ABSTRACT
Severe acute malnutrition (SAM) is a major challenge in low-income countries and gut microbiota (GM) dysbiosis may play a role in its etiology. Here, we determined the GM evolution during rehabilitation from SAM and the impact of probiotics (Lactobacillus rhamnosus GG and Bifidobacterium animalis subsp. lactis BB-12) supplementation. The GM (16S rRNA gene amplicon sequencing) of children admitted to hospital with SAM showed distinct composition over admission (e.g. Klebsiella spp., and Enterobacteriaceae spp.), discharge (e.g. Clostridiaceae spp., Veilonella dispar) and follow-up (e.g. Lactobacillus ruminis, Blautia spp., Faecalibacterium prausnitzii), reaching similar β- and α-diversity as healthy individuals. Children with diarrhea had reduced distribution of Bacteroidaceae, Lachnospiraceae, increased Enterobacteriaceae and Moraxellaceae, and lower α-diversity. Children suffering from edematous SAM had diminished proportion of Prevotellaceae, Lachnospiraceae, Ruminoccaceae and a higher α-diversity when compared to non-edematous SAM. Supplementation of probiotics did not influence β-diversity upon discharge or follow-up, but it increased (p < .05) the number of observed species [SE: > 4.5]. Children where the probiotic species were detected had lower cumulative incidence (p < .001) of diarrhea during the follow-up period compared to children receiving placebo and children receiving probiotics, but where the probiotics were not detected. The GM of children with non-edematous and edematous SAM differ in composition, which might have implications for future GM targeted treatments. Probiotics treatment reduced the cumulative incidence of diarrhea during the outpatient phase, with the strongest effect in children where the administered probiotics could be detected in the GM.
Introduction
Malnutrition remains a global challenge with 45% of childhood deaths being attributed to undernutrition.Citation1 In addition, undernutrition in early life is associated with long-term sequelae, including stunting, less schooling, and reduced economic productivity later in life.Citation2 Recent research indicates that gut microbiota (GM) dysbiosis may be involved in development or maintenance of acute malnutrition.Citation3–Citation6
Malnutrition seems to be associated with reduced GM diversity and maturity as determined by metagenomics and culturomics approaches.Citation3–Citation5,Citation7–Citation9 In Bangladesh, a birth cohort of children from urban slum was followed until 2 years of age with frequent analyses of GM composition.Citation4 Based on age-discriminatory bacterial taxa microbiota maturity scores were developed. Children with severe acute malnutrition (SAM) showed significant GM immaturity compared to well-nourished peers. Nutritional interventions, including Ready-to-Use Therapeutic Food (RUTF), only partially and temporarily improved the GM maturity. Using the same GM maturity models on samples from other Bangladeshi children, reduced maturity was found in stool samples during and 1 month after diarrhea episodes.Citation4 Findings of reduced GM maturity or diversity in malnourished children, and temporary improvement of GM during RUTF treatment has also been observed in Malawian children.Citation3 In Uganda, a cross-sectional study found differences in the GM composition of children with non-edematous and edematous SAM and reported lower numbers of observed species in the GM of children with non-edematous compared to edematous SAM.Citation10
It has been suggested that disruption of normal development of the GM may be causally related to development of malnutrition.Citation3,Citation5,Citation7-Citation9 This was first shown in the GM from a Malawian cohort of twin pairs who became discordant for kwashiorkor (SAM with edema).Citation3 When the GM of twin pairs discordant for kwashiorkor was transferred to germ-free mice, only mice receiving GM from a child with kwashiorkor became malnourished when fed a diet similar to the diet of Malawian children. Likewise, children with SAM in Niger and Senegal were reported to have GM composition depleted in oxygen-sensitive bacteria and being enriched in putatively pathogenic Proteobacteria, Fusobacteria, and Streptococcus gallolyticus.Citation8,Citation9 Moreover, animal studies have confirmed that transfer of GM originating from undernourished children to germ-free mice impair growth.Citation7
If early life GM dysbiosis can contribute to malnutrition, microbial interventions may be able to support repairing or normalization of the GM. Several bacterial species (e.g. species of Lactobacillus, Bifidobacterium adolescentis, and Bacteroides salyersiae) have been previously proposed as probiotic candidates as an alternative to fecal transplantation to address children suffering from SAM.Citation9 Meta-analyses have found that probiotics reduce the duration of acute diarrhea by 1 day and reduce the risk of acute diarrhea lasting 4 days or more.Citation11 However, most studies were performed in high-income countries and in well-nourished children and the knowledge of the impact of probiotic treatment in children with SAM is scarce.Citation12 The PRONUT study investigated the effect of a synbiotic mixture, a combination of four lactic acid bacteria in a total dose of 1011 colony-forming units in total per day and four fermentable fiber sources. No effect on the primary outcome (nutritional cure) was observed, but a near-significant effect (relative risk = 0.65, p = .06) on overall mortality was found.Citation12 The ProbiSAM study investigated the effect of administrating a combination of Lactobacillus rhamnosus GG (LGG) and Bifidobacterium animalis subsp. lactis BB-12 (BB-12) to children hospitalized with SAM.Citation13 It was found that administration of the probiotics did not influence days with diarrhea during in-patient treatment, but a significant 26% reduction in days with diarrhea during the outpatient treatment (8–12 weeks after hospitalization) was observed in the probiotic group.
