?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Loss of response to therapy in inflammatory bowel disease (IBD) has led to a surge in research focusing on precision medicine. Three systematic reviews have been published investigating the associations between gut microbiota and disease activity or IBD therapy. We performed a systematic review to investigate the microbiome predictors of response to advanced therapy in IBD. Unlike previous studies, our review focused on predictors of response to therapy; so the included studies assessed microbiome predictors before the proposed time of response or remission. We also provide an update of the available data on mycobiomes and viromes. We highlight key themes in the literature that may serve as future biomarkers of treatment response: the abundance of fecal SCFA-producing bacteria and opportunistic bacteria, metabolic pathways related to butyrate synthesis, and non-butyrate metabolomic predictors, including bile acids (BAs), amino acids, and lipids, as well as mycobiome predictors of response.
Introduction
Background
Inflammatory bowel disease (IBD) is a chronic and relapsing inflammatory condition that affects the gastrointestinal (GI) tract. Incidence is increasing and the etiopathogenesis is considered to be a combination of a genetic predisposition, environmental and dietary factors and the complex interaction between the host immune response and the gut microbiome.Citation1 DNA sequencing and multiomics have dramatically advanced research into the host microbiome; alterations in which have been demonstrated in patients with IBD compared with healthy controls (HCs), termed as ‘dysbiosis’. Patients with IBD have 25% fewer taxa compared to healthy controlsCitation2,Citation3 and the functional roles of the bacterial communities in IBD often favor a pro-inflammatory state.Citation4
Typically, patients with IBD have reduced biodiversity with a decrease in Bacillota (previously Firmicutes [e.g. Faecalibacterium prausnitzii and Roseburia hominis]) and an increase in Enterobacteriaceae. Microbiota profiles differ according to IBD subtype, disease phenotype,Citation5 and disease activity.Citation2,Citation5–8 As such, individualized alterations in microbial composition have been associated with future disease flare or prognostic outcomes,Citation9–12 and several studies have demonstrated normalization of fecal microbiota toward that of HCs after treatment (even after adjusting for baseline degree of dysbiosis and antibiotic use).Citation13–15 In the IBD mycobiome, increased Basidiomycota and reduced Ascomycota (particularly Sacchromyces cerevisiae) have been associated with disease activity. Differential ratios of these two phyla have been observed in disease remission, disease activity and HCs, and it has been suggested that this ratio may be a marker of fungal dysbiosis. Increased abundance of Candida albicans has also been observed in IBDCitation16,Citation17 with normalization after infliximab (IFX) treatment to levels observed in HCs.Citation16 The mycobiome has also been correlated to the bacteriome, more so in UC than CD, and also to single nucleotide polymorphisms associated with IBD (e.g. Card9).Citation17 Viruses are more prevalent in the microbiome than bacteria yet even fewer studies have investigated the IBD virome. Although the bacteriome appears to reflect IBD disease activity more accurately than the virome, several viruses have been associated with CD (e.g. Retroviridae). In addition, since certain viruses can modulate bacteriome activity, an understanding of the IBD virome is likely required to appreciate the relationship between microbial networks and their role in IBD pathophysiology and response to therapy.Citation18,Citation19
The loss of response to therapy in IBD has led to a surge in research on precision medicine. Several studies have investigated the association between the microbiome and disease activity, or response to therapy, in the hope that these associations may serve as biomarkers to predict therapeutic response. To date, three systematic reviews have been published investigating associations between the gut microbiota and response to therapy in IBD.Citation20–22 Estevinho et al included 10 studies published between 2014–2018 largely describing the longitudinal changes in the microbiome during treatment.Citation20 Radhakrishnan and colleagues reported associations between the microbiome and IBD therapy and included 19 studies, 25% of which had no baseline microbiome analysis.Citation22 Jagt and colleagues systematically reviewed fecal metabolomics in pediatric IBD and only 3 of the 19 included studies compared changes dependent on response to therapy.Citation21 Ananthakrishnan et al have also published a review article on microbiome biomarkers in IBD in 2020.Citation23 From the available data, individual studies largely investigate the bacteriome with, or without, additional functional analysis. Since metabolomic shifts account for nearly 70% of microbiome variance, functional and metabolomic analyses provide an indirect way of quantifying microbial activity.Citation24 The most commonly reported metabolic changes include a reduction in short-chain fatty acids (SCFAs), including butyrate, which coincides with the reduction in Bacillota (comprising many butyrate producing microbes),Citation2 and changes in bile and amino acid profiles.Citation21 There is scant data with regards to the mycobiome and virome in IBD, and even fewer studies investigating the association with disease activity or response to therapy.
Purpose of the review
None of the described reviews specifically focused on microbiome predictors of response to advanced therapy. Sixteen of the studies included in our review,Citation25–40 largely published in the last two years, were not included in the aforementioned reviews. In addition, this review focuses on the predictors of response to advanced therapy, including studies that have assessed microbiome predictors before the proposed time of response or remission. Therefore, several studies presented in previous reviews that only described associations at the time of response have been excluded. We also provide an update on the available data regarding the mycobiome and virome, which were not included in prior reviews. Therefore, this systematic review summarizes the available data on microbiome-associated predictors of response to advanced therapy for IBD.
Methods
Aims and primary outcome
The primary outcome was the identification of microbiome-associated predictors of response or remission to advanced therapy at any time point. We aimed to provide a qualitative summary of the literature in this area and identify the study limitations and areas for further research.
Search strategy
We performed a systematic review of the medical literature from inception to February 2023 using Medline and Embase and searched the OVID platform. We aimed to report the microbiome predictors of response to advanced medical therapy. Search terms using subject headings and keywords included, but were not limited to, the following: ulcerative colitis, Crohn’s disease, inflammatory bowel disease, generic names for advanced IBD therapies or mechanism of action (including thiopurines, methotrexate, and licensed biologic therapies or small molecules), dietary interventions, synonyms relating to the gut microbiome and treatment outcome, response, or remission. The full details of the search string are provided in Supplementary Information 1. Hand searching of the reference lists was also performed to obtain additional studies.
Inclusion/exclusion criteria
We included studies investigating i) patients with IBD of any age, diagnosed by conventional means, ii) with microbiome analysis (performed by any method) at baseline or before the time point for prediction, iii) related to IBD advanced therapy, iv) with clear definitions of therapeutic response, and v) comparison of responders and non-responders. We excluded abstracts, articles unavailable in English, studies investigating non-established medical therapy or ileal pouch anal anastomoses, and animal studies.
Study selection and data extraction
Study selection was performed in two stages: title and abstract screening, and full-text review. Any discrepancies encountered during the search were resolved by the senior author (GL). Advanced therapy was defined as any of the following licensed therapies for IBD: thiopurines, methotrexate, anti-TNF therapy, anti-integrin therapy, ustekinumab, risankizumab, JAK-inhibitors, and sphingosine-1-phosphate receptor modulators. We did not include studies investigating the use of antibiotics, 5-aminosalicylic-acid (5ASA) therapy or corticosteroids in our search. 5ASA therapy is low-risk, and thus the decision to use this first line when indicated is unlikely to be altered by predictive models since if treatment is successful, immunosuppression is avoided. Antibiotics and corticosteroids are used in specific circumstances either prior to or alongside advanced therapies. Therefore, predictive markers of response have the most clinical use for advanced therapies.
Data were extracted from the selected manuscripts by the primary author (SM) using a predefined data-capture form (Supplementary information 2).
Quality assessment
Two authors (JLCK, CM) independently assessed the methodological quality of the studies using the National Institute of Health study quality assessment tool (Supplementary information 3).Citation41 The included studies were largely case series or single-arm cohort studies and therefore tools to assess for risk of bias in cohort studies with a comparator or control arm were not used. Study quality was assessed subjectively, as per the recommended guidance, based on the outcomes to 12 pre-defined questions and the overall impression after critical appraisal. If there were significant disagreement in the scoring, the authors discussed the results and reached a consensus with involvement of the primary author (SM).
Results
Summary of included studies
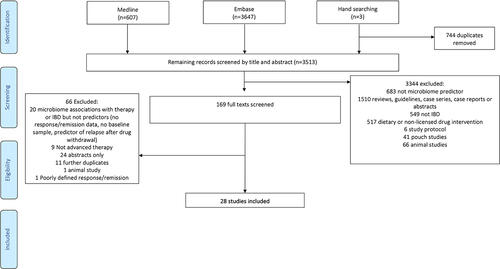
The PRISMA flow diagram displaying the study selection process is shown in . After removal of duplicates, the Medline, Embase, and manual search of references identified 3513 articles. After screening and full-text review, 28 studies were included in the qualitative analysis.
A summary of the included studies is presented in (with more details in Table S1). Table S2 outlines the reasons for the exclusion of certain studies that are relevant to the field. The 28 studies included one randomized controlled trial, two multicenter prospective observational cohorts, one multicenter prospective observational cohort combined with a retrospective cohort, 22 single-center prospective observational cohorts, and two retrospective observational studies.
Table 1. Summary of included studies.
