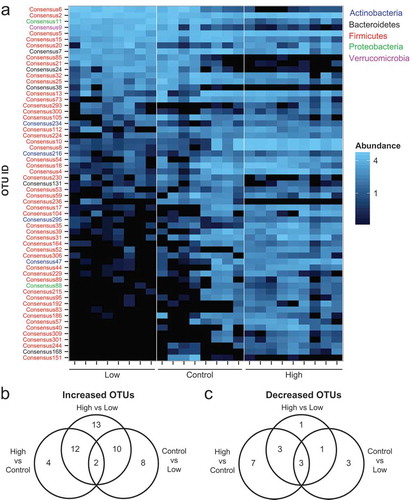
Because the compositional differences by iron diet were more pronounced in the feces, we next evaluated the impact of dietary iron supplementation and depletion on specific taxa within the fecal microbiota. A heat map was generated to highlight OTUs that significantly differed in relative abundance ()) (Table S2). Statistical analysis using a mixed linear model revealed that the relative abundances of 59 OTUs significantly differed at an FDR-corrected p-value < 0.05 between at least two dietary treatments. Iron supplementation resulted in the enrichment of 18 OTUs relative to the control diet and 37 OTUs relative to the low iron diet (), Table S3). In contrast, dietary iron restriction decreased the relative abundances of 7 OTUs compared to the control diet and 8 OTUs compared to the high iron diet (), Table S3). These data suggest that increased intestinal iron availability during community assembly corresponds with an alteration of specific bacterial taxa within the fecal microbiota with predominant enrichment.
Figure 2. Dietary iron enhances the relative fecal abundances of numerous bacterial taxa. (a) Heat map of OTUs that are significantly different in abundance with variable iron diets (FDR-corrected p< 0.05). Each column represents an individual WT mouse. Each row represents individual OTUs, color coded by phylum, that are significant between at least two diet groups as determined using a mixed linear model. The colors of the heat map represent the mean relative abundance (normalized and log transformed) of each OTU. B + C) Venn diagrams of OTUs that are significantly (b) increased or (c) decreased in relative abundance (FDR-corrected p< 0.05) in the feces as determined using a mixed linear model.
Dietary iron restriction promotes a bloom of Enterobacteriaceae and predicted bacterial iron uptake systems
Given that increases in Enterobacteriaceae are associated with inflammation-induced dysbiosis,Citation38,Citation39 we assessed the impact of dietary iron restriction or supplementation on Enterobacteriaceae abundance during community assembly using the Ribosomal Database Project (RDP) pipeline. In the control and low iron diet groups, the relative fecal abundances of Proteobacteria phylum and Enterobacteriaceae family members including Escherichia coli were significantly higher relative to the high iron diet group (Figure S4, –)). This suggests that increased intestinal dietary iron concentrations as a result of iron supplementation may accelerate negative selection against Enterobacteriaceae during community assembly.
Figure 3. Dietary iron restriction promotes a bloom of Enterobacteriaceae and predicted bacterial iron uptake systems. (a) Relative fecal abundance of Enterobacteriaceae. (b) Abundance of fecal E. coli 16S sequences relative to total bacteria 16S sequences as determined by quantitative PCR. Data are presented as the fold change relative to the low iron diet group. Each point represents an individual mouse, n = 8-9 mice per group. Lines are at the medians and p values were determined by pairwise comparisons by the Kruskal–Wallis test. (c) Differences in the least square mean between diet groups of log10 normalized counts of predicted genes involved in iron acquisition as determined by PICRUSt. (a) Box and whisker plots show the minimum, first quartile, median, third quartile, and maximum relative abundance. (a, c) FDR-corrected p-values were determined using a mixed linear model. * p< 0.05, ** p < 0.01, *** p < 0.001.
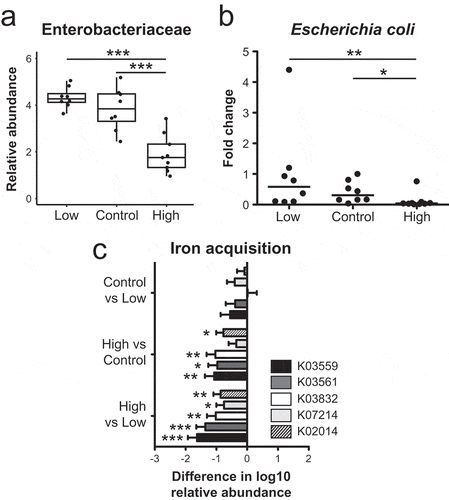
Because dietary iron restriction decreases fecal iron concentrations, we investigated whether low luminal iron promotes an increase presence of microbial genes involved in scavenging iron within the fecal microbiota. Using the metagenomic inference tool PICRUSt, we observed a significant increase in the predicted presence of tonB (K03832), exbB (K03561) and exbD (K03559) in mice maintained on the low iron or control diets when compared to high iron diet mice (), Table S4). These three genes are required for bacterial import of siderophore-bound iron.Citation40 Also increased was the relative abundance of additional genes involved in siderophore-mediated iron acquisition and utilization, including a predicted enterochelin esterase (K07214) that hydrolyzes ferric enterobactinCitation41 and a predicted iron complex outer membrane receptor (K02014). Transcript levels of these and other E. coli iron acquisition genes are increased in vitro under iron-limiting conditions,Citation42,Citation43 including in a resident intestinal E. coli strain (Table S5). Moreover, these findings generated by PICRUSt are consistent with the increased abundance of Enterobacteriaceae observed using the RDP pipelines. Taken together, these data suggest that decreased intestinal iron concentrations observed with the low iron and control diets relative to the iron supplemented diet select for siderophilic bacteria including E. coli with a greater capacity to scavenge iron.