In the present study, we investigate GM development during rehabilitation from SAM, whether probiotics supplementation has any effects on the GM during inpatient and outpatient treatment of SAM, and whether GM composition and development is linked to the observed reduction in days with diarrhea when administered probiotics during the randomized controlled ProbiSAM trial.
Results
Cohort overview and sequencing
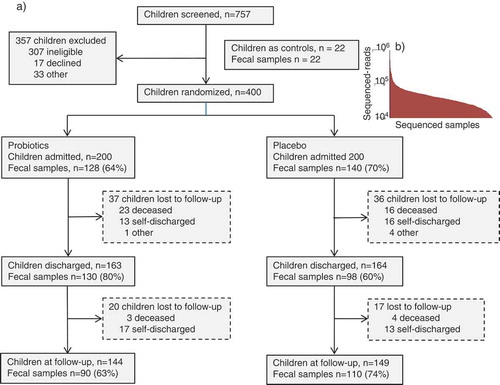
The study enrolled 400 children with a mean (±SD) age of 17.0 (±8.5) months. Males constituted 58% of the population, 14% were HIV seropositive, 66% suffered from edematous malnutrition and 61% had diarrhea at admission. Fecal samples were collected at hospital admission, at discharge and 8 weeks post-discharge (follow-up). The proportion of individuals from which a fecal sample was obtained at admission to hospital, discharge, and after 8 weeks post-discharge varied between 60% and 80% at the different time points (). Control children included 22 apparently healthy children aged 6–59 months living in communities similar to the children admitted with SAM ().
Sequencing of DNA extracted from fecal samples generated 34.5 million reads derived from the 16S rRNA gene V3-V4-region with an average of 47,996 (max: 532,852, min: 10,289) sequences per subject (). The analysis of amplicon-sequencing data generated 44,808 OTU phylotypes (representative sequences clustered at 97% sequence similarity) that were summarized over 365 bacterial species.
GM variation during treatment of SAM
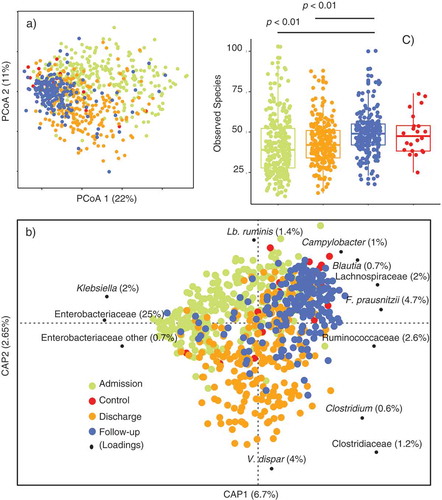
Constrained and un-constrained analyses of Bray–Curtis dissimilarity metrics (-b) displayed GM compositional signatures associated with changes from admission to discharge and follow-up and explaining up to 13% (adonis p < .001) of the total variance. At follow-up, the GM of children treated for SAM was indistinguishable from that of healthy individuals from the same-community setting ( and b). Based on Constrained Analysis of Principal coordinates (CAP), the most discriminatory GM members () at admission were associated with Enterobacteriaceae members (Klebsiella and Enterobacteriaceae other). At discharge, the relative abundance of Clostridiaceae members (Clostridium uncl., Veilonella dispar (Clostridium cluster IXCitation14) and other Clostridiaceae) had increased and at follow-up, the GM composition was enriched with Lactobacillaceae (Lactobacillus ruminis), Campylobacteraceae (Campylobacter uncl.), and several members of the Clostridium cluster IV and XIVaCitation15 (such as Blautia, Lachnospiraceae, Ruminoccoaceae and Faecalibacterium prausnitzii).