The studies included a total of 3447 patients; 2658 (77%) were IBD patients (52% CD), and the rest were control subjects. Of the 28 studies, sixteen analyzed IBD phenotypes (CD, UC and IBD-unclassified) together as one cohort,Citation5,Citation14,Citation25,Citation27,Citation32–39,Citation42,Citation44,Citation45,Citation48 and tenCitation13,Citation15,Citation26,Citation28,Citation29,Citation31,Citation40,Citation43,Citation47,Citation49 and twoCitation30,Citation46 independently reported CD and UC, respectively. There were 20 adultCitation5,Citation14,Citation25,Citation26,Citation28,Citation29,Citation31,Citation33–40,Citation42–44,Citation46,Citation47 and 8 pediatricCitation13,Citation15,Citation27,Citation30,Citation32,Citation45,Citation48,Citation49 studies. Many studies performed a microbiome sub-analysis. Therefore, the data specific to microbiome predictors of response to therapy included 1232 cases who had baseline microbial analysis as well as data on therapeutic response, accounting for 46% of the study population.
Several studies investigated more than one advanced therapy, with the specific therapies investigated including anti-TNF (23 studies),Citation5,Citation13–15,Citation25,Citation26,Citation28–30,Citation32–40,Citation44–47,Citation49 vedolizumab (3 studies),Citation27,Citation33,Citation42ustekinumab (2 studies),Citation33,Citation43 azathioprine (1 study)Citation44 and non-specified advanced therapy (biologic/immunomodulator) in 2 studies.Citation31,Citation48
Four studies did not report specific exclusion criteriaCitation14,Citation42,Citation45,Citation46 whilst the remaining studies excluded various patient groups (summarized in Table S1). Patients did not receive antibiotics prior to inclusion in 13 studiesCitation5,Citation15,Citation25,Citation26,Citation29,Citation31,Citation36,Citation37,Citation40,Citation44,Citation47,Citation49,Citation50 and in 12 studies antibiotic use was not an exclusion to enrollment.Citation27,Citation28,Citation30,Citation32–35,Citation38,Citation39,Citation43,Citation46,Citation48 In addition to antibiotics, six studies also excluded probiotics (plus prebiotics in one study)Citation49 if they were received between one and three months prior to enrollment.Citation5,Citation29,Citation36,Citation40,Citation47,Citation49
Microbiome analysis
Various techniques were used to evaluate the microbiome (Table S3). The bacteriome was evaluated in all included studiesCitation5,Citation13–15,Citation25–40,Citation42–49 and the mycobiome in two studies (via internal transcribed spacer [ITS] amplicon sequencing).Citation38,Citation51 Fecal metagenomic sequencing was performed via 16S ribosomal ribonucleic acid (rRNA)Citation5,Citation14,Citation15,Citation29–32,Citation34–36,Citation38–40,Citation43,Citation44,Citation47–49,Citation52 or whole genome shotgun metagenomic sequencingCitation13,Citation27,Citation28,Citation33,Citation42 while other studies used quantitative PCR to analyze the fecal microbiota.Citation25,Citation46 Additionally, four studies utilized rectalCitation29,Citation30 or colonic biopsies.Citation35,Citation39 Amongst studies performing microbial analysis, all assessed relative abundance with one study evaluating absolute abundance.Citation32 Salivary amplicon sequencing in addition to 16S rRNA abundance in extracellular vesicles in fecal, saliva, urine and serum samples were performed in one study.Citation36 Other studies assessed fecal relative abundance using fluorescent signal strength (FSS – a pre-determined primer based methodology)Citation37 or HITChip phylogenetic microarray.Citation45 Eight studies performed functional microbial analysesCitation14,Citation26,Citation27,Citation29,Citation33,Citation40,Citation42,Citation44 and four studies investigated the metabolome.Citation14,Citation28,Citation33,Citation49
Assessment of study quality
Overall the studies were of fair quality and were individually rated as good (n = 2), fair (n = 25) or poor (n = 1). (Supplementary information 3). Lower ratings were largely due to small sample sizes, the absence of blinding and a lack of serial outcome measurements.
Synthesis of results
Table S4 summarizes the microbiota and metabolomic predictors of response to therapy.
Microbial diversity
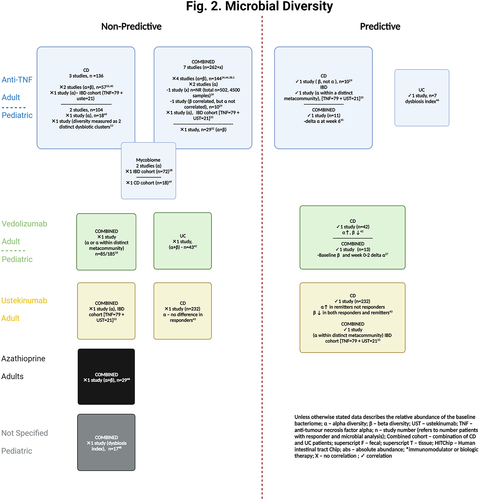
Several studies have investigated whether alpha or beta diversity at baseline predicts response to therapy ( and Table S5). Alpha diversity relates to variance or biodiversity within a particular sample and is often referred to as ‘evenness’ or ‘richness’. Beta diversity refers to similarities or dissimilarities in microbial communities between samples. Although fecal microbiota diversity has been shown to differ between patients with IBD and HCsCitation14,Citation15,Citation28,Citation29,Citation36,Citation39,Citation45,Citation53 and longitudinal changes in diversity are frequently seen after therapeutic intervention, toward eubiosis,Citation5,Citation13–15,Citation26,Citation36,Citation38,Citation40,Citation53 this is not always the case.Citation37,Citation44,Citation54,Citation55
Diversity in the fecal microbiota at baseline did not predict response to therapy in 12 studies.Citation5,Citation13,Citation14,Citation26,Citation32,Citation34,Citation38–40,Citation44,Citation48,Citation49 This included one study in 19 newly diagnosed pediatric patients with IBD where remission at 1 year was endoscopically defined. The degree of dysbiosis improved over time but did not match that of HCs at final follow-up.Citation48 The association between diversity and treatment response was not reported in nine studies.Citation15,Citation25,Citation28,Citation30,Citation31,Citation35–37,Citation47 Only seven studies (mostly with small cohort sizes) observed an association, with all studies assessing the fecal microbiome. Analysis of fecal microbiota in two studies demonstrated increased alpha and reduced beta diversity at baseline. These observations predicted week-14 response to vedolizumab therapy in CD (but not UC) in 85 patients from the PRISM (Prospective Registry of IBD Study at Massachusetts General Hospital)Citation42 registry (analyzed with shotgun metagenomic sequencing) and 6-week remission after ustekinumab induction in a sub-study of 232 CD patients enrolled in CERTFI (a randomized-controlled trial investigating the efficacy of ustekinumab in moderate to severe CD)[analyzed by qPCR].Citation43 A further analysis of the PRISM registry demonstrating differential response to biologic therapy based on microbial richness within metacommunites at baseline is discussed in detail below.Citation33 Conversely to the findings in UC patients above, Magnusson et al demonstrated that the baseline microbial diversity, evaluated with the dysbiosis index, did predict therapeutic response to anti-TNF at week-6. This study only analyzed seven baseline fecal samples.Citation46 Examination of 16S rRNA from rectal tissue of 10 patients with CD prior to infliximab (IFX) induction, demonstrated differential beta diversity in responders and non-responders in both pre- and post-treatment samples.Citation29 In a pediatric cohort of 68 IBD patients receiving anti-TNF therapy, Kolho et al did not directly compare responders with non-responders but demonstrated that the shift toward the eubiosis of HCs at 6 weeks (evaluated with HITChip) was predictive of biochemical remission (fecal calprotectin [fcal] <200mcg/g) 3 months later.Citation45 Lastly, Colman et al demonstrated that the delta in alpha diversity between week 0–2 and the baseline beta diversity (as measured with shotgun metagenomic sequencing) predicted week-14 vedolizumab trough level. The latter was associated with corticosteroid-free remission (CSFR) at week-14. The value of this indirect association is unclear given the limited data on the benefit of vedolizumab therapeutic drug monitoring in improving long-term outcomes.Citation56 Park et al published the first study investigating next-generation sequencing analysis of saliva and nanoparticle analysis of extracellular vesicles in feces, urine, saliva, and serum. No differences in diversity were observed when compared to HCs. The correlation between baseline diversity and therapeutic response was not assessed.Citation36
When comparing studies based on allowance of antibiotic use at enrollment, a similar proportion of studies in each group found no association between diversity and response to therapy (6/13 and 5/12 respectively). Compared to the 18 studies that did not specifically exclude pro- or pre-biotics a similar proportion of studies found no association between diversity and response to therapy (3/6 and 8/18, respectively). Clearly, it is hard to draw conclusions from this given the heterogeneity of the individual studies, interplay of other confounding factors and lack of documentation as to whether antibiotics or probiotics were actually received in patients involved in studies where they were not an exclusion to study entry.
Given the inconsistencies in the literature and the small number of positive studies, baseline diversity does not appear to have a strong correlation with future therapeutic response. No clear trends were observed with regards to the type of diversity analysis performed and this may be at least in part due to the multiple alpha and beta diversity metrices used, reducing the ability to directly compare results between studies. Two larger studies have identified microbial networks that are associated with response to therapy.Citation33,Citation39 One of the microbial networks identified (Group 1) by Lee et al, characterized by a more diverse community profile and higher abundances of the Bacillota phyla (including Faecalibacterium, Eubacterium, Ruminococcus and Roseburia), was associated with higher rates of clinical remission.Citation33 Similarily, Yilmaz et al demonstrated that a reduction in the members of cluster CDA was associated with worse outcomes and poorer response to therapy.Citation39 Although similar taxa were observed between cluster CDA and Group 1 (Faecalibacterium, Ruminococcus, Blautia and Roseburia), microbial diversity was not observed to be higher in cluster CDA. Therefore, microbial interactions and the functional capacity of specific microbial networks rather than microbial diversity alone may be more important factors to focus on in future research.