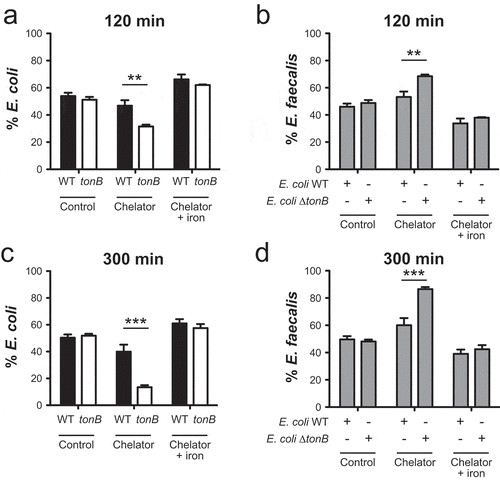
Because the predicted abundance of tonB (which encodes a protein known to be required for importing siderophore-bound ironCitation44) was among the iron acquisition genes that were increased in the low iron and control diet groups, we next determined whether TonB-mediated iron acquisition provides resident intestinal E. coli a relative growth advantage when iron availability is restricted. To address this, we created an isogenic tonB deletion mutant in E. coli to simultaneously inactivate all siderophore-mediated iron import.Citation45 The growth of the parental strain exceeded that of the tonB mutant in the presence of the membrane impermeable iron chelator DTPA (Figure S5A). Addition of exogenous iron to the chelated cultures rescued growth of the tonB mutant (Figure S5A), demonstrating that siderophore-mediated iron scavenging limits the impact of extracellular iron chelation on E. coli growth. We then competed the parental or tonBmutant E. coli strain with a non-siderophilic bacterium, Enterococcus faecalis,that exhibits minimal iron requirements IFigure S5B).Citation24 When iron was chelated, the relative abundance of the parental E. coli strain – but not the tonB mutant – was significantly increased over co-cultured E. faecalis (,)), which corresponded with an enrichment of E. faecalis in the presence of the tonB mutant (,)). When exogenous iron was added to the chelated cultures, the tonB-mediated enrichment of E. coli over E. faecalis reverted to baseline values (, Figure S5D-G). Consistent with these in vitro observations, our sequencing data revealed minimal changes in the relative fecal abundance of Enterococcaceae family members as intestinal iron concentrations were altered (Figure S5C). Taken together, these data suggest that TonB-dependent iron acquisition may help promote the in vivo bloom of E. coli via efficient iron scavenging through siderophore-mediated iron acquisition when luminal intestinal iron availability is decreased.
Figure 4. TonB-dependent iron acquisition enhances the relative abundance of E. coli when iron availability is restricted. E. coli WT or the tonB-deficient mutant were grown in the presence of E. faecalis in rich medium (control), in rich medium with the iron chelator DTPA (chelator) or in rich medium with DTPA and ferrous iron (chelator + iron). Abundance of bacteria was determined by quantitative selective plating. The % abundance of (a, c) E. coli WT or the tonB mutant in the presence of E. faecalis and (b, d) the % abundance of E. faecalis in the presence of E. coli WT or the tonB mutant after 120 or 300 min of growth. Data are represented as the mean ± SEM of at least three independent experiments. P-values were determined by two-way ANOVA. ** p < 0.01, *** p < 0.001.
Dietary iron impacts microbiota structure in colitis-susceptible Il10−/- mice
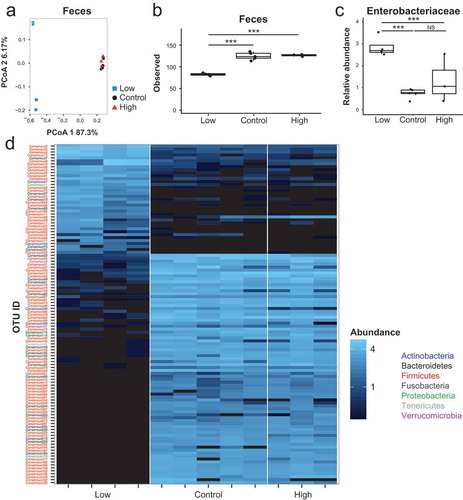
We previously observed expanded Enterobacteriaceae during the onset of colitis in inflammation-susceptible, ex-GF Il10−/- mice following conventionalization.Citation32 Because we observed that dietary iron restriction also promotes a bloom of Enterobacteriaceae in uninflamed WT mice, we investigated the impact of iron restriction on the microbiota and colitis development in ex-GF Il10−/- mice. As was the case with the WT cohort, fecal communities assembled by iron availability in Il10−/- mice (), PCoA axis 1; FDR-corrected p-value = 2.7x10−Citation6). However, in contrast to WT mice, PCoA demonstrated that fecal communities from mice fed the control and high iron diets clustered together. Similar to WT mice, microbial richness was reduced ()) and the relative abundance of Enterobacteriaceae was enhanced ()) with dietary iron restriction. While 112 taxa were different when comparing the low iron and control groups and 106 taxa when comparing the low and high iron groups, few taxa (11 OTUs) were significantly different between the control and high diet groups (), Table S6). PCoA analysis also revealed modest clustering of the mucosal microbiota by diet (Figure S6A). This did not achieve statistical significance in contrast to WT mice ()). However, as observed with WT mice, iron supplementation corresponded with increased microbial richness and abundances of numerous OTU in the mucosal microbiota relative to control diet (Figure S6B-C). Together, these data suggest that while dietary iron restriction alters the microbial community in Il10−/- mice, additional iron supplementation does not further alter the structure of the Il10−/- fecal microbiota relative tothe control diet.
Figure 5.: Fecal community composition is insensitive to additional dietary iron supplementation in Il10−/- mice. (a) Principal coordinate analysis based on Bray–Curtis metrics for the fecal microbiota in ex-GF Il10−/- mice. (b) Microbial richness as measured by observed OTUs. (C) Relative fecal abundance of Enterobacteriaceae. (b, c) Box and whisker plots show the minimum, first quartile, median, third quartile, and maximum relative abundance. Each symbol represents an individual mouse, n = 3–5 mice per group. FDR-corrected p-values were determined using a mixed linear model. (d) Heat map of OTUs that significantly differ in relative fecal abundance (FDR-corrected p < 0.05) as determined using a mixed linear model. Each column represents an individual Il10−/- mouse. Each row represents individual OTUs, color-coded by phylum. The colors of the heat map represent the mean relative abundance (normalized and log transformed) of each OTU.
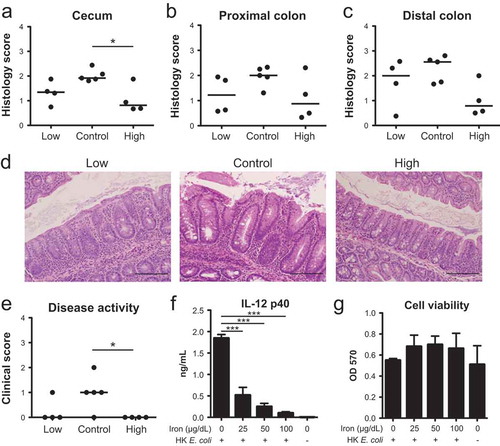
To determine the functional consequences of these changes to the Il10−/- microbiota, we characterized the development of colitis in Il10−/- mice subjected to iron supplementation or restriction. Four weeks post-conventionalization, high iron diet mice exhibited a modest but significant decrease in colitis severity compared to control mice as assessed by histological inflammation (–)) and clinical disease activity ()). We also compared the spontaneous secretion of IL-12 p40 by colonic explant cultures and serum IL-12 p40 levels between experimental groups as increased levels of this cytokine correspond with more severe colitis in this model.Citation46 A trend towards decreased IL-12 p40 in the colon (Figure S7A) and in the serum (Figure S7B) was observed in both the high iron and low iron versus the control group (, , , )). Similarly, a trend towards reduced histopathology and disease activity was observed in low iron diet mice compared to the control group. Taken together, our results indicate that both iron restriction and supplementation modestly attenuate colitis development in the Il10−/- colitis model.