In relation to mean α-diversity (), the lowest number of observed species was determined at admission (41.4 ± 17.5), followed by an increase as treatment progressed. Upon discharge and follow-up, the mean α-diversity was 42.7 ± 12.7 and 49.3 ± 12.2 (p < .01 relative to both admission and discharge), respectively. As also observed with respect to β-diversity ( and b), no significant differences between healthy subjects and at follow-up for children treated for SAM were observed regarding the number of observed species (42.7 ± 12.7) ().
Diarrhea and edema on admission
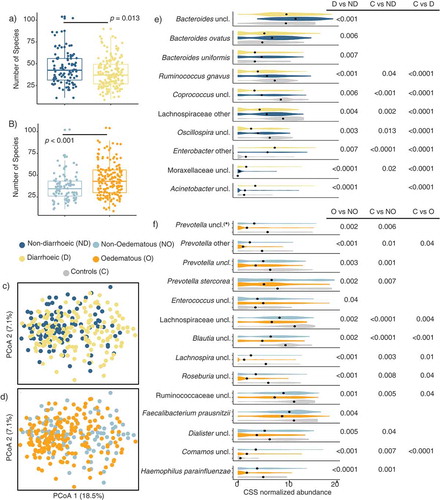
At admission, significant differences in the number of observed species were observed between children with and without diarrhea (39 ± 16 vs. 46 ± 19, p = .013), with diarrhea being associated with a lower number of observed species. Similarly, non-edematous SAM was associated with a lower number of observed species relative to edematous SAM (36 ± 15 vs. 45 ± 18, p < .001, those with any grade of edema) (-b). Likewise, Bray–Curtis dissimilarity analysis demonstrated significant compositional differences between children admitted with vs. without diarrhea (adonis, p = .001, R2= 0.018), and edematous vs. non-edematous SAM (adonis, p = .001, R2= 0.019) (-d). No significant interaction effects of diarrhea and edema on α – (two-way ANOVA p = .98) nor β -diversity (CAP p = .24) were observed indicating no mutual dependence between the two conditions with respect to GM composition.
The GM of children with diarrhea at admission was characterized by lower relative abundance of Bacteroidaceae (Bacteroides spp., B. ovatus and B. uniformis), Lachnospiraceae (R. gnavus, Coprococcus and Oscillospira uncl.), higher Moraxellaceae (Moraxellaceae uncl. and Acinetobacter uncl.), and Enterobacteriaceae (Enterobacter) relative to children admitted without diarrhea and/or the healthy children (). At admission the GM of children admitted with non-edematous SAM had lower abundance of Prevotellaceae members (Prevotella spp. including Prevotella stercorea), Lachnospiraceae (Blautia, Lachnospira, and Roseburia spp.), Ruminoccoaceae, Clostridiaceae (F. prausnitzii), Veillonelaceae (Dialister spp.), Comamonadaceae (Comamos uncl.), as well as Pasteurellaceae (H. parainfluenzae) () compared to the children admitted with edematous SAM and healthy subjects. Only the abundance of Enterococcus uncl. () was significantly higher in children with non-edematous SM compared to edematous SAM.
Probiotics establishment increases α-diversity and reduces days of diarrhea
Probiotic administration did not influence (adonis, p > .05) the β-diversity profiles at discharge () nor follow-up (), and on average the proportion of reads mapping the probiotic strains was 0.33% at discharge and 0.02% at follow-up (as determined by amplicons with >97% similarity to the relevant 16S rRNA gene fragment of Lb. rhamnosus and B. animalis). Furthermore, in the probiotic group recovery of the two probiotic strains from fecal samples was not evenly distributed among the children (). Children in which at least one probiotic strain was detected were defined as individuals with high level of probiotic response (responders), whereas children where the probiotic strains could not be detected were defined as individuals with low level of probiotic response (non-responders) ( and ). Interestingly, at discharge, the average number of observed species in the children defined as responders was 4.8 (p ≤ 0.05) higher than in the children that received placebo. Likewise, at follow-up, the number of species in responders was 6.4 (p ≤ 0.01) and 4.5 (p ≤ 0.03) higher than in the children defined as non-responders or receiving placebo, respectively ( and ), after correcting for the confounding effect of age. There were no significant associations between age, sex, presence of diarrhea or edema at admission, GM composition at admission, HIV status, weight-for-length or duration of hospitalization and being a responder/non-responder.
Figure 4. Probiotic administration and establishment in relation to α-diversity and cumulative incidence of diarrhea.