Bacteria composition and functional capacity
Several studies have investigated the differential abundance of microbial taxa at baseline and evaluated whether they can predict response to therapy (Table S6). Where taxonomic changes do not predict response, some studies have performed functional and/or metabolomic analysis since the measurement of fecal metabolites as microbial functional properties are reported to be more consistent between individuals than the abundance of certain taxa.Citation14 These additional analyses also attempt to infer causation rather than association, particularly where all identified correlations are found to be significant (i.e. taxonomic, functional, and metabolomic analyses).
Opportunistic organisms
In addition to the increase in Clostridia/SCFA-producing bacteria observed with therapy, several studies have observed trends in the abundance of opportunistic organisms (those with the potential to cause infection in the presence of immune dysregulation). Increased levels of Fusobacterium and Escherischia-Shigella have been noted in CD when compared to HCs in both fecalCitation15,Citation29,Citation49 and salivary samples.Citation36 Reduced relative abundance of Enterobacter, Fusobacterium and Escherichia-Shigella have been observed during IFX therapy compared with baseline.Citation40 Fusobacterium have been associated with increased rates of endoscopic disease activity.Citation31 Specific to anti-TNF therapy, Fusobacterium has been observed to fall in overall abundance after treatment,Citation26 and more so in responders than non-responders.Citation29 Small studies in patients with CD treated with adalimumab have observed increased abundance of Escherichia-Shigella in non-responders at 12 weeksCitation26 and reduced abundance in responders (compared to stable abundance in non-responders) at 6 months.Citation47 Effenberger et al noted an increase in abundance of Klebsiella in patients failing to respond to azathioprine.Citation44 All of these studies observed changes at the time of the reported response with none observing baseline changes that could serve as potential predictors. Lower baseline abundance of Escherichia-Shigella was noted in a subset of patients achieving 6-week remission in the CERTIFI trial.Citation43 An increase in baseline absolute abundance of Actinomyces, another potentially pathogenic organism, has also been reported in a small study of IBD patients (n = 29) who did not respond to IFX.Citation32 In other small studies, the increased differential abundance of Enterobacter and Fusobacterium in non-responders to advanced therapy did not reach significanceCitation48 or conversely, an increase in Eshcerichia-Shigella in responders was observed after treatment (without predicting response to therapy).Citation29
The mycobiome and virome.
The potential of the mycobiome to predict the response to therapy has been explored in two studies. Ventin-Holmberg et al evaluated mycobiome predictors of response to IFX at 12 weeks in 68 patients with IBD. Baseline fungal diversity did not predict the therapeutic response, but an increase in Candida at baseline was observed in non-responders. In addition, the relationship between selected genera in the bacteriome and mycobiome correlated with response.Citation38 In a smaller study including 18 patients who received IFX, disease severity correlated with certain fungal genera, but significant differences in abundance were only demonstrated after therapy.Citation49 Neither of these studies replicated the findings from Sokol et al with regards to an increased Basidiomycota:Ascomycota ratio, all three of which analyzed the mycobiome via amplicon sequencing.
Only one study evaluated the virome, in addition to the bacteriome, and found no difference between IBD vs HCs.Citation13 No studies were identified that have evaluated virome predictors of response to therapy.
SCFA-producing organisms and butyrate synthesis pathways
In a prospective observational study,Citation14 16S rRNA sequencing and fecal metabolomic profiles were examined in a discovery cohort (encompassing 12 IBD patients receiving anti-TNF therapy). Fourteen indicator phylotypes differentiated HCs from active IBD patients at baseline. Coprococcus and Roseburia inulinivorans (both SCFA producers)Citation57 were the top baseline indicators, progressing from reduced abundance at baseline to no significant difference from HCs at week 30. These taxa were not associated with the response to therapy. However, in silico metabolomic analysis (the use of computational modeling to identify the predicted alterations in metabolomic pathways based on the measured abundance of bacterial species) demonstrated significant differences between IBD patients (and their therapeutic response) and HCs. In particular, a baseline reduction in metabolic interchange (a marker of metabolic exchange between organisms) was observed in anti-TNF non-remitters and remained significant after treatment. These findings were replicated in a validation cohort in which metabolomic analysis was performed on prospectively collected baseline fecal samples (23 treatment-naïve patients receiving either anti-TNF [n = 10] or vedolizumab therapy [n = 13]). Importantly, the findings persisted even after adjusting for baseline disease activity with both clinical and objective inflammatory markers. The sample size was too small to assess whether the observations were a class effect or also occurred with vedolizumab treatment. In silico meta-analysis of microbial metabolites from both the discovery and validation cohorts identified 10 baseline metabolites associated with subsequent non-remission after anti-TNF therapy when compared to HCs, seven of which were specifically reduced in non-remitters (but not remitters) at baseline (acetaldehyde, L-Arginine, Butyrate, L-lactate, ammonium, ornithine, and carbon dioxide). There was also an 81% reduction in butyrate synthesis at baseline in anti-TNF non-remitters versus remitters. To further test the hypothesis generated from the in silico modeling, they screened 50 fecal metabolites in nine patients prior to the initiation of anti-TNF therapy. The specific findings are outlined in Table S6 but highlight an increase in baseline fecal butyrate in 14-week remitters.Citation14 The 12 patients with IBD (4 UC, 8 CD) in the discovery cohort were treated with etanercept and 22% remitters and 33% non-remitters were maintained on 20 mg prednisolone. The IBD validation cohort received licensed medications (IFX or vedolizumab), but 26% of the remitters and 50% of the non-remitters continued with varying doses of steroids at 14-weeks. Since etanercept has not been shown to be effective in CD clinical trials,Citation58 the discovery cohort results may represent a response to steroids rather than to anti-TNF therapy. The validity of extrapolating data from an unlicensed drug used in the discovery cohort to a licensed drug in the validation cohort, albeit of the same therapeutic class, is uncertain. Effenberger et al also employed in silico metabolomic modelling in 65 patients with IBD and similarly identified a higher abundance of butyrate production at baseline (1.7-fold) in remitters than in non-remitters at weeks 12 and 30 (defined by CDAI < 150, normal C-reactive protein [CRP], and fcal < 150mcg/g). This was significant in the subgroup of patients with CD treated with azathioprine and not with anti-TNF therapy. No adjustment was made for baseline disease severity and therefore it is possible that increased baseline butyrate represents selection bias where certain patients with potentially milder disease activity were selected to initiate azathioprine rather than anti-TNF therapy.Citation44
The IBD team at Massachusetts General Hospital has developed two prospective databases (IBD [PRSIM] and endoscopy [GI disease and endoscopy registry]) with the aim of advancing translational research in IBD with a particular focus on genetic and microbial alterations at diagnosis and also for prediction of relapse or response to therapy.Citation59 Two important studies have been published based on analysis of the PRISM registry providing insights into how the microbiome may predict response to therapy. The first studyCitation42 evaluated data from 85 patients undergoing vedolizumab therapy and demonstrated the significance of the abundance of Roseburia inulinivorans, and also of Burkholderiales, which were increased at baseline in week-14 remitters (compared to non-remitters). Paired samples were available for 41 patients at both baseline and follow-up (10/24 of CD in remission and 11/17 of UC in remission). Longitudinally in CD, there was a significant reduction in Roseburia inulinovorans in remitters and, conversely, an increase in non-remitters. Differential abundance of several taxa were also observed after treatment, depending on the response to therapy, with associated changes in several functional pathways linked to oxidative stress, colonic inflammation, and/or host immunity (Table S6). In the few patients who had samples at all time points (baseline, 14, 30 and 54 weeks; 5 CD, 8 UC), week-14 remitters showed persistent microbiome changes at week 30 and week 54 suggesting an early microbiome ‘response’ to therapy may be associated with a durable outcome.Citation42 The findings were validated in an anti-TNF cohort of 20 patients suggesting these findings were not specific to a particular therapy. The reduction in abundance of Roseburia inulinovorans longitudinally is at odds with the hypothesis that SCFA-producing bacteria may be beneficial. The authors postulate that this may be related to the ability of some strains of Roseburia to produce pro-inflammatory proteins that stimulate IL-8. This further serves to highlight the complexities in microbiome research. This same group went on to evaluate a larger cohort from the same database. 185 IBD patients underwent fecal and serum sampling prior to induction with anti-TNF, ustekinumab or vedolizumab therapy.Citation33 Using microbiota profile modeling, two distinct bacterial metacommunities were identified and predicted response to therapy. Group 1 was more diverse and included F.prausnitzii and Ruminococcus bromii whereas group 2 was dominated by Bacteroides ovatus, Bacteroides thetaiotaomicron and Veillonella parvula. Increased microbial richness at baseline (group 1) predicted clinical remission at 1-year in patients receiving ustekinumab or anti-TNF therapy, while the same was not true for patients receiving vedolizumab. Significantly higher rates of clinical (67% vs. 36%) but not endoscopic (65% vs. 36%) remission were observed at week-52 in group 1 (compared to group 2). Group 1 patients were more likely to respond to ustekinumab/anti-TNF therapy than to vedolizumab therapy, independent of baseline disease severity or IBD subtype. Several microbial species at baseline (many of which were SCFA-producers) were associated with clinical remission at week-14 with weaker or non-significant associations noted at week-52 (Table S6). The microbial diversity of Group 1 (compared to Group 2) was negatively associated with cytokines linked to inflammatory cascades and treatment resistance, such as IL10RB and IL12RB1. When comparing anti-cytokine to anti-integrin therapy, differential serum proteins (including caspase 8 and IFNLR1) were associated with remission.Citation33