Figure 6.: Dietary iron impacts the development of colitis in Il10−/- mice. Ex-GF Il10−/- mice were maintained on an iron-deficient (low), control or iron supplemented (high) diet. (A–C) Histology scores (0–4) of the (A) cecum, (B) proximal colon and (C) distal colon. (D) Representative H&E histology at 200x of the ceca. Scale bar = 20 μm. (E) Clinical activity scores (0–4) after 4 weeks. (F + G) Macrophages were stimulated with heat-killed E. coli and cultured with iron for 8 h to assess: (F) IL-12 p40 secretion and (G) macrophage viability by MTT assay. Data are represented as the mean ± SEM of three independent experiments. P-values were determined by one-way ANOVA. * p< 0.05, ** p < 0.01, *** p < 0.001.
The lack of significant compositional changes between the fecal microbiotas of control Il10−/- mice with increased colitis versus Il10−/- mice fed a high iron diet suggests that host factors may also contribute to the impact of dietary iron supplementation on colitis development. Given that iron homeostasis and macrophage function are intimately linked,Citation47 we tested whether iron impacts in vitro macrophage production of IL-12 p40 as a marker for proinflammatory macrophage activity. Consistent with our IL-12 p40 in vivo observations (Figure S7), iron reduced macrophage IL-12 p40 secretion ()) without impacting viability ()). Taken together, these results support previous findings that iron impacts innate immune function and may contribute to the protective effect of dietary iron supplementation on colitis development in the Il10−/- mouse model. Furthermore, these findings suggest that the protective effects of low iron on colitis development in this model likely occur through a different mechanism.
Dietary iron modulation differentially impacts the WT and Il10−/- fecal microbiotas
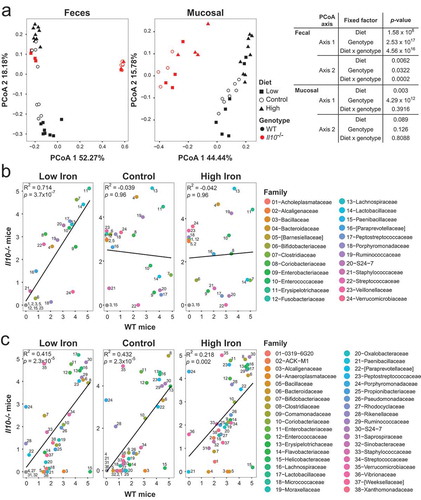
Given that dietary iron supplementation compositionally alters the microbiotas of WT and Il10−/- mice in distinct ways, we directly compared the microbiotas of both genotypes under conditions of dietary iron restriction and supplementation. PCoA and our mixed linear model revealed that genotype and genotype x diet interactions strongly associated with microbiota composition in the feces (), S8). In the mucosa, however, only genotype strongly associated with community composition. We next determined whether bacterial taxa respond to iron availability in similar ways regardless of host genotype. In the fecal community, there was a significant positive correlation between bacterial abundance in WT and Il10−/- mice maintained on the low iron diet ()), demonstrating a similarity in biological response of the luminal microbiota to iron restriction. In contrast, there was no correlation between bacterial abundance in WT and Il10−/- mice maintained on the control or high iron diets, suggesting that inflammation or other intrinsic factors associated with the Il10−/- genotype cause the fecal microbiota of these mice to respond differently to increased iron availability than in WT mice. By contrast, in the mucosa, bacterial abundance in WT and Il10−/- mice was positively correlated regardless of the diet group ()). Consistent with our mixed linear results ()), this suggests that niche selection pressure may be a stronger driver in modulating the abundance of specific bacterial taxa in the mucosa rather than genotype or dietary iron content. More broadly, diet and host genetics strongly impact community composition in the lumen, whereas only host genetics strongly influences the mucosal community.
Figure 7. Dietary iron modulation differentially impacts the WT and Il10−/- fecal microbiotas. (a) Principal coordinate analysis based on Bray–Curtis metrics in WT and Il10-/- mice for the fecal and mucosal microbiota. Each symbol represents an individual mouse, n= 3–9 mice per group. FDR corrected p-values were determined using a mixed linear model, evaluating the null hypothesis that the fixed factor indicated in each table does not impact the coordinate. (b, c) Correlations between the median log normalized counts of bacterial taxa at the family level of WT and Il10-/- mice in the (b) fecal and (c) mucosal microbiota.
Discussion
Treatment of iron deficiency through oral iron supplements or iron-fortified diets can modulate luminal intestinal iron concentrationsCitation13,Citation37 and likely influences bacterial iron availability, consequent bacterial growth, and community structure. We, therefore, investigated the ecological impact of dietary iron restriction and supplementation on the intestinal microbiota. Our findings demonstrate that dietary iron restriction during community assembly resulted in significant compositional changes frequently associated with a dysbiotic state. Other 16S rRNA community profiling studies report complementary findings in established intestinal microbial communities.Citation36,Citation48 For example, dietary iron restriction initiated at weaning-induced compositional changes to the fecal microbiota, including decreased microbial richness relative to both the baseline community at weaning and the control diet.Citation36 Interestingly, subsequent dietary iron repletion did not fully restore microbial richness or community composition.Citation36 In contrast, in adult mice with an established microbiota, microbial richness was not affected by the dietary iron restriction.Citation11 Thus, the early assembly of the intestinal microbiota may be uniquely susceptible to iron restriction.
Our findings demonstrate that dietary iron restriction that is the result of administering a diet with trace iron levels is associated with the reduced luminal abundance of numerous taxa within the Firmicutes and an increase in endogenous Enterobacteriaceae. The abundance of resident Enterobacteriaceae did not differ upon iron restriction in other rodent studies harboring an established microbiota.Citation11,Citation36 However, consistent with our results, dietary iron restriction initiated at weaning results in a bloom of endogenous Enterobacteriaceae that was reversed with subsequent dietary iron repletion.Citation35 Decreased Enterobacteriaceae was similarly observed in human infants with low initial intestinal pathogen burden receiving iron-fortified foods.Citation49 Taken together, these findings suggest that dietary iron restriction may perturb the assembly of the luminal intestinal bacterial community, promoting compositional changes, including increased resident Enterobacteriaceae, that have been linked to numerous microbially driven diseases. This may be mediated through the creation of an intestinal environment that favors the growth of early colonizers with multiple inducible iron acquisition systems such as E. coli. However, an important distinction between resident and pathogenic Enterobacteriaceae should be noted. In contrast to the effects of iron supplementation on endogenous E. coli, increased oral iron intake enhances the abundance of pathogenic enterobacteria including Enterotoxigenic E. coli (ETEC) and Salmonella.Citation5,Citation7,Citation50 Taken together, the initial composition of the intestinal microbiota is an important factor that affects how dietary iron will ecologically alter the microbiota, with important implications for identifying IDA patients that may respond favorably to oral iron supplementation regimens.