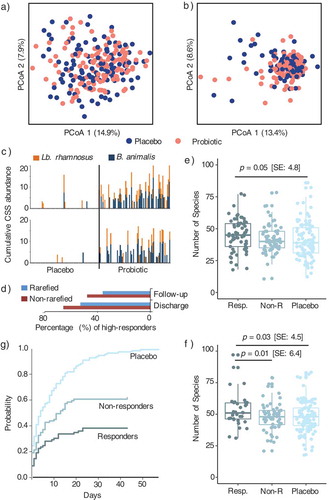
Throughout the trial, diarrhea incidence (minimum of 1 day) during outpatient treatment was assessed over a median (IQR) period of 56 (56:58) days. Cumulative incidence function (CIF) analysis on outpatient data found that the probability of having more days with diarrhea was significantly higher (p < .001) among placebo subjects as compared to the probiotic group (). Interestingly, differences between responders and non-responders (p < .001) were also found, where the probability of having more days with diarrhea was significantly reduced (~two fold) in the responders as compared to non-responders ().
Discussion
GM changes during rehabilitation from SAM
Here we show that children with SAM have significantly reduced number of observed species and major compositional differences (β-diversity) compared to healthy subjects. The number of observed species increased during the course of treatment with the lowest number found at admission increasing until it no longer differed from the healthy individuals at follow-up. In line with this, malnutrition in children has previously been associated with reduced diversity and maturity of the gut microbiota.Citation3–Citation5,Citation7
Distinct GM compositional signatures were observed during the treatment of the children admitted with SAM (i.e. admission, discharge and follow-up). Between admission and discharge, a beneficial shift with less Enterobacteriaceae and increasing Veillonela and Clostridiales abundance were seen. A development that continued between discharge and follow-up, where Clostridiales members (Faecalibacterium, Blautia, and other unclassified members of Ruminococcaceae and Lachnospiraceae) became more abundant, similar to the GM of the healthy subjects. This is also in agreement with previous observations where several obligate anaerobes related to Clostridiales, such as C. butyricum, R. bromii, and R. intestinalis, were reported to be enriched in healthy children as compared to those suffering from kwashiorkor in Western Africa.Citation9 Similarly, B. breve has also found to be enriched in kwashiorkor cases,Citation9 but in the present study, no significant (p = .36) differences were found among the healthy controls and children with SAM.
The observed normalization of the GM is likely due to improved nutritionCitation3 in response to the therapeutic foods given during in- and outpatient treatment that contain high-quality protein and high amounts of micronutrients to replenish micronutrient deficiencies and support catch-up growth of the children. The micronutrients may also support development of a GM with a composition beneficial for the child.Citation16 A study from Bangladesh in 64 children with SAMCitation4 found their GM maturity index to be reduced compared to healthy subjects. Treatment with therapeutic foods improved the GM maturity. However, it did not reach the level of healthy children and eventually regressed after cessation of therapeutic food treatment.Citation4 Similarly, in a detailed study of 13 Malawian twin pairs discordant for edematous malnutrition (Kwashiorkor) it was found that treatment with ready-to-use therapeutic food (RUTF) resulted in a transient maturation of the GM.Citation3 These changes in GM in children with SAM treated with RUTF could provide a protective barrier to readmission from SAM in the future, but it remains to be investigated how stable the changes are.
Diarrhea, edema, and GM
Diarrhea is a well-known morbidity factor and indicator of poor prognosis in children with SAM.Citation17–Citation19 We found the presence of diarrhea at hospital admission to be associated with reduced number of observed species, as well as reduced relative abundance of species belonging to Bacteriodaceae and Lachnospiraceae and higher relative abundance of Moraxellaceaea and Enterobacteriaceae. Others have reported transiently reduced GM maturity in children with diarrhea in BangladeshCitation4 and an association with moderate to severe diarrhea and lower GM diversity in children below 5 years in four low-income countries.Citation20
Distinct features of the GM were associated with non-edematous compared to edematous SAM at admission, with non-edematous children having significantly fewer observed species. A similar finding has been made in an earlier cohort of children with SAM from the same hospital.Citation10 Several factors could explain the lowered number of observed species seen in relation to non-edematous SAM. It is hypothesized that longer starvation of the GM in non-edematous SAM may lead to a lower GM diversity. The non-edematous children also tend to have more infections when they are admitted to hospital.Citation21,Citation22 They may, therefore, have been treated with more antibiotics from health clinics before hospital admission leading to a lower number of observed species. The abundance of taxa normally associated with Sub-Saharan African children not suffering from disease such as Prevotellaceae membersCitation23 was also significantly higher in children admitted with edematous SAM compared to non-edematous SAM and closer to the abundance observed in the healthy subjects.