The largest study to date was a retrospective analysis of 502 patients with IBD (composed of two independent cohorts). The authors identified consistent clusters of organisms associated with poor treatment response. The favorable cluster labeled CDA included several SFCA-producing Clostridia (Table S6). Taxa specifically associated with response to anti-TNF therapy included Bifidobacterium, Collinsella, Lachnospira, Lachnospiraceae, Roseburia, and Eggerthella. Phascolarctobacterium was correlated with non-response. These findings were replicated in both cohorts. Differential abundance of taxa with regard to response to corticosteroids was also observed in both cohorts but could not be replicated in the opposite cohort. Among the 83 taxa analyzed, several were associated with disease activity (rather than prognosis): Enterobacteriaceae and Klebsiella were associated with active disease in CD, and Prevotella and Ruminococcus in UC. There was no association between CDA clusters and clinical disease scores or fecal calprotectin levels. This may suggest that these parameters were poorly correlated with clinical disease activity in this study or that cluster CDA could serve as a biomarker for protective physiology against disease relapse but not for disease activity.Citation39
In addition to the aforementioned large landmark studies, several smaller studies have corroborated the association between SCFA-producing bacteria and response to therapy. In small pediatric studies, the presence of various genera of Clostridia in CD or combined CD/UC cohorts were associated with response to anti-TNF therapy, either clinically (pediatric CD activity index [PCDAI] 10 after up to 6 infusions)Citation15,Citation49 or, additionally, biochemically defined (3-fold reduction or normalization of fcal at week-6).Citation45 In this latter study, severe inflammation was characterized by a reduction in butyrate-producing organisms, gram-positive bacteria (particularly Clostridium clusters IV and XIVa) and diversity.Citation45 Wang et al also performed metabolomic analysis demonstrating that reduced levels of SCFA-producing bacteria were associated with low levels of fecal SCFAs (acetic, butyric, and propanol acids).Citation49 Consistent trends have been observed in two adult studies with endoscopic assessment at the final follow-up, thus providing an association with the gold standard for objectively defined remission. A reduction of SCFA-producing bacteria predicted non-response to IFX in 72 patients with IBD (majority UC in this study) who were followed for a year (87% underwent endoscopy at final follow-up).Citation38 The converse was demonstrated in 49 patients with CD, where the incremental abundance of Clostridiales from week 6 (but not baseline abundance) predicted clinical response to IFX at weeks 14 and 30.Citation40 One pediatric cohort of newly diagnosed patients with IBD and endoscopically defined end points demonstrated that the differential abundance of four operational taxonomic units (OTUs) were associated with response to advanced therapy. In non-responders at baseline, Coprococcus, Adlercreutzia and Dialister were reduced and an unnamed genus of Enterobacteriaceae was increased. Of note, none of the UC patients responded to therapy in this study.Citation48
F.prausnitizii, an SCFA-producing bacterium, has been investigated in several studies and deserves specific mention. Analysis of baseline fecal samples from just seven patients with UC demonstrated more pronounced dysbiosis in anti-TNF non-responders. Increased levels of F. prausnitzii (both at baseline and up-trending during induction) were associated with favorable outcomes. The absence of an increase in the abundance F. prausnitzii longitudinally during induction was associated with non-response. Additionally microbial change preceded the fall in fCal levels.Citation46 Whilst F.prausnitzii could be a potential early biomarker for response, this finding has not been consistently replicated in other larger studies.Citation14,Citation25 The correlation has, however, been demonstrated in a large non-anti-TNF cohort. In a subgroup analysis of the CERTIFI trial investigating the efficacy of ustekinumab in the maintenance of moderate to severe CD, increased relative abundance of Faecalibacterium at baseline was observed in 6-week remitters and was also present at induction in every patient who entered remission at 6 weeks.Citation43 In the aforementioned study from the PRISM registry, F.prausnitizii belonged to the metacommunity correlated with favorable response to both anti-TNF and ustekinumab.Citation33 Colman and colleagues correlated the presence of Faecalibacterium at week-2, along with several other SCFA producing bacteria (Table S6), with week-14 vedolizumab trough levels, which were in turn associated with week-14 CSFR. In silico functional analysis in this study demonstrated the enrichment of two predominant butyrate biosynthesis pathways, which were also independently correlated with week-14 vedolizumab trough levels. They postulated that reduced levels of SCFA-producing bacteria, which are known to be associated with mucosal barrier function, may lead to increased GI drug loss. Notably, disease activity (raised erythrocyte sedimentation rate [ESR], low albumin) was also associated with drug clearance.Citation27
These studies collectively investigated the connection between SCFA-producing bacteria and/or butyrate-producing pathways and their relationship with treatment response in patients with IBD. The majority of these studies indicated that reduced metabolic exchange between organisms,Citation14 lower fecal butyrate concentrationsCitation14,Citation44 and decreased relative abundance of SCFA- and butyrate-producing taxaCitation14,Citation15,Citation33,Citation38,Citation39,Citation42,Citation45,Citation49 at baseline may be predictive of non-response to advance therapies. However, inconsistency exist among the studies, particularly in the context of F.prausnitzii, and do not provide definitive evidence to support the use of these microbial factors as biomarkers of response. Additionally, most studies primarily focus on differences in specific taxa rather than assessing the levels of specific metabolites such as butyrate. Metabolic redundancy within the gut microbiota makes it challenging to infer that increases in SCFA- or buyrate-producing taxa directly correspond with increased levels of these metabolites without direct measurement.Citation60 Future studies aiming to gain a deeper understanding of the gut microbiome’s influence on therapeutic response should extend their focus beyond taxa and consider specific metabolic pathways and the production of specific metabolites, as these microbial factors are likely to have a more direct impact on therapeutic outcomes.
Additional metabolomic or functional analyses
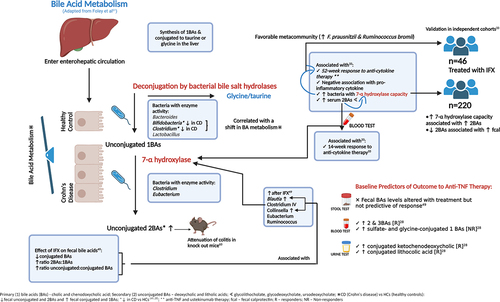
Functional and metabolomic analyses are reported to be more consistent between individuals than the differential abundance of taxa and therefore may provide a better insight into the pathogenesis of disease.Citation14 In the aforementioned analysis of the PRISM registry, Lee et al performed additional functional microbiome profiling to understand the changes associated with remission. They highlighted the potential role of microbial enzymes such as glycoside hydrolases and enzymes associated with secondary bile acid (BA) synthesis. Several baseline serum secondary BAs were associated with week-14 remission and glycerophosphoethanolamines and diacylglycerols were associated with non-remission. The abundance of secondary BAs was associated with the microbiota with known 7a/B-dihydroxylation capacity (largely mediated by Clostridia). The abundance of these microbes was higher in the metacommunity which was associated with the response to ustekinumab/anti-TNF therapy. The association between response to therapy and the prevalence of microbiota with 7a/B-dehydroxylation capacity was further validated in 46 patients receiving IFX in two combined historic cohorts. Another independent cohort of 220 patients was used to corroborate the metabolomic link between raised secondary BAs, 7a/B-deyhdroxylation capacity and disease activity. Paired fecal metagenomic and metabolomic data demonstrated that patients with 7a/B-deyhdroxylation capacity had higher fecal secondary BAs. Conversely, patients with active disease had lower fecal secondary BAs, suggesting that they may be protective against inflammation.Citation33
In addition to measuring fecal butyrate metabolites, Wang et al also demonstrated that CD patients at baseline compared to HCs had reduced fecal levels of unconjugated and secondary BAs and increased levels of conjugated and primary BAs (but similar overall BA levels). These findings were associated with a reduction in the abundance of Bifidobacterium and Clostridium clusters IV and XI; bacteria which are associated with bile salt hydrolase capacity, which may lead to a reduction in deconjugation. Although IFX treatment was associated with an increase in bacteria with 7a-hydroxylase activity which was associated with an increased ratio of unconjugated to conjugated BAs, fecal BAs were not predictive of response to therapy in this smaller study. The relationship between bile salt metabolism and microbiome predictors of response to therapy is summarized in .Citation28,Citation33,Citation49,Citation61–63 Differential changes according to treatment response were also noted in the amino acids of the fecal metabolome (Table S6).Citation49
Ding et al investigated the taxonomic and metabolomic predictors of the response to anti-TNF therapy in 86 patients with IBD. Despite no significant taxonomic changes, fecal, serum, and urinary BAs differed between responders versus non-responders, with different microbial signatures demonstrated in each sample type ( & Table S6). Increased serum secondary and tertiary BAs were similarly observed in anti-TNF responders, whereas higher levels of sulfate- and glycine-conjugated primary BAs were noted in non-responders. Including the three most predictive BAs in a composite model provided an AUC of 0.81 (±0.17) for predicting anti-TNF response. Fecal and serum histidine, urinary cysteine, and differential levels of lipid markers also predicted response. In responders, serum phosphatidylcholine, ceramide, sphingomyelin, and triglyceride levels were reduced, whereas fecal phosphocholines and triglycerides were increased. Fecal lipid profiling provided an AUC of 0.94 (±0.10) for response to therapy.Citation28
Table 2. Predictive models.