In response to low iron availability, bacteria upregulate iron acquisition genes to scavenge iron from the environment. Bacterial taxa such as Enterobacteriaceae encode numerous siderophore-mediated iron transport systems that enhance their competitive advantage when iron availability is restricted. Indeed, controlled in vitro growth competitions between E. coli and non-siderophilic E. faecalis demonstrated that TonB-mediated iron acquisition enhanced the relative fitness of E. coli when the iron was sequestered by the extracellular chelator DTPA. While DTPA can sequester other metals, the addition of exogenous iron reversed the TonB-mediated fitness advantage of E. coli over E. faecalis, suggesting that these observations are likely the result of DTPA chelation of iron rather than other metals. However, it should be noted that these in vitro growth competitions were performed under aerobic conditions. Under steady state conditions, the gut lumen is an anaerobic environment, but becomes increasingly aerobic with inflammation, although not to the same extent as a shaking culture.Citation51 Nonetheless, consistent with our findings, iron chelation in colonic fermentators inoculated with a fecal microbiota and subjected to anaerobic growth results in a similar expansion of Enterobacteriaceae.Citation52 Thus, decreasing iron availability in the intestine likely provides a fitness advantage for resident bacteria that are more effective at scavenging iron. Supporting this model, metagenomic predictions using the PICRUSt algorithm suggest an increased capacity for siderophore-mediated iron acquisition in the microbiotas of mice administered an iron-deficient diet. Furthermore, the presence of genes involved in flagella biosynthesis and assembly, an iron-dependent process, as well as zinc, manganese and hemoglobin utilization is increased with decreased dietary iron consumption. However, while PICRUSt only predicts functional capacity using 16S profiling data, the PICRUSt algorithm has been shown to recapitulate metagenomic results with approximately 80% accuracy when applied to data obtained from the Human Microbiome Project.Citation53 Nonetheless, further studies assessing transcript levels of iron acquisition genes and in vivo growth competitions utilizing siderophore deficient mutants are necessary to confirm this ecological model.
Intestinal inflammation is associated with compositional changes to the intestinal microbiota, including an expansion of Enterobacteriaceae.Citation38,Citation39,Citation54–Citation56 Because dietary iron restriction induced similar perturbations to the fecal microbiota, we investigated the impact of dietary iron on the development of colitis in ex-GF Il10−/- mice. Interestingly, dietary iron supplementation modestly limited the development of inflammation. However, an important limitation of our study is that iron supplementation was provided to iron replete hosts rather than iron-deficient hosts, as often occurs in IBD patients. However, many IBD patients with anemia of chronic disease are encouraged to take supplemental iron despite normal iron stores, and IBD patients are routinely encouraged to take multi-vitamins that contain iron, regardless of their iron status. Nonetheless, in agreement with our observations, Dostal and colleagues reported higher baseline inflammation in the ilea and ceca of rats on a control diet compared to iron-deficient rats receiving iron supplementation.Citation48 Similarly, dietary iron restriction has also been reported to minimize intestinal inflammation.Citation11,Citation48 In chemically induced models of colitis, increased dietary iron exacerbated colitis.Citation10,Citation12 However, in many of these studies, dietary iron was administered at doses approximately 100 times higher than what is present in normal mouse chow and may therefore not reflect amounts consumed with iron fortification or oral iron supplements. Indeed, similar to our study, oral iron supplementation ranging from 2 to 10 fold higher than iron levels in the control diets protected against TNBS-induced and DSS-induced colitis.Citation57,Citation58 Administration of an iron-fortified diet in infants with high initial pathogen burden also resulted in increased fecal markers of inflammation.Citation7 However, two other studies demonstrated no impact of dietary iron supplementation on inflammation markers in infants or children with low pathogen burden.Citation49,Citation59 Interestingly, dietary iron restriction has been reported to minimize intestinal inflammation,Citation11,Citation48 consistent with our statistically insignificant trends. Finally, it should also be noted that a recent study demonstrated that the formulation of iron also impacts the effect of oral iron supplementation on colitis severity, where ferrous iron and ferrous bisglycinate both ameliorated DSS-induced colitis and ferric-EDTA had the opposite effect.Citation58 Together, these studies provide evidence that dietary iron influences colitis development, albeit in a complex manner that depends on the iron formulation as well as microbial and host factors that remain to be fully defined.
In Il10−/- mice, fecal communities clustered by diet, suggesting that intestinal iron concentrations had a stronger impact in shaping fecal community structure than colitis severity, while mucosally associated bacterial communities were influenced more by colonic inflammation. Interestingly, dietary iron restriction in Il10−/- mice did not exacerbate colitis relative to the control group despite compositional changes to the fecal microbiota frequently associated with inflammation.Citation27,Citation38,Citation39,Citation55,Citation56 Consistent with our findings, oral iron supplementation induced compositional changes to the fecal microbiota in UC and CD patients that were not associated with altered disease activity.Citation60 This suggests that in our model, the impact of iron supplementation on the development of colitis may not be mediated through compositional changes to the luminal microbiota, but rather through direct effects on the host. Indeed, oral iron supplementation increases the generation of free radicals in the intestines and serum markers of lipid peroxidation.Citation11,Citation61 Cellular iron status is also intricately linked with epithelial and macrophage function including secretion of proinflammatory cytokines and susceptibility to infection.Citation62–Citation64 Similarly, we also observed that iron modulates E. coli-stimulated macrophage production of the proinflammatory cytokine IL-12 p40, which correlates with severity of colitis in the Il10−/- mouse model.Citation46 It should be noted that the iron concentrations used for the macrophage stimulations are lower than total fecal iron concentrations. However, it is also unclear what fraction of total iron in the colonic lumen is bioavailable to both microbe and host. Finally, these findings do not exclude the possibility that dietary iron may impact the course of inflammation by altering the function of resident bacteria. Indeed, decreasing iron availability reduces the fermentative activity of the intestinal microbiota,Citation35,Citation48,Citation52 a functional change that has been associated with dysbiosis. Oral iron supplementation has also been shown to alter the fecal metabolome in anemic and IBD patients,Citation60 which may reflect altered microbial function. Thus, longitudinal studies investigating the impact of dietary iron on the function of the intestinal microbiota prior to, during, and after the onset of inflammation are warranted to further elucidate the contribution of the intestinal microbiota on iron-mediated effects on colitis. Together our data support a model where iron-mediated modulation of the intestinal microbiome and host immune function both contribute to colitis development. Given the high prevalence of IDA in individuals with gastrointestinal inflammatory disorders such as IBD, the precise mechanisms underlying the effects of iron on colitis development should be explored in future studies.