Responders vs non-responders
The probiotic strains LGG and BB-12 have in previous studies been found to colonize the gut transiently after oral administration. Recovery of both strains depends on the dose administered and vary between individuals.Citation24,Citation25 We detected each strain in approximately half of the fecal samples from children randomized to receiving the probiotics. The observed recovery is in line with recovery obtained with BB-12Citation24 and slightly lower recovery than previously observed for LGGCitation25 in previous studies.
There were no significant differences in β-diversity among individuals considered as probiotic high- and non-responders. However, the mean number of observed species was 4.8 higher among responders compared to placebo at discharge. At follow-up, the number of observed species in responders was 6.4 and 4.5 higher compared to non-responders and placebo, respectively. This indicates that in the case of SAM, LGG, and BB-12 may influence GM α-diversity. Responders had lower cumulative incidence of diarrhea during outpatient treatment compared to probiotic non-responders, which could indicate a better gut colonization and stronger interaction with the immune system.Citation26 Yet, we were unable to identify malnutrition-related factors associated with being a responder/non-responder. As described above, non-responders are also reported in healthy volunteers for both LGG and BB-12 and being a non-responder, may therefore, be very common and not related to malnutrition.Citation24,Citation25
Strengths and limitations
A particular strength of the present study is the large sample size compared to other studies investigating the effect of re-feeding and probiotic administration to children suffering from SAM. However, in this context, it is a limitation, that the number of included children without disease (healthy subjects) is relatively small. At the different sampling time points we obtained samples from 60-80% of the enrolled children. Possibly the mothers/caregivers with the most ill children were those with lowest compliance which might skew the sample set toward the most ill children being under-represented. However, it should be noted that most missing samples are due to failure to pass stool/collect stool within the specified timeframe.
Conclusion
GM diversity and composition change over the course of rehabilitation from SAM and approach the GM of apparently healthy subjects as treatment progresses. Further, our study supports that non-edematous and edematous SAM are associated with GM compositional differences, which might have implications for future GM targeted treatments. Finally, using probiotics alongside the standard treatment protocol for SAM reduces the incidence of days with diarrhea after discharge. This may be partly mediated by the observed increase in the number of observed species seen in the children, where the administered probiotics could be detected at discharge and follow-up (“Responders”). Although the effect of probiotics on the GM was modest and previous studies have shown that nutritional interventions may only lead to transient improvements of the GM, the results indicate a potential direction for future research and management of SAM.
Materials and methods
Ethics statement
Before study initiation, ethical approval was obtained from the School of Medicine Research and Ethics Committee at Makerere University, Kampala, and a consultative approval was provided by The National Committee of Health Research Ethics in Denmark. Written informed consent was obtained from all caregivers on behalf of their children. In addition, clearance to conduct the study was given by the Uganda National Council of Science and Technology and the Ugandan National Drug Authority. Further details have been described elsewhere [11]. The study was registered at www.isrctn.com, ISRCTN16454889.
Study design, patients, and study procedures
The study is a prospective study nested in a randomized, double-blind, placebo-controlled trial assessing the effect of probiotics on diarrhea among children with SAM. The trial was conducted at Mwanamugimu Nutrition Unit, Mulago National Referral Hospital, Kampala, Uganda between March 2014 and September 2015. Children admitted to Mulago Hospital with SAM generally have multiple medical complications and the case fatality rates are high (approx. 20%). Children between 6 and 59 months with SAM were eligible for the probiotic trial. SAM was defined as mid-upper-arm circumference (MUAC) < 11.5 cm, weight-for-height z-score (WHZ) < −3 SD or bipedal pitting edema. Caregivers also had to provide written informed consent and be willing to return for follow-up. Children were excluded if they were in shock, had severe respiratory distress, an admission weight below 4.0 kg, obvious congenital anomalies or if they had been admitted with SAM the previous 6 months. Controls included apparently healthy 22 children aged 6–59 months with WHZ > −1 and living in communities similar to the children admitted with SAM.