Studies showing no association between the gut microbiome and response to therapy
Several studies have not observed that microbial abundance is capable of predicting the response to therapy. These findings may be true, or due to several factors: the small sample size,Citation26,Citation47 how the response was defined, cohort heterogeneity or the methodology. Busquets et al included cessation due to adverse event in their definition of response and found no individual taxa associated with response to therapy.Citation25 Vatn et al performed a multicentre study in Europe recruiting newly diagnosed patients with IBD to identify fecal microbiota signatures associated with IBD and their phenotypes. The secondary outcome was the identification of signatures associated with disease course and response to therapy. A total of 158 patients had information on their medical trajectory, with patients being followed for at least 12 months, 41 of whom required treatment escalation for disease flare (biologic, cyclosporine, or surgery). Disease severity at baseline according to CRP or fcal was associated with the need for escalation, but no microbial predictors were identified. This was the only study to evaluate bacterial abundance based on the FSS of 54 pre-determined bacterial DNA markers. FSS was available in 24/29 patients who escalated to anti-TNF therapy, eight of whom achieved clinical and biochemical remission. No differences were seen in this subgroup.Citation37 Several of the negative studies, in terms of taxa predicting response to therapy, combined IBD subtypesCitation14,Citation28,Citation37,Citation44 or had a more stringent definition of clinical response to therapy requiring either at least one objective marker of inflammation or corticosteroid free remission.Citation26,Citation28,Citation44,Citation47 In one study, all of the non-responders (7/20) were in clinical remission but failed to meet the criteria for response due to ongoing corticosteroid therapy.Citation47 While this study is likely underpowered, the inflammatory burden in each group was not compared objectively and differential abundance of bacteria has been observed with corticosteroid exposure.Citation32,Citation43 In addition, samples were taken at baseline and 6 months.Citation47 Larger studies performing sequencing more frequently have demonstrated a predictive value at 14- but not 52-weeks with regard to specific taxa.Citation33
Predictive models
The correlation between microbial biomarkers at baseline and subsequent outcomes after treatment are just associations. As previously described, true predictors need to be compared between remitters and non-remitters and performance should be evaluated as predictors (sensitivity, specificity, positive and negative predictive values) rather than just a significant p-value for association.Citation64
In addition to the metabolomic model from Ding et al,Citation28 ten other studies have developed predictive scores for potential microbiome predictors of the response to therapy. These results are outlined in detail in . Four studies demonstrated that microbiome predictors alone could predict the response to therapy, with an AUC of 77% or higher. In three small studies, the relative abundance of varying numbers of genera (often Clostridia) predicted the clinical response to anti-TNF during, or shortly after induction,Citation25,Citation38,Citation45,Citation48 (although in one of these studies, the time point of the predictor is unclear).Citation25 One study included specific fungal genera with differences in abundance between IBD subtypes, thus including Candida in the UC model. This improved the accuracy for predicting 12-week response to IFX to 93% for CD and 82% for UC.Citation38
Five studies combined clinical parameters (three of which incorporated fecal calprotectinCitation5,Citation43,Citation45) with microbial variables and demonstrated that the addition or sole use of microbial variables predicts response to therapy better than using clinical data alone. Doherty et al were able to predict 6-week response to ustekinumab in the CERTIFI sub-study.Citation43 In the PRISM registry a combined microbial model (‘vedoNet’) that included 40 variables (taxa and metabolomic pathways) predicted 14-week clinical response to vedolizumab (AUC 87% and a false discovery rate of less than 25%). This was validated in a prospective cohort of 20 patients treated with anti-TNF and correctly predicted response in 11/13 cases.Citation42 In the extended analysis of the PRISM registry data 21 patients with available clinical, metagenomic, metabolomic and proteomic results were evaluated. Their proposed model predicted response to anti-cytokine therapy with AUC 0.96 (95% CI, 0.88–1.00). Limiting factors include the use of clinical remission rather than objective markers in over 50% patients, the combination of ustekinumab/anti-TNF as ‘cytokine responders’ and only 21/185 patients being included in the formation of their predictive model.Citation33 In 49 patients with CD treated with IFX, the trend in microbial composition (particularly Blautia and Lachnospiraceae) from baseline to 6 weeks predicted clinical remission at weeks 14 and 30 (accuracy 83% [71–96%] and 84% [72–97%], respectively) and endoscopic response at week 30 (accuracy 89% [79–99%]). No differences were observed between responders and non-responders at baseline.Citation40 In a subset of 16 patients with baseline and follow-up microbial profiling, restoration of Clostridiales toward that of HCs was associated with clinical remission, and patients with a higher abundance at baseline responded better to therapy (accuracy of 87%, improving to 94% if combined with clinical parameters).Citation5
Lastly, Hattori et al investigated the association of the microbiome with mucosal healing (MH) and whether the microbiome could predict prognosis. They performed baseline fecal 16S rRNA sequencing in patients undergoing entire small bowel (SB) endoscopic assessment (either by video capsule endoscopy or double balloon enteroscopy) in 38 patients with SB CD receiving immunomodulators or biologic therapy. Patients with active colonic inflammation or perianal disease were excluded from this study. A low relative abundance of Bacteroidetes and higher Fusobacterium was observed in patients with active endoscopic SB disease. Faecalibacterium, Lachnospira, Paraprevotella, Dialister, Streptococcus and Clostridium were reduced in patients with ulceration at baseline. A microbial predictive score (MPS) for MH was developed as a point system, using these six genera. The AUC for predicting MH was 0.80 (sensitivity, 0.64; specificity, 0.92). Multivariate regression analysis demonstrated that MPS was the strongest predictor of MH (OR 37.5 [3.41–411.99]). Patients with a higher abundance of these bacteria were also significantly less likely to relapse over 11–13 months follow-up, with no relapses at the final follow-up in patients scoring 5 (0 vs. 40% relapse rate for MPS
5 and < 5, respectively). Combining MPS < 5 with serum albumin (threshold <41.5 g/L) differentiated patients further into moderate and high risk of relapse at final follow up (20% vs 40%, respectively). These findings were not related to a specific therapy, but demonstrated that lack of eubiosis may serve as a treatment target for early escalation of therapy.Citation31
Discussion
In this systematic review, we summarized the current literature on microbiome predictors of response to advanced therapies for IBD. The included studies encompass articles reporting baseline or interval microbiome analyses that putatively predicted, or were associated, with a future treatment response. We have highlighted key themes in the literature that may serve as future biomarkers of treatment response, namely, the favorable abundance of fecal SCFA-producing bacteria and metabolic pathways related to butyrate synthesis; the abundance of fecal opportunistic bacteria associated with non-remission; non-butyrate metabolomic predictors from various sample types (including BAs, amino acids, and lipids); and mycobiome predictors of response. Several predictive indices (using baseline or delta values) have been described, demonstrating that microbial predictors of the response to therapy are more accurate than clinical or biochemical biomarkers alone. Only one study to date provides a predictive model, with independent validation, that may enable the selection of one biologic therapy over another.Citation33 All other studies present associations that have not been externally validated, and in one studyCitation42 the predictive score was not specific to a particular therapy. It may be that certain microbial milieus at baseline, or the failure to achieve a specific shift in microbial parameters from baseline, could help us to select patients with more refractory disease who may benefit from enhanced monitoring or early treatment escalation. Further data are required to establish whether this practice would lead to better outcomes. The identification of microbial biomarkers that are most predictive of response would enable targeted analysis, and thus the utilization of less expensive techniques, making it more applicable to everyday clinical care.
Data regarding the mechanisms underlying the proposed associations are largely derived from in vitro and animal studies. SCFAs (acetate, propionate, and butyrate) are produced by several bacteria, particularly Bacillota, of the order Clostridia. SCFAs are a carbon source for colonic epithelial cells and play an important role in mucosal barrier function.Citation65,Citation66 Their anti-inflammatory properties include boosting extrathymic production of regulatory T cells in mice,Citation67 reduction in oxidative stress via inhibition of NF-kB,Citation68 inhibition of IFNɣ,Citation69 induction of IL-10 (F. prausnitzii),Citation70 upregulation of antimicrobial peptides (Roseburia)Citation71 and they have also been associated with immune surveillance.Citation66 Bifidobacterium, a SCFA-producing organism associated with response to therapy, has also been linked to mucosal barrier function via promotion and maturation of the colonic epithelium.Citation72 However, supplementation of observed microbiome deficiencies has generally not been helpful. Butyrate therapy has not been shown to be efficacious in IBDCitation73–75 and there is insufficient evidence that restoration of a dysbiotic microbiome with pro-, pre-, or synbiotic therapy is routinely beneficial.Citation76 Bifidobacterium longum isolated from the feces of healthy individuals attenuated DSS-induced colitis in mice and augmented the efficacy of IFX. This was associated with an increase in fecal secondary BAs.Citation77 However, replenishing bacterial strains known to be relatively deficient in patients with quiescent UC in vivo has not been associated with a reduced risk of flare.Citation78
The interaction between BAs, microbiota, and luminal inflammation has been previously reviewed.Citation79 Lee and colleagues have corroborated this data and, in addition, demonstrated a link between microbial diversity, BA synthesis, and a favorable response to anti-cytokine over vedolizumab therapy and validated their hypothesis in independent cohorts. This study is the first to demonstrate an association between microbiota composition, metabolomic markers and a differential response to advanced therapy.Citation33 Less is understood about the putative mechanistic associations between response to therapy and other metabolites discussed in this manuscript.Citation28 Similarly, there is minimal data regarding the mycobiome. A recent RCT randomized consecutive patients with fecal Candida (28% of 242 patients screened) with active mild-to-moderate UC to standard therapy plus placebo (n = 30) or fluconazole (n = 31). The primary endpoint of endoscopic response at 4-weeks was not reached, but secondary endpoints were nominally significant, favoring antifungal therapy (reduction in Robart’s histological score [74% vs. 33%] and fCal [84% vs. 37%]). The presence of fecal Candida was associated with baseline disease severity but the groups were poorly matched, with more patients in the placebo group taking corticosteroids at enrollment.Citation80 Further studies are required to establish whether fungal dysbiosis is relevant to therapeutic response or simply a marker of disease severity.