Materials and methods
Mice and experimental design
Il10−/- and WT 129S6/SvEV mice were maintained in GF conditions at the National Gnotobiotic Rodent Resource Center at UNC-Chapel Hill. Three groups of age-matched, 8–14 week-old GF WT (n = 8–9, 3 cages/group) and Il10−/- mice (n= 4–5, 2–3 cages/group) were transferred to conventional housing and placed on iron-deficient (<10 ppm iron/kg diet, n= 5 males, 3 females for WT mice; n= 3 males, 1 female for Il10−/- mice), control (35 ppm iron/kg diet, n= 5 males, 3 females for WT mice; n= 3 males, 2 females for Il10−/- mice) or iron supplemented diets (200 ppm iron/kg diet, n= 5 males, 4 females for WT mice; n= 1 male, 3 females for Il10−/- mice). The following day, mice were colonized by oral/rectal swab with fecal slurry from a WT SPF 129S6/SvEV mouse housed in the same facility. At 2 and 4 weeks of dietary intervention, fecal samples were collected with fecal iron measurements. After 4 weeks of dietary intervention, mice were killed and fresh feces and cecal tissue were collected and processed for 16S rRNA sequencing as previously described.Citation32,Citation38 One WT low iron and Il10−/- control mucosal sample was excluded from the analysis because of low DNA yield or similar technical reasons. The WT and Il10−/- cohorts were run at different times and were therefore colonized with different donors; thus subsequent 16S rRNA sequencing analyses were run separately for both cohorts. Mice on iron-deficient diets received 5 μg/g body weight of dextran iron sulfate (D8517, Sigma) i.p. Mice on control and iron supplemented diets received PBS i.p. as a vehicle control. Fecal iron content was assessed in a separate cohort of WT and Il10−/- mice (Figure S2). Envigo produced all diets, based on the TD.99397 iron-deficient diet. Diets differed only in iron content, added as ferrous sulfate and ferric citrate at 1:1 ratio. All animal protocols were approved by the UNC-Chapel Hill Institutional Animal Care and Use Committee.
Assessment of colitis and disease activity
At necropsy, colonic segments were fixed in 10% neutral buffered formalin. Histological inflammation scores (0–4) of cecal, proximal and distal colonic sections were blindly assessed as described.Citation46 Clinical disease activity (0–4) was assessed as described.Citation65 Unstimulated colonic explant cultures were prepared to assess spontaneous cytokine secretion as described.Citation66
Bacteria strains and growth conditions
Bacteria strains are listed in Table S1. Bacteria were grown overnight in brain heart infusion (BHI) medium at 37°C prior to inoculation into experimental conditions: BHI control, BHI with 250 μM of iron chelator diethylene triamine pentaacetic acid (DTPA; D6518, Sigma), or BHI with 250 μM DTPA and 250 μM ferrous sulfate (I146, Fisher Scientific). Bacterial growth was assessed by spectrophotometry (OD600) and quantitative plating. For dual cultures, selective media was utilized to distinguish E. faecalis (BHI agar with 50 μg/mL kanamycin) and E. coli (MacConkey agar). Deletion mutants in E. coli were created using the λ-red recombinase system as described.Citation67
Quantification of iron in fecal and liver samples
Iron contents in liver and feces were measured using atomic absorption spectrophotometry (AA) as describedCitation68 with modifications: briefly, samples were weighed, desiccated in a 90°C oven overnight and weighed again to obtain wet and dry sample weights. Samples were digested in 50% HNO3 and acid was evaporated for 24 h at room temperature. Samples were diluted in 2% HNO3 and analyzed by AA.
Quantitative PCR
Quantitative PCR was performed on fecal DNA to quantify the abundance of 16S rRNA sequences from E. coli and all bacteria using previously reported primers.Citation32 Sensifast SYBR No-ROX Kit (Bioline) was utilized with the following PCR conditions: 95°C for 2 min, followed by 40 cycles at 95°C for 5 s, 60°C for 10 s and 72°C for 20 s.
Bacterial RNA isolation and microarray hybridization
E. coli NC101 was grown in M9 medium with the indicated concentrations of ferrous sulfate. After 1 h, aliquots were collected for RNA isolation, which was isolated using the RNAeasy isolation kit (Qiagen) following the manufacturer’s instructions. Purified RNA was treated with on-column DNase (Qiagen) and DNA-free DNase (Ambion) following the manufacturer’s instructions. RNA samples were prepared for microarray hybridization using Affymetrix E. coli Genome 2.0 arrays as previously describedCitation69 at the UNC Functional Genomics Core. Statistical analysis was performed using GeneSpring 7.2 software.
Bone marrow-derived macrophage (BMDM) cultures
Bone marrow cells were isolated as described.Citation70 Conditioned medium from murine fibroblast line L929 served as a source of M-CSF for macrophage differentiation.Citation71 BMDMs were seeded in 24-well plates and maintained in RPMI 1640 (Gibco) with 10% heat-inactivated fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin/antimycotic (Gibco) at 37°C, 5% CO2. To assess the impact of iron on BMDM cytokine production, BMDMs were stimulated with heat-killed E. coli NC101 (MOI = 10) following the addition of the indicated amounts of ferrous sulfate to cultures. After 8 hrs, supernatants were collected and stored at −20°C to assess cytokine production. BMDM viability was assessed via the reduction of tetrazolium dye, MTT, using spectrophotometry.
Quantification of cytokine production
IL-12 p40 and interferon gamma were quantified by enzyme-linked immunosorbent assay (ELISA; BD Biosciences).
Illumina 16S library construction, sequencing, and processing
Amplification of the hypervariable V6 region of the 16S rRNA gene via a two-step PCR strategy was performed as described.Citation38 DNA concentrations were quantified using a NanoDrop spectrophotometer and equal amounts of all samples were pooled together and subjected to paired-end Illumina Hi-Seq sequencing. A total of 97,286,872 raw reads were generated for a total of 74 samples. Raw reads were preprocessed as described.Citation31,Citation72 Briefly, forward and reverse reads were merged if they satisfied having 70 bases overlap with 100% similarity. Successfully merged reads were quality trimmed at Q Score 20 followed by length check. Reads shorter than 50 bases were discarded. This resulted in 45,486,399 high quality reads that were incorporated into the subsequent analysis. These reads were then clustered into 1,301 operational taxonomic units (OTUs) using AbundantOTU+ v.0.93b (http://omics.informatics.indiana.edu/AbundantOTU/) (command option “-abundantonly” was used) at 97% similarity incorporating 99.84% of the input sequences. OTUs were checked for chimeric sequences using uchime v. 4.2.40 (http://www.drive5.com/uchime/)Citation73 utilizing the GOLD database. Uchime identified nine OTUs as chimeras, which were excluded from further analysis. Taxonomy was assigned through the Quantitative Insight into Microbial Ecology (QIIME) (v. 1.8.0)Citation74 using uclust consensus taxon assigner. Finally, we retained OTUs that have ≥0.005% of the total number of sequences according to Bokulich and colleaguesCitation75 and excluded one sample due to technical reasons, resulting in a final dataset of 73 samples with an average of 611,168.6 reads/sample (median = 491,900; min = 299,100; max = 1,072,000 reads/sample).