Standard treatment was provided to all children according to the Integrated Management of Acute Malnutrition guidelines for UgandaCitation27 with adaptation from the WHO guidelines.Citation28 In addition, one daily dose of a combination of two probiotic strains BB-12 and LGG or placebo was given. The total probiotic dose was 10 billion colony-forming units [CFU] per day with half of each strain (Chr. Hansen A/S, Hørsholm, Denmark). The probiotic/placebo supplement was administered from hospital admission to discharge and throughout an outpatient treatment period of 8–12 weeks, depending on the nutritional recovery rate of each child. More detailed information about the study is reported elsewhere.Citation13
Sample collection, processing, and DNA extraction
Fecal samples were collected at admission, discharge, and after 8 weeks of outpatient treatment. Admission samples were collected from the time of admission to day 3 of hospitalization; discharge and 8-week follow-up samples were collected on the day of discharge/follow-up or the day before. During hospitalization, stool was collected in plastic bags and stored for maximum 1 h before the contents of the stool bags were emptied into 2-ml DNAse free cryotubes. The cryotubes were immediately frozen in liquid nitrogen. Outpatient samples were collected in the children’s home up to one day before a follow-up visit. Caregivers transferred stool to a lidded 10-ml plastic vial containing 5-ml RNA-later (Qiagen GmbH, Hilden, Germany). Caregivers were asked to fill stool up to a mark ensuring an approximate 5:1 ratio of RNA-later and stool. When samples were received, the 10-ml plastic vial was centrifuged at 1,300–2,200 × g for 10 min. RNA-later was discarded and the stool was re-suspended in 2-ml TE buffer. One ml was transferred to a 2-ml DNAse free cryotube and immediately frozen in liquid nitrogen and stored at −80°C. Samples were shipped on dry ice to Denmark for further processing and analysis.
Fecal samples were centrifuged at 13,000 × g for 10 min at room temperature and ~200 mg of the fecal pellet was used for DNA extraction using the PowerSoil® DNA Isolation Kit (MOBIO Laboratories, Carlsbad, CA, USA), following the instructions of the manufacturer, but with minor modifications. Briefly, prior DNA extraction, samples were placed into the PowerBead tubes and heat treated at 65°C for 10 min and then at 95°C for 10 min. Subsequently, solution C1 was added and bead-beating performed in FastPrep (MP Biomedicals, Santa Ana, CA, USA) using 3 cycles of 15 s each, at a speed of 6.5 m.−1 The remaining DNA extraction procedure followed the manufacturer’s instructions.
High-throughput 16S rRNA gene amplicon sequencing
GM composition was determined by high-throughput 16S rRNA gene amplicon sequencing. The primers designed with adapters Nextera Index Kit® (Illumina, CA, USA) targeted the V3-V4 region (~466 bp) and the amplicon library preparation, purification and sequencing were performed as previously described.Citation29 Briefly, the amplification profile (1st PCR) followed: Denaturation at 95°C for 2 min; 33 cycles of 95°C for 15 s, 55°C for 15s and 68°C for 40 s; followed by final elongation at 68°C for 5 min, while barcoding (2nd PCR) was performed at 98°C for 1 min; 12 cycles of 98°C for 10 s, 55°C for 20 s and 72°C for 20 s; elongation at 72°C for 5 min. The amplified fragments with adapters and tags were purified using AMPure XP beads (Beckman Coulter Genomic, CA, USA). Prior to library pooling clean constructs were quantified using a Qubit Fluorometer (Invitrogen, Carlsbad, CA, USA) and mixed in approximately equal concentrations to ensure even representation of reads per sample followed 250 bp pair-ended MiSeq (Illumina, CA, USA) sequencing.
Processing of HTS data
The raw dataset containing pair-ended reads with corresponding quality scores were merged and trimmed using settings previously described.Citation29 De-replicating, purging from chimeric reads and constructing de novo Operational Taxonomic Units (OTU, with 97% similarity) was conducted using the UPARSE pipeline.Citation30 The green genes (v13.8) 16S rRNA gene collection was used as a reference database.Citation31
Statistical analyses
For abundance-based analyses (β-diversity), contingency tables (based on OTUs/phylotypes summarized to species level) were normalized with cumulative sum scaling (CSSCitation32). The influence of explanatory variables over GM composition was evaluated through the Constrained Analysis of Principal Coordinates (CAP) on Bray–Curtis distances, while using the ANOVA-like permutation tests (1,000) to determine the significance of each effect.Citation33 Through CAP, top discriminant GM features were selected based on a minimum absolute index of 0.15 along the canonical axes; (i) mean differences between such discriminant features was performed with two-tailed Student’s t-test and corrected for Type I error with False Discovery Rate (FDR).
For mean species diversity (α-diversity) samples were randomly rarefied to 10,000 sequences each, and the number of observed species determined as a function of sequence depth. Differences in α-diversity were determined by either (i) non-parametric using 999 Monte Carlo permutations or (ii) by mixed linear models (MLM) in order to correct for the co-founding effect of age.
Days with diarrhea during outpatient treatment were evaluated using cumulative incidence function (CIF)Citation34 within placebo, probiotic responders and probiotic non-responders subjects, whereas differences in cumulative incidence rates were assessed with the Gray’s test.Citation35
Data availability
The accession number of the sequencing-data reported in this paper is ENA: PRJEB29297. Sequencing metadata are available on request.