Limitations
The available data are heterogeneous and have several limitations. Many of the included studies were small (some with opposing resultsCitation26,Citation29) and their findings were not validated in independent cohorts. Additionally, the approach to microbiome analysis varies. The majority evaluated the relative, rather than absolute, microbial abundance. The former is influenced by the total microbial count and therefore may be less sensitive in identifying alterations in specific microbes that may be associated with the outcome assessed. Amplicons, rather than whole genome sequencing, assess the hypervariable region of the 16S rRNA gene, which may be unable to identify closely related species or strains sharing similar genetic makeup that may be relevant to disease pathogenesis or trajectory.Citation23 The majority of studies incorporate combined cohorts of both UC and CD.Citation5,Citation14,Citation25,Citation27,Citation29,Citation32–36,Citation38,Citation42,Citation44,Citation45,Citation48 Large cohort studies with over 500 patients have shown microbiota differences between UC and CD; in particular, reduced diversity in CD with an overall loss of beneficial species such as Faecalibacterium, Oscillospira, Bifidobacterium and Ruminococcus. Citation39 Differential abundances between UC and CD have also been demonstrated in smaller studies in both the bacteriome and the mycobiome,Citation32,Citation38,Citation45 with disease locationsCitation39,Citation81 and/or phenotypeCitation5 likely influencing microbial signatures. The degree to which each of these factors may influence microbial predictive markers is unclear, but stratification of the PRISM dataset by IBD type did not alter the predictive value of VedoNet.Citation42 Other factors such as cohort age (pediatric vs adult)Citation82 as well as sample type (fecal vs mucosal vs saliva)Citation83 are also likely to contribute to heterogeneity in results between studies. Once additional studies focusing on pediatric cohorts and sample types other than feces have been undertaken, microbial biomarkers of response which differ between pediatric and adult cohorts as well as fecal and other sample types may be able to be assessed. However, due to the limited pediatric and non-fecal sample studies at this time these comparisons have not been made in this review.
Definitions of response and remission vary significantly with many studies using clinical parameters alone to define response or remissionCitation14,Citation15,Citation30,Citation42,Citation43,Citation49 which are known to correlate poorly with endoscopic healing.Citation84 Where endoscopic assessments have been used at baseline and follow-up, studies are small,Citation26,Citation29,Citation35 although one study did achieve baseline and follow-up assessments in 49 CD patients with objective scoring assessed and proposed a predictive score for endoscopic healing at week 30.Citation40 In other studies, using endoscopy, there is a lack of clarity regarding the number of patients in whom it was performedCitation25,Citation28,Citation36,Citation37 or objective scores were not employed or defined.Citation35,Citation48 Two studies did present endoscopic outcomes in reasonably sized cohortsCitation33,Citation38 but neither were endoscopically assessed at baseline, without which it is not possible to truly understand the influence of disease severity at baseline, which may confound the results. Other studies used a combination of clinical, biochemical, and/or endoscopic parameters with three, including CSFR, in their definition of response.Citation27,Citation37,Citation47 In an area with so much scientific uncertainty, it seems sensible to define remission with the gold standard of endoscopy performed at baseline and follow-up, although this more invasive method may hinder clinical trial enrollment.
Zhou et al performed a meta-analysis combining their Chinese cohort with data from the RISK and PRISM registries (USA) and showed similarities in IBD microbiome profiling across ethnicities.Citation5 However, geographical differences in microbiota profiles have previously been described.Citation85,Citation86 Potential confounders were not routinely addressed in the included studies (e.g., dietary intake, exercise,Citation87 smoking,Citation88 alcohol consumption, age, BMICitation39). This is of particular relevance because in the 502 IBD patients investigated by Yilmaz et al, BMI and age exerted more significant changes in microbial composition than any disease or treatment-related factor.Citation39 The exclusion of antibiotics prior to enrollment in the majority of studies is perhaps necessary at this stage of our understanding of the microbiome. However, future studies including these groups will be required, since they are commonly used in everyday clinical practice, particularly in CD.
We did not include studies investigating the response to 5-ASA or corticosteroid therapy for the reasons outlined in our methods. Thus, one large multicenter study (PROTECT) investigating predictors of response in newly diagnosed children with UC was excluded.Citation89 It is worth mentioning that Ruminococcaceae and Sutterella were associated with CSFR at week 52 even after adjusting for clinical predictors. In combination with a rectal gene signature (lower antimicrobial peptide gene expression [PC1]), reduced Suterella and increased Ruminococcaceae at baseline were associated with CSFR at 52 weeks. Based on these data, our results should not be evaluated in isolation. Further work is required to differentiate microbial predictors associated with refractoriness to any therapy in patients who may require more aggressive treatment algorithms, from those in whom particular therapies should be avoided due to the low likelihood of response.
We have also not delved deeply into the mechanisms behind the associations described, which is outside the scope of this review. Several of the included studies also correlated the demonstrated microbial predictors with differential proteomic and genomic findings that will be relevant to future research.Citation29,Citation30,Citation33,Citation35 Aside from multiomics, the interaction between microbiome predictors and drug pharmacokinetics deserves further evaluation. Microbial composition has been associated with both IFXCitation90 and vedolizumabCitation27 levels and in the epi-IIRN database, the use of antibiotics (particularly cephalosporins and penicillins) was associated with immunogenicity to IFX. The latter group replicated increased rates of immunogenicity in antibiotic-exposed mice, while germ-free mice (harboring no microorganisms) did not develop anti-drug antibodies.Citation91
Concluding remarks
Research in this area is challenging, with the key question remaining unanswered: Does the inflammatory environment and associated epithelial disruption lead to dysbiosis, or is microbial dysbiosis the instigator of a pro-inflammatory state? HCs have been frequently enrolled in studies to identify the parameters of health and disease. However, using data derived from HCs to create predictive modelsCitation14 is potentially flawed if patients with IBD are unable to reach eubiosis even in the presence of MH.Citation48
Despite the data limitations, we present an updated summary of microbiome predictors of response to advanced therapy in IBD. Future research should focus on collaborative, multiomic analyses with clearly defined homogenous cohorts and definitions of treatment response. New multicentre studies recruiting cohorts in different geographical locations are already underway.Citation92,Citation93 The aim would be to move toward interventional studies where predictive scores alter treatment pathways in the hope that we can improve long-term patient outcomes.
Author contributions
SM, GL, and BB: review conception, SM systematic review, data collation, analysis, and manuscript drafting; JLCK, CM risk of bias review; SM, GK figure content; SM, GL, ES, JLCK, CM, GK and BB: manuscript review and final approval
Supplemental Material
Download MS Word (564.5 KB)Disclosure statement
BB: Advisor/speaker: Ferring, Janssen, Abbvie, Takeda, Pfizer, Novartis, BMS, Merck, Sandoz, Organon, and Lifelabs. Advisor: Alimentiv, Gilead, Iterative Scopes, AMT, Celgene, Merck, Amgen, Pendopharm, Genentech, BMS, Allergan, Protagonist, Fresenius Kabi, Mylan, Viatris, Bausch Health, Celltrion Healthcare, BioJamp Pharma, and Eupraxia. Research support: Janssen, Abbvie, GSK, BMS, Amgen, Genentech, and Merck. Stock Options: Qu Biological. SM: received speaker fees from Falk Pharma. JLCK, CM, GK, GL nil to declare.
Data availability statement
All available data is presented in the submitted work.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2023.2287073.
Additional information
Funding
References
- Zhang YZ, Li YY. Inflammatory bowel disease: pathogenesis. World J Gastroenterol. 2014;20:91–29. doi:10.3748/wjg.v20.i1.91.
- Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655–662. doi:10.1038/s41586-019-1237-9.
- Moustafa A, Li W, Anderson EL, Wong EHM, Dulai PS, Sandborn WJ, Biggs W, Yooseph S, Jones MB, Venter CJ, et al. Genetic risk, dysbiosis, and treatment stratification using host genome and gut microbiome in inflammatory bowel disease. Clin Transl Gastroenterol. 2018;9(1):e132. doi:10.1038/ctg.2017.58.
- Joossens M, Huys G, Cnockaert M, et al. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60:631. doi:10.1136/gut.2010.223263.