Functional prediction
We used Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PiCRUST)Citation53 to predict metagenomic functional content from our 16S rRNA survey. Briefly, the same reads fed to AbundantOTU+ were used to produce close reference OTUs using QIIME (v. 1.8.0) and GreenGenes dataset (gg_13_5). PiCRUST software was applied to the resulting biom file to generate KEGG orthologs and metagenomic predictions.
Statistical analysis
OTU, phylum, family or KEGG ortholog raw counts were normalized and log10 transformed according to the following formula:
PCoA plots were produced from Bray–Curtis dissimilarity statistic on the normalized and log10 transformed reads. Alpha diversity (Observed OTU estimate and Shannon diversity index) was calculated after rarefying the raw counts to a depth of the minimum count in all samples. We used a mixed linear model utilizing SAS (v. 9.3) software (SAS Institute Inc, Cary, NC) for analysis including accounting for possible effects from co-housing.Citation31
The mixed linear model, in which dietary iron level (low, control, or high) and source (mucosal or fecal) are fixed effects and the cage is a random effect, is formulated as follows:
where Yijkl represents either PCoA axis value, log10 normalized OTU count, log10 normalized phylum count, log10 normalized family count, log10 normalized KEGG ortholog count or richness value for dietary iron level i, source j, cage k and replicate l. Di.is the effect of the ith dietary iron level. Sj is the effect from the jth source. (SD)ij is the interaction effect between dietary iron level i and source j. Ck(i) is the effect from the kth cage that is nested within the ith dietary iron level and εijkl denotes the error associated with measuring Yijkl. For ), fixed terms Sj and (SD)ij denote the jth Genotype and interaction between dietary iron level i and Genotype j, respectively. Parallel analysis using QIIME (v. 1.8.0) close reference OTU picking approach with the GreenGenes dataset (gg_13_5) and employing UniFrac distance metric after OTU count normalization and log10 transformation (as described above) or after rarefying the counts produced similar results to those obtained from AbundantOTU+ (Figure S7). We controlled for false discovery rate (FDR) by correcting P-values using Benjamini and Hochberg (BH) approach.Citation76
For in vitro experiments, p-values were calculated using one-way ANOVA with Tukey’s multiple comparison post-test for ≥3 experimental groups or two-way ANOVA with Bonferroni multiple comparison post-test when ≥2 variables were compared. For animal experiments, p-values were determined using a non-parametric Kruskal–Wallis test.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Additional Information
The sequences will be available upon acceptance/publication of the manuscript.
Supplemental Material
Download Zip (1.1 MB)Acknowledgments
We acknowledge the Histology, Gnotobiotic and Advanced Analytics Cores at the UNC Center for Gastrointestinal Biology and Disease (supported by NIH P30DK34987) and the National Gnotobiotic Rodent Resource Center (supported by NIH P40 OD01995). This work was supported by grants to RB Sartor from the Crohn’s and Colitis Foundation of America (Microbiome Consortium and Gnotobiotic Facility), NIH P40 OD01995, NIH P30 DK34987, PODK094779, and NIH R01 DK053347; to JC Arthur (NIH K01 DK103952); to M Ellermann from the UNC Graduate School (Dissertation Completion Fellowship) and Howard Hughes Medical Institute (Translational Medicine Fellowship); and to N Maharshak from Crohn’s and Colitis Foundation of America (Research Fellowship Award).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Berglund S, Domellöf M. Meeting iron needs for infants and children. Curr Opin Clin Nutr Metab Care. 2014;17:267–272. doi:10.1097/MCO.0000000000000043.
- Adu-Afarwuah S, Lartey A, Brown KH, Zlotkin S, Briend A, Dewey KG. Home fortification of complementary foods with micronutrient supplements is well accepted and has positive effects on infant iron status in Ghana. Am J Clin Nutr. 2008;87:929–938. doi:10.1093/ajcn/87.4.929.
- Domellöf M, Braegger C, Campoy C, Colomb V, Decsi T, Fewtrell M, Hojsak I, Mihatsch W, Molgaard C, Shamir R, et al. Iron Requirements of Infants and Toddlers. J Pediatr Gastroenterol Nutr. 2014;58:119–129. doi:10.1097/MPG.0000000000000206.
- Gera T, Sachdev HPS. Effect of iron supplementation on incidence of infectious illness in children: systematic review. BMJ. 2002;325:1142. doi:10.1136/bmj.325.7373.1142.
- Zimmermann MB, Chassard C, Rohner F, N‘Goran EK, Nindjin C, Dostal A, Utzinger J, Ghattas H, Lacroix C, Hurrell RF. The effects of iron fortification on the gut microbiota in African children: a randomized controlled trial in Cote d‘Ivoire. Am J Clin Nutr. 2010;92:1406–1415. doi:10.3945/ajcn.110.004564.
- Soofi S, Cousens S, Iqbal SP, Akhund T, Khan J, Ahmed I, Zaidi AKM, Bhutta ZA. Effect of provision of daily zinc and iron with several micronutrients on growth and morbidity among young children in Pakistan: a cluster-randomised trial. Lancet. 2013;382:29–40. doi:10.1016/S0140-6736(13)60437-7.
- Jaeggi T, Gam K, Moretti D, Chassard C, Holding P, Dostal A, Boekhorst J, Hm T, Dw S, Tjalsma H, et al. Iron fortification adversely affects the gut microbiome, increases pathogen abundance and induces intestinal inflammation in Kenyan infants. Gut. 2015;64:731–742. doi:10.1136/gutjnl-2014-307720.
- Abhyankar A, Moss AC. Iron replacement in patients with inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:1976–1981. doi:10.1097/MIB.0000000000000386.
- Erichsen K, Ulvik RJ, Nysaeter G, Johansen J, Ostborg J, Berstad A, Berge RK, Hausken T. Oral ferrous fumarate or intravenous iron sucrose for patients with inflammatory bowel disease. Scand J Gastroenterol. 2005;40:1058–1065. doi:10.1080/00365520510023198.
- Kulnigg S, Gasche C. Systematic review: managing anaemia in Crohn‘s disease. Aliment Pharmacol Ther. 2006;24:1507–1523. doi:10.1111/apt.2006.24.issue-11-12.
- Werner T, Wagner SJ, Martínez I, Walter J, Chang J-S, Clavel T, Kisling S, Schuemann K, Haller D. Depletion of luminal iron alters the gut microbiota and prevents Crohn‘s disease-like ileitis. Gut. 2011;60:325–333. doi:10.1136/gut.2011.237727.