Disclosure of potential conflicts of interest
Chr. Hansen A/S sponsored parts of the gut microbiome analysis. The company did not play any role in data analysis, data interpretation or manuscript writing.
References
- Black RE, Victora CG, Walker SP, Bhutta ZA, Christian P, De Onis M, Ezzati M. Maternal and child nutrition 1 maternal and child undernutrition and overweight in low-income and middle-income countries. Lancet. 2013;832:427–451. doi:10.1016/S0140-6736(13)60937-X.
- Victora CG, Adair L, Fall C, Hallal PC, Martorell R, Richter L, Sachdev HS. Maternal and child undernutrition 2 maternal and child undernutrition : consequences for adult health and human capital. Lancet. 2008;371:340–357. doi:10.1016/S0140-6736(07)61692-4.
- Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Liu J, Houpt E, J V L, Holmes E, Nicholson J, et al. Gut microbiomes of Malawian Twin. Science. 2013;339:548–554. doi:10.1126/science.1229000.
- Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, Destefano J, Meier MF, Muegge BD, et al. Persistent gut microbiota immaturity in. Nature. 2014;510:417–421. doi:10.1038/nature13421.
- Blanton LV, Barratt MJ, Charbonneau MR, Ahmed T, Gordon JI. Childhood undernutrition, the gut microbiota, and microbiota-directed therapeutics. Science. 2016;352:1533. doi:10.1126/science.aad9359.
- Ahmed T, Auble D, Berkley JA, Black R, Ahern PP, Hossain M, Hsieh A, Ireen S, Arabi M, Gordon JI. An evolving perspective about the origins of childhood undernutrition and nutritional interventions that includes the gut microbiome. Ann N Y Acad Sci. 2014;1332:22–38. doi:10.1111/nyas.12487.
- Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Venkatesh S, Ilkaveya O, Subramanian S, Manary MJ, Trehan I, Jorgensen JM, et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016;351:pii: aad3311. doi:10.1126/science.aad3311.
- Pham TP, Tidjani Alou M, Bachar D, Levasseur A, Brah S, Alhousseini D, Sokhna C, Diallo A. Gut microbiota alteration is characterized by a proteobacteria and fusobacteria bloom in Kwashiorkor and a bacteroidetes paucity in Marasmus. Sci Rep. 2019;9:9084. doi:10.1038/s41598-019-45611-3.
- Tidjani Alou M, Million M, Traore SI, Mouelhi D, Khelaifia S, Bachar D, Caputo A, Delerce J, Brah S, Alhousseini D, et al. Gut bacteria missing in severe acute malnutrition, can we identify potential probiotics by culturomics? Front Microbiol. 2017;8:899. doi:10.3389/fmicb.2017.00899.
- Hee K, Kristensen S, Wiese M, Johanne M, Rytter H. Gut microbiota in children hospitalized with oedematous and non-oedematous severe acute malnutrition in Uganda. PLoS Negl Trop Dis. 2016;10:e0004369. doi:10.1371/journal.pntd.0004369.
- Sj A, Eg M, Gv G, Lf D. Probiotics for treating acute infectious diarrhoea (Review). Cochrane Database Syst Rev. 2010;10:CD003048. doi:10.1002/14651858.CD003048.pub3.
- Kerac M, Bunn J, Seal A, Thindwa M, Tomkins A, Sadler K, Bahwere P, Collins S. Probiotics and prebiotics for severe acute malnutrition (PRONUT study): a double-blind efficacy randomised controlled trial in Malawi. Lancet. 2009;374:136–144. doi:10.1016/S0140-6736(09)60884-9.
- Grenov B, Namusoke H, Lanyero B, Nabukeera-barungi N, Ritz C, Friis H, Michaelsen KF. Effect of probiotics on diarrhea in children with severe acute malnutrition : a randomized controlled study in Uganda. Gastroenterology. 2017;64:396–403. doi:10.1097/MPG.0000000000001515.
- Tetro J, Allen-Vercoe E. The human microbiome handbook, 1st ed. Lancaster (PA): DEStech Publications; 2016.
- Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A. Commensal clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog. 2013;5:23. doi:10.1186/1757-4749-5-23.
- Hibberd MC, Wu M, Rodionov DA, Li X, Cheng J, Griffin NW, Barratt MJ, Giannone RJ, Hettich RL, Osterman AL, et al. The effects of micronutrient deficiencies on bacterial species from the human gut microbiota. Sci Tranlational Med. 2017;9:pii: eaal4069. doi:10.1126/scitranslmed.aal4069.