- Zhou Y, Xu ZZ, He Y, Yang Y, Liu L, Lin Q, Nie Y, Li M, Zhi F, Liu S, et al. Gut microbiota offers universal biomarkers across ethnicity in inflammatory bowel disease diagnosis and infliximab response prediction. mSystems. 2018;3:3. doi:10.1128/mSystems.00188-17.
- Ankersen DV, Weimers P, Marker D, Johannesen T, Iversen S, Lilje B, Kristoffersen AB, Saboori S, Paridaens K, Skytt Andersen P, et al. eHealth: disease activity measures are related to the faecal gut microbiota in adult patients with ulcerative colitis. Scand J Gastroenterol. 2020;55(11):1291–1300. doi:10.1080/00365521.2020.1829031.
- Klaassen MAY, Imhann F, Collij V, et al. Anti-inflammatory gut microbial pathways are decreased during Crohn’s disease exacerbations. J Crohn’s Colitis. 2019;13:1439–1449. doi:10.1093/ecco-jcc/jjz077.
- Taylor H, Serrano-Contreras JI, McDonald JAK, Epstein J, Fell JM, Seoane RC, Li JV, Marchesi JR, Hart AL. Multiomic features associated with mucosal healing and inflammation in paediatric Crohn’s disease. Aliment Pharmacol Ther. 2020;52:1491–1502. doi:10.1111/apt.16086.
- Andoh A, Kuzuoka H, Tsujikawa T, Nakamura S, Hirai F, Suzuki Y, Matsui T, Fujiyama Y, Matsumoto T. Multicenter analysis of fecal microbiota profiles in Japanese patients with Crohn’s disease. J Gastroenterol. 2012;47:1298–1307. doi:10.1007/s00535-012-0605-0.
- Braun T, Di Segni A, BenShoshan M, Neuman S, Levhar N, Bubis M, Picard O, Sosnovski K, Efroni G, Farage Barhom S, et al. Individualized dynamics in the gut microbiota precede Crohn’s disease flares. Am J Gastroenterol. 2019;114(7):1142–1151. doi:10.14309/ajg.0000000000000136.
- Kugathasan S, Denson LA, Walters TD, Kim M-O, Marigorta UM, Schirmer M, Mondal K, Liu C, Griffiths A, Noe JD, et al. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet. 2017;389(10080):1710–1718. doi:10.1016/S0140-6736(17)30317-3.
- Park SK, Kim HN, Choi CH, Im JP, Cha JM, Eun CS, Kim T-O, Kang S-B, Bang KB, Kim HG, et al. Differentially abundant bacterial taxa associated with prognostic variables of Crohn’s disease: results from the IMPACT study. J Clin Med. 2020;9(6):1748. doi:10.3390/jcm9061748.
- Lewis JD, Chen EZ, Baldassano RN, Otley A, Griffiths A, Lee D, Bittinger K, Bailey A, Friedman E, Hoffmann C, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host & Microbe. 2015;18(4):489–500. doi:10.1016/j.chom.2015.09.008.
- Aden K, Rehman A, Waschina S, Pan W-H, Walker A, Lucio M, Nunez AM, Bharti R, Zimmerman J, Bethge J, et al. Metabolic functions of gut microbes associate with efficacy of tumor necrosis factor antagonists in patients with inflammatory bowel diseases. Gastroenterology. 2019;157(5):1279–92.e11. doi:10.1053/j.gastro.2019.07.025.
- Wang Y, Gao X, Ghozlane A, Hu H, Li X, Xiao Y, Li D, Yu G, Zhang T. Characteristics of Faecal microbiota in paediatric Crohn’s disease and their dynamic changes during infliximab therapy. J Crohns Colitis. 2018;12:337–346. doi:10.1093/ecco-jcc/jjx153.
- Kowalska-Duplaga K, Krawczyk A, Sroka-Oleksiak A, SALAMON D, Wędrychowicz A, FYDEREK K, GOSIEWSKI T. Dependence of colonization of the large intestine by candida on the treatment of Crohn’s disease. Pol J Microbiol. 2019;68:121–126. doi:10.21307/pjm-2019-014.
- Sokol H, Leducq V, Aschard H, Pham H-P, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, et al. Fungal microbiota dysbiosis in IBD. Gut. 2017;66(6):1039–1048. doi:10.1136/gutjnl-2015-310746.
- Pérez-Brocal V, García-López R, Nos P, Beltrán B, Moret I, Moya A. Metagenomic analysis of Crohn’s disease patients identifies changes in the virome and microbiome related to disease status and therapy, and detects potential interactions and biomarkers. Inflamm Bowel Dis. 2015;21:2515–2532. doi:10.1097/MIB.0000000000000549.
- Ungaro F, Massimino L, D’Alessio S, Danese S. The gut virome in inflammatory bowel disease pathogenesis: from metagenomics to novel therapeutic approaches. United European Gastroenterol J. 2019;7:999–1007. doi:10.1177/2050640619876787.
- Estevinho MM, Rocha C, Correia L, Lago P, Ministro P, Portela F, Trindade E, Afonso J, Peyrin-Biroulet L, Magro F, et al. Features of fecal and colon microbiomes associate with responses to biologic therapies for inflammatory bowel diseases: a systematic review. Clin Gastroenterol Hepatol. 2020;18(5):1054–1069. doi:10.1016/j.cgh.2019.08.063.
- Jagt JZ, Verburgt CM, de Vries R, de Boer NKH, Benninga MA, de Jonge WJ, van Limbergen JE, de Meij TGJ. Faecal metabolomics in Paediatric inflammatory bowel disease: a systematic review. J Crohns Colitis. 2022;16:1777–1790. doi:10.1093/ecco-jcc/jjac079.
- Radhakrishnan ST, Alexander JLCK, Mullish BH, Gallagher K, Powell N, Hicks LC, Hart AL, Li J, Marchesi JR, Williams HRT, et al. Systematic review: the association between the gut microbiota and medical therapies in inflammatory bowel disease. Aliment Pharmacol Ther. 2022;55(1):26–48. doi:10.1111/apt.16656.
- Ananthakrishnan AN. Microbiome-based biomarkers for IBD. Inflamm Bowel Dis. 2020;26(10):1463–1469. doi:10.1093/ibd/izaa071.
- Zierer J, Jackson MA, Kastenmüller G, Mangino M, Long T, Telenti A, Mohney RP, Small KS, Bell JT, Steves CJ, et al. The fecal metabolome as a functional readout of the gut microbiome. Nat Genet. 2018;50(6):790–795. doi:10.1038/s41588-018-0135-7.
- Busquets D, Oliver L, Amoedo J, Ramió-Pujol S, Malagón M, Serrano M, Bahí A, Capdevila M, Lluansí A, Torrealba L, et al. RAID prediction: pilot study of fecal microbial signature with capacity to predict response to anti-TNF treatment. Inflamm Bowel Dis. 2021;27:S63–s6. doi:10.1093/ibd/izab273.
- Chen L, Lu Z, Kang D, Li G, Sun M, Liu Z, Wu W, Fang L. Distinct alterations of fecal microbiota refer to the efficacy of adalimumab in Crohn’s disease. Front Pharmacol. 2022;13:913720. doi:10.3389/fphar.2022.913720.
- Colman RJ, Mizuno T, Fukushima K, Haslam DB, Hyams JS, Boyle B, Noe JD, D’Haens GR, Van Limbergen J, Chun K, et al. Real world population pharmacokinetic study in children and young adults with inflammatory bowel disease discovers novel blood and stool microbial predictors of vedolizumab clearance. Aliment Pharmacol Ther. 2022;57:524–539. doi:10.1111/apt.17277.
- Ding NS, McDonald JAK, Perdones-Montero A, Rees DN, Adegbola SO, Misra R, Hendy P, Penez L, Marchesi JR, Holmes E, et al. Metabonomics and the gut microbiome associated with primary response to anti-TNF therapy in Crohn’s disease. J Crohns Colitis. 2020;14(8):1090–1102. doi:10.1093/ecco-jcc/jjaa039.
- Dovrolis N, Michalopoulos G, Theodoropoulos GE, Arvanitidis K, Kolios G, Sechi LA, Eliopoulos AG, Gazouli M. The Interplay between mucosal microbiota composition and host gene-expression is linked with infliximab response in inflammatory bowel diseases. Microorganisms. 2020;8:438. doi:10.3390/microorganisms8030438.
- Haberman Y, Karns R, Dexheimer PJ, Schirmer M, Somekh J, Jurickova I, Braun T, Novak E, Bauman L, Collins MH, et al. Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response. Nat Commun. 2019;10(1):38. doi:10.1038/s41467-018-07841-3.
- Hattori S, Nakamura M, Yamamura T, Maeda K, Sawada T, Mizutani Y, Yamamoto K, Ishikawa T, Furukawa K, Ohno E, et al. The microbiome can predict mucosal healing in small intestine in patients with Crohn’s disease. J Gastroenterol. 2020;55(12):1138–1149. doi:10.1007/s00535-020-01728-1.
- Höyhtyä M, Korpela K, Saqib S, Junkkari S, Nissilä E, Nikkonen A, Dikareva E, Salonen A, de Vos WM, Kolho K-L, et al. Quantitative fecal microbiota profiles relate to therapy response during induction with tumor necrosis factor α antagonist infliximab in pediatric inflammatory bowel disease. Inflamm Bowel Dis. 2022;29:116–124. doi:10.1093/ibd/izac182.