- Chua ACG, Klopcic BRS, Ho DS, Fu SK, Forrest CH, Croft KD, Olynyk JK, Lawrance IC, Trinder D. Dietary iron enhances colonic inflammation and IL-6/IL-11-stat3 signaling promoting colonic tumor development in mice. PLoS One. 2013;8:e78850. doi:10.1371/journal.pone.0078850.
- Lund EK, Wharf SG, Fairweather-Tait SJ, Johnson IT. Oral ferrous sulfate supplements increase the free radical-generating capacity of feces from healthy volunteers. Am J Clin Nutr. 1999;69:250–255. doi:10.1093/ajcn/69.2.250.
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen -Y-Y, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi:10.1126/science.1208344.
- Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Henrissat B, Knight R, Gordon JI. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi:10.1126/science.332.6026.173-c.
- LA D, Cf M, Rn C, Db G, Je B, Be W, Av L, As D, Varma Y, Ma F, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2013;505:559–563.
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi:10.1038/nature05376.
- Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487:104–108. doi:10.1038/nature11225.
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi:10.1038/nature12331.
- Ojeda P, Bobe A, Dolan K, Leone V, Martinez K. Nutritional modulation of gut microbiota - the impact on metabolic disease pathophysiology. J Nutr Biochem. 2016;28:191–200. doi:10.1016/j.jnutbio.2015.08.013.
- Ellermann M, Arthur JC. Siderophore-mediated iron acquisition and modulation of host-bacterial interactions. Free Radic Biol Med. 2017;105:68–78. doi:10.1016/j.freeradbiomed.2016.10.489.
- Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. doi:10.1038/nature25973.
- Caldwell DR, Arcand C. Inorganic and metal-organic growth requirements of the genus Bacteroides. J Bacteriol. 1974;120:322–333.
- Marcelis JH, Den HJ D-S, Hoogkamp-Korstanje JA. Iron requirement and chelator production of staphylococci, Streptococcus faecalis and enterobacteriaceae. Antonie Van Leeuwenhoek. 1978;44:257–267. doi:10.1007/BF00394304.
- Imbert M, Blondeau R. On the iron requirement of lactobacilli grown in chemically defined medium. Curr Microbiol. 1998;37:64–66. doi:10.1007/s002849900339.
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi:10.1016/S0168-6445(03)00055-X.
- Martinez C, Antolín M, Santos J, Torrejon A, Casellas F, Borruel N, Guarner F, Malagelada J-R. Unstable composition of the fecal microbiota in ulcerative colitis during clinical remission. Am J Gastroenterol. 2008;103:643–648. doi:10.1111/j.1572-0241.2007.01592.x.
- Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci. 2011;108(Suppl 1):4578–4585. doi:10.1073/pnas.1000081107.
- Pantoja-Feliciano IG, Clemente JC, Costello EK, Perez ME, Blaser MJ, Knight R, Dominguez-Bello MG. Biphasic assembly of the murine intestinal microbiota during early development. ISME J. 2013;7:1112–1115. doi:10.1038/ismej.2013.15.
- Carvalho FA, Koren O, Goodrich JK, Johansson MEV, Nalbantoglu I, Aitken JD, Su Y, Chassaing B, Walters WA, González A, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139–152. doi:10.1016/j.chom.2012.07.004.
- McCafferty J, Muhlbauer M, Gharaibeh RZ, Arthur JC, Perez-Chanona E, Sha W, Jobin C, Fodor AA. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J. 2013;7:2116–2125. doi:10.1038/ismej.2013.106.
- Maharshak N, Packey CD, Ellermann M, Manick S, Siddle JP, Huh EY, Plevy S, Sartor RB, Carroll IM. Altered enteric microbiota ecology in interleukin 10-deficient mice during development and progression of intestinal inflammation. Gut Microbes. 2013;4:316–324. doi:10.4161/gmic.25486.
- Balamurugan R, Mary RR, Chittaranjan S, Jancy H, Shobana Devi R, Ramakrishna BS. Low levels of faecal lactobacilli in women with iron-deficiency anaemia in south India. Br J Nutr. 2010;104:931–934. doi:10.1017/S0007114510001637.
- Shanmugam NKN, Trebicka E, Fu -L-L, Shi HN, Cherayil BJ. Intestinal inflammation modulates expression of the iron-regulating hormone hepcidin depending on erythropoietic activity and the commensal microbiota. J Immunol. 2014;193:1398–1407. doi:10.4049/jimmunol.1400278.
- Dostal A, Chassard C, Hilty FM, Zimmermann MB, Jaeggi T, Rossi S, Lacroix C. Iron depletion and repletion with ferrous sulfate or electrolytic iron modifies the composition and metabolic activity of the gut microbiota in rats. J Nutr. 2012;142:271–277. doi:10.3945/jn.111.148635.
- Pereira DIA, Aslam MF, Frazer DM, Schmidt A, Walton GE, McCartney AL, Gibson GR, Anderson GJ, Powell JJ. Dietary iron depletion at weaning imprints low microbiome diversity and this is not recovered with oral nano Fe(III). Microbiol Open. 2014;4:12–27. doi:10.1002/mbo3.213.
- Carrier J, Aghdassi E, Platt I, Cullen J, Allard JP. Effect of oral iron supplementation on oxidative stress and colonic inflammation in rats with induced colitis. Aliment Pharmacol Ther. 2001;15:1989–1999. doi:10.1046/j.1365-2036.2001.01113.x.
- Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan T-J, Campbell BJ, Abujamel T, Dogan B, Rogers AB, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi:10.1126/science.1224820.
- Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, et al. Host-derived nitrate boosts growth of E. Science. 2013;339:708–711.
- Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol. 2010;64:43–60. doi:10.1146/annurev.micro.112408.134247.
- Bryce GF, Weller R, Brot N. Studies on the enzymatic synthesis of 2,3-dihydroxy-N-benzoyl-L-serine in Escherichia coli. Biochem Biophys Res Commun. 1971;42:871–879. doi:10.1016/0006-291X(71)90511-0.
- McHugh JP, Rodríguez-Quinoñes F, Abdul-Tehrani H, Svistunenko DA, Poole RK, Cooper CE, Andrews SC. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J Biol Chem. 2003;278:29478–29486. doi:10.1074/jbc.M303381200.
- Seo SW, Kim D, Latif H, O‘Brien EJ, Szubin R, Palsson BO. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat Commun. 2014;5:4910. doi:10.1038/ncomms5972.
- Letain TE, Postle K. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in Escherichia coli. Mol Microbiol. 1997;24:271–283.
- Braun V, Hantke K. Recent insights into iron import by bacteria. Curr Opin Chem Biol. 2011;15:328–334. doi:10.1016/j.cbpa.2011.01.005.
- Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J, Huycke MM, Sartor RB. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology. 2005;128:891–906. doi:10.1053/j.gastro.2005.02.009.
- Cherayil BJ, Ellenbogen S, Shanmugam NN. Iron and intestinal immunity. Curr Opin Gastroenterol. 2011;27:523–528. doi:10.1097/MOG.0b013e32834a4cd1.
- Dostal A, Lacroix C, Pham VT, Zimmermann MB, Del‘Homme C, Bernalier-Donadille A, Chassard C. Iron supplementation promotes gut microbiota metabolic activity but not colitis markers in human gut microbiota-associated rats. Br J Nutr. 2014;111:2135–2145. doi:10.1017/S0007114513003401.
- Krebs NF, Sherlock LG, Westcott J, Culbertson D, Hambidge KM, Feazel LM, Robertson CE, Frank DN. Effects of different complementary feeding regimens on iron status and enteric microbiota in breastfed infants. J Pediatr. 2013;163:416–423. doi:10.1016/j.jpeds.2013.06.055.
- Paganini D, Uyoga MA, Kortman GAM, Cercamondi CI, Moretti D, Barth-Jaeggi T, Schwab C, Boekhorst J, Timmerman HM, Lacroix C, et al. Prebiotic galacto-oligosaccharides mitigate the adverse effects of iron fortification on the gut microbiome: a randomised controlled study in Kenyan infants. Gut. 2017;66:1956–1967. doi:10.1136/gutjnl-2017-314418.
- Lopez CA, Miller BM, Rivera-Chávez F, Velazquez EM, Byndloss MX, Chávez-Arroyo A, Lokken KL, Tsolis RM, Winter SE, Bäumler AJ. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration coli in the Inflamed Gut. Science. 2016;353:1249–1253. doi:10.1126/science.aaf7934.
- Dostal A, Fehlbaum S, Chassard C, Zimmermann MB, Lacroix C. Low iron availability in continuous in vitro colonic fermentations induces strong dysbiosis of the child gut microbial consortium and a decrease in main metabolites. FEMS Microbiol Ecol. 2013;83:161–175. doi:10.1111/fem.2012.83.issue-1.
- Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi:10.1038/nbt.2595.
- Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. The treatment-naive microbiome in new-onset Crohn‘s disease. Cell Host Microbe. 2014;15:382–392. doi:10.1016/j.chom.2014.02.005.
- Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology. 2017;152:327–339.e4. doi:10.1053/j.gastro.2016.10.012.
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi:10.1073/pnas.0706625104.
- Ettreiki C. Juvenile ferric iron prevents microbiota dysbiosis and colitis in adult rodents. World J Gastroenterol. 2012;18:2619. doi:10.3748/wjg.v18.i21.2619.
- Constante M, Fragoso G, Lupien-Meilleur J, Calvé A, Santos MM. Iron supplements modulate colon microbiota composition and potentiate the protective effects of probiotics in dextran sodium sulfate-induced colitis. Inflamm Bowel Dis. 2017;23:753–766. doi:10.1097/MIB.0000000000001089.
- Dostal A, Baumgartner J, Riesen N, Chassard C, Smuts CM, Zimmermann MB, Lacroix C. Effects of iron supplementation on dominant bacterial groups in the gut, faecal SCFA and gut inflammation: a randomised, placebo-controlled intervention trial in South African children. Br J Nutr. 2014;112:547–556. doi:10.1017/S0007114514001160.
- Lee T, Clavel T, Smirnov K, Schmidt A, Lagkouvardos I, Walker A, Lucio M, Michalke B, Schmitt-Kopplin P, Fedorak R, et al. Oral versus intravenous iron replacement therapy distinctly alters the gut microbiota and metabolome in patients with IBD. Gut. 2017;66:863–871. doi:10.1136/gutjnl-2015-309940.
- Erichsen K, Ulvik RJ, Grimstad T, Berstad A, Berge RK, Hausken T. Effects of ferrous sulphate and non-ionic iron-polymaltose complex on markers of oxidative tissue damage in patients with inflammatory bowel disease. Aliment Pharmacol Ther. 2005;22:831–838. doi:10.1111/apt.2005.22.issue-9.
- Werner T, Hoermannsperger G, Schuemann K, Hoelzlwimmer G, Tsuji S, Haller D. Intestinal epithelial cell proteome from wild-type and TNFDeltaARE/WT mice: effect of iron on the development of chronic ileitis. J Proteome Res. 2009;8:3252–3264. doi:10.1021/pr800772b.
- Wang L, Johnson EE, Shi HN, Walker WA, Wessling-Resnick M, Cherayil BJ. Attenuated inflammatory responses in hemochromatosis reveal a role for iron in the regulation of macrophage cytokine translation. J Immunol. 2008;181:2723–2731.
- Wang L, Harrington L, Trebicka E, Shi HN, Kagan JC, Hong CC, Lin HY, Babitt JL, Cherayil BJ. Selective modulation of TLR4-activated inflammatory responses by altered iron homeostasis in mice. J Clin Invest. 2009;119:3322–3328.
- Scheinin T, Butler DM, Salway F, Scallon B, Feldmann M. Validation of the interleukin-10 knockout mouse model of colitis: antitumour necrosis factor-antibodies suppress the progression of colitis. ClinExp Immunol. 2003;133:38–43. doi:10.1046/j.1365-2249.2003.02193.x.
- Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, Rennick DM, Sartor RB. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–5231.
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi:10.1073/pnas.120163297.
- Chen J, Shen H, Chen C, Wang W, Yu S, Zhao M, Li M. The effect of psychological stress on iron absorption in rats. BMC Gastroenterol. 2009;9:83. doi:10.1186/1471-230X-9-83.
- Patwa LG, Fan T-J, Tchaptchet S, Liu Y, Lussier YA, Sartor RB, Hansen JJ. Chronic intestinal inflammation induces stress-response genes in commensal Escherichia coli. Gastroenterology. 2011;141:1842–51.e1–10. doi:10.1053/j.gastro.2011.06.064.
- Lutz MB, Kukutsch N, Ogilvie AL, Rössner S, Koch F, Romani N, Schuler G. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods. 1999;223:77–92.
- Stanley ER, Heard PM. Factors regulating macrophage production and growth. Purification and some properties of the colony stimulating factor from medium conditioned by mouse L cells. J Biol Chem. 1977;252:4305–4312.
- Arthur JC, Gharaibeh RZ, Mühlbauer M, Perez-Chanona E, Uronis JM, McCafferty J, Fodor AA, Jobin C. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun. 2014;5:4724. doi:10.1038/ncomms5972.
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi:10.1093/bioinformatics/btr381.
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010;7:335–336. doi:10.1038/nmeth.f.303.
- Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Meth. 2012;10:57–59. doi:10.1038/nmeth.2276.
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol). 1995;57:289–300. doi:10.1111/rssb.1995.57.issue-1.