- Talbert A, Thuo N, Karisa J, Chesaro C, Ohuma E, Ignas J, Berkley JA, Toromo C, Atkinson S, Maitland K. Diarrhoea complicating severe acute malnutrition in Kenyan Children : A prospective descriptive study of risk factors and outcome. PLoS One. 2012;7:e38321. doi:10.1371/journal.pone.0038321.
- Nabukeera-barungi N, Grenov B, Lanyero B, Namusoke H, Mupere E, Christensen VB, Michaelsen KF, Mølgaard C, Rytter MJ, Friis H. Predictors of mortality among hospitalized children with severe acute malnutrition : a prospective study from Uganda. Pediatr Res. 2018;84:92–98. doi:10.1038/s41390-018-0016-x.
- Irena AH, Mwambazi M, Mulenga V. Diarrhea is a major killer of children with severe acute malnutrition admitted to inpatient set-up in Lusaka, Zambia. Nutr J. 2011;10:110. doi:10.1186/1475-2891-10-110.
- Pop M, Walker AW, Paulson J, Lindsay B, Antonio M, Hossain MA, Oundo J, Tamboura B, Mai V, Astrovskaya I, et al. Diarrhea in young children from low-income countries leads to large-scale alterations in intestinal microbiota composition. Genome Biol. 2014;15:R76. doi:10.1186/gb-2014-15-6-r76.
- Rytter JMH, Namusoke H, Babirekere-iriso E, Kæstel P, Girma T, Christensen VB, Michaelsen KF, Friis H. Social, dietary and clinical correlates of oedema in children with severe acute malnutrition : a cross-sectional study. BMC Pediatr. 2015;15:1–9. doi:10.1186/s12887-015-0341-8.
- Girma T, Kæstel P, Mølgaard C, Michaelsen KF, Hother A, Friis H. Predictors of oedema among children hospitalized with severe acute malnutrition in Jimma University Hospital, Ethiopia: a cross sectional study. BMC Pediatr. 2013;13:204. doi:10.1186/1471-2431-13-204.
- De Filippo C, Cavalieri D, Di M, Ramazzotti M, Baptiste J. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi:10.1073/pnas.1005963107.
- Larsen CN, Nielsen S, Kæstel P, Brockmann E, Bennedsen M, Christensen HR, Eskesen DC. Dose – response study of probiotic bacteria Bifidobacterium animalis subsp lactis BB-12 and Lactobacillus paracasei subsp paracasei CRL-341 in healthy young adults. Eur J Clin Nutr. 2006;60:1284–1293. doi:10.1038/sj.ejcn.1602450.
- Petschow BW, Figueroa R, Harris CL, Beck LB, Ascp B, Ziegler E, Goldin B. Effects of feeding an infant formula containing lactobacillus GG on the colonization of the intestine a dose-response study in healthy infants. J Clin Gastroenterol. 2005;39:786–790. doi:10.1097/01.mcg.0000177245.53753.86.
- Sanders ME. Impact of probiotics on colonizing microbiota of the gut. J Clin Gastroenterol. 2011;45:115–119. doi:10.1097/MCG.0b013e318227414a.
- MHU. Integrated management of acute malnutrition guidelines. Kampala (Uganda): Ministry of Health; 2010.
- WHO. Guideline: updates on the management of severe acute malnutrition in infants and children. Geneva (Switzerland): World Health Organization; 2013.
- Krych Ł, Kot W, Bendtsen KMB, Hansen AK, Vogensen FK, Nielsen DS. Have you tried spermine? A rapid and cost-e ff ective method to eliminate dextran sodium sulfate inhibition of PCR and RT-PCR. J Microbiol Methods. 2018;144:1–7. doi:10.1016/j.mimet.2017.10.015.
- Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Meth. 2013;10:996–998. doi:10.1038/nmeth.2604.
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. Isme J. 2012;6:610–618. doi:10.1038/nmeth.2604.
- Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10:1200–1202. doi:10.1038/nmeth.2658.
- Oksanen AJ, Blanchet FG, Kindt R, Legendre P, Minchin PR, Hara RBO, Simpson GL, Solymos P, Stevens MHH, Wagner H. Vegan: community ecology package. R packag. 2015.
- Scrucca L, Santucci A, Aversa F. Competing risk analysis using R: an easy guide for clinicians. Bone Marrow Transplant. 2007;40:381–387. doi:10.1038/sj.bmt.1705727.
- Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat. 1988;16:1141–1154. doi:10.1214/aos/1176350951.