- Lee JWJ, Plichta D, Hogstrom L, Borren NZ, Lau H, Gregory SM, Tan W, Khalili H, Clish C, Vlamakis H, et al. Multi-omics reveal microbial determinants impacting responses to biologic therapies in inflammatory bowel disease. Cell Host & Microbe. 2021;29(8):1294–304.e4. doi:10.1016/j.chom.2021.06.019.
- Ditto MC, Parisi S, Landolfi G, Borrelli R, Realmuto C, Finucci A, Caviglia GP, Ribaldone DG, Astegiano M, Zanetti A, et al. Intestinal microbiota changes induced by TNF-inhibitors in IBD-related spondyloarthritis. RMD Open. 2021;7(3):e001755. doi:10.1136/rmdopen-2021-001755.
- Mavragani CP, Nezos A, Dovrolis N, Andreou N-P, Legaki E, Sechi LA, Bamias G, Gazouli M. Type I and II interferon signatures can predict the response to anti-TNF agents in inflammatory bowel disease patients: involvement of the microbiota. Inflamm Bowel Dis. 2020;26:1543–1553. doi:10.1093/ibd/izaa216.
- Park YE, Moon HS, Yong D, Seo H, Yang J, Shin T-S, Kim Y-K, Kim JR, Lee YN, Kim Y-H, et al. Microbial changes in stool, saliva, serum, and urine before and after anti-TNF-α therapy in patients with inflammatory bowel diseases. Sci Rep. 2022;12(1):6359. doi:10.1038/s41598-022-10450-2.
- Vatn S, Carstens A, Kristoffersen AB, Bergemalm D, Casén C, Moen AEF, Tannaes TM, Lindstrøm J, Detlie TE, Olbjørn C, et al. Faecal microbiota signatures of IBD and their relation to diagnosis, disease phenotype, inflammation, treatment escalation and anti-TNF response in a European multicentre study (IBD-Character). Scand J Gastroenterol. 2020;55(10):1146–1156. doi:10.1080/00365521.2020.1803396.
- Ventin-Holmberg R, Eberl A, Saqib S, et al. Bacterial and fungal profiles as markers of infliximab drug response in inflammatory bowel disease. J Crohn’s Colitis. 2021;15:1019–1031. doi:10.1093/ecco-jcc/jjaa252.
- Yilmaz B, Juillerat P, Øyås O, Ramon C, Bravo FD, Franc Y, Fournier N, Michetti P, Mueller C, Geuking M, et al. Microbial network disturbances in relapsing refractory Crohn’s disease. Nat Med. 2019;25(2):323–336. doi:10.1038/s41591-018-0308-z.
- Zhuang X, Tian Z, Feng R, Li M, Li T, Zhou G, Qiu Y, Chen B, He Y, Chen M, et al. Fecal microbiota alterations associated with clinical and endoscopic response to infliximab therapy in Crohn’s disease. Inflamm Bowel Dis. 2020;26(11):1636–1647. doi:10.1093/ibd/izaa253.
- Quality assessment tool for before-after (pre-post) studies with no control group. https://www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools. July 2021.
- Ananthakrishnan AN, Luo C, Yajnik V, Khalili H, Garber JJ, Stevens BW, Cleland T, Xavier RJ. Gut microbiome function predicts response to anti-integrin biologic therapy in inflammatory bowel diseases. Cell Host & Microbe. 2017;21:603–10.e3. doi:10.1016/j.chom.2017.04.010.
- Doherty MK, Ding T, Koumpouras C, Telesco SE, Monast C, Das A, Brodmerkel C, Schloss PD. Fecal microbiota signatures are associated with response to ustekinumab therapy among Crohn’s disease patients. mBio. 2018;9. doi:10.1128/mBio.02120-17.
- Effenberger M, Reider S, Waschina S, et al. Microbial butyrate synthesis indicates therapeutic efficacy of azathioprine in IBD patients. J Crohn’s Colitis. 2021;15:88–98. doi:10.1093/ecco-jcc/jjaa152.
- Kolho KL, Korpela K, Jaakkola T, Pichai MVA, Zoetendal EG, Salonen A, de Vos WM. Fecal microbiota in pediatric inflammatory bowel disease and Its relation to inflammation. Am J Gastroenterol. 2015;110:921–930. doi:10.1038/ajg.2015.149.
- Magnusson MK, Strid H, Sapnara M, Lasson A, Bajor A, Ung K-A, Öhman L. Anti-TNF therapy response in patients with ulcerative colitis is associated with colonic antimicrobial peptide expression and microbiota composition. J Crohns Colitis. 2016;10:943–952. doi:10.1093/ecco-jcc/jjw051.
- Ribaldone DG, Caviglia GP, Abdulle A, Pellicano R, Ditto MC, Morino M, Fusaro E, Saracco GM, Bugianesi E, Astegiano M, et al. Adalimumab therapy improves intestinal dysbiosis in Crohn’s disease. J Clin Med. 2019;8(10):1646. doi:10.3390/jcm8101646.
- Shaw KA, Bertha M, Hofmekler T, Chopra P, Vatanen T, Srivatsa A, Prince J, Kumar A, Sauer C, Zwick ME, et al. Dysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome Med. 2016;8(1):75. doi:10.1186/s13073-016-0331-y.
- Wang Y, Gao X, Zhang X, Yang Y, Bindelle J, Ran C, Zhou Z. Intestinal Cetobacterium and acetate modify glucose homeostasis via parasympathetic activation in zebrafish. Gut Microbes. 2021;13:1–18. doi:10.1080/19490976.2021.1900996.
- Lewis JD, Sandler RS, Brotherton C, Brensinger C, Li H, Kappelman MD, Daniel SG, Bittinger K, Albenberg L, Valentine JF, et al. A randomized trial comparing the specific carbohydrate diet to a mediterranean diet in adults with Crohn’s disease. Gastroenterology. 2021;161(3):837–52.e9. doi:10.1053/j.gastro.2021.05.047.
- Wang L, Liao Y, Yang R, Zhu Z, Zhang L, Wu Z, Sun X. An engineered probiotic secreting Sj16 ameliorates colitis via ruminococcaceae/butyrate/retinoic acid axis. Bioengine & Transl Medi. 2021;6:e10219. doi:10.1002/btm2.10219.
- Chen H, Li H, Liu Z. Interplay of intestinal microbiota and mucosal immunity in inflammatory bowel disease: a relationship of frenemies. Therap Adv Gastroenterol. 2020;13:1756284820935188. doi:10.1177/1756284820935188.
- Sanchis-Artero L, Martínez-Blanch JF, Manresa-Vera S, Cortés-Castell E, Rodriguez-Morales J, Cortés-Rizo X. Evaluation of changes in gut microbiota in patients with Crohn’s disease after anti-Tnfα treatment: prospective multicenter observational study. Int J Env Res Pub He. 2020;17(14):5120. doi:10.3390/ijerph17145120.
- Lopez-Siles M, Martinez-Medina M, Busquets D, Sabat-Mir M, Duncan SH, Flint HJ, Aldeguer X, Garcia-Gil LJ. Mucosa-associated Faecalibacterium prausnitzii and Escherichia coli co-abundance can distinguish irritable bowel syndrome and inflammatory bowel disease phenotypes. Int J Med Microbiol. 2014;304:464–475. doi:10.1016/j.ijmm.2014.02.009.
- Olbjørn C, Cvancarova Småstuen M, Thiis-Evensen E, Nakstad B, Vatn MH, Jahnsen J, Ricanek P, Vatn S, Moen AE, Tannæs TM, et al. Fecal microbiota profiles in treatment-naïve pediatric inflammatory bowel disease – associations with disease phenotype, treatment, and outcome. Clin Exp Gastroenterol. 2019;12:37–49. doi:10.2147/CEG.S186235.
- Hyams JS, Turner D, Cohen SA, Szakos E, Kowalska-Duplaga K, Ruemmele F, Croft NM, Korczowski B, Lawrence P, Bhatia S, et al. Pharmacokinetics, safety and efficacy of intravenous vedolizumab in Paediatric patients with ulcerative colitis or Crohn’s disease: results from the phase 2 HUBBLE study. J Crohns Colitis. 2022;16(8):1243–1254. doi:10.1093/ecco-jcc/jjac036.
- Flint HJ, Duncan SH, Scott KP, Louis P. Links between diet, gut microbiota composition and gut metabolism. Proc Nutr Soc. 2015;74(1):13–22. doi:10.1017/S0029665114001463.
- Sandborn WJ, Hanauer SB, Katz S, et al. Etanercept for active Crohn’s disease: a randomized, double-blind, placebo-controlled trial. Gastroenterology. 2001;121:1088–1094. doi:10.1053/gast.2001.28674.
- https://www.massgeneral.org/medicine/gastroenterology/csibd/corelabs/clinical-core.
- Blakeley-Ruiz JA, Erickson AR, Cantarel BL, Xiong W, Adams R, Jansson JK, Fraser CM, Hettich RL. Metaproteomics reveals persistent and phylum-redundant metabolic functional stability in adult human gut microbiomes of Crohn’s remission patients despite temporal variations in microbial taxa, genomes, and proteomes. Microbiome. 2019;7:18. doi:10.1186/s40168-019-0631-8.
- Foley MH, O’Flaherty S, Barrangou R, Theriot CM, Knoll LJ. Bile salt hydrolases: gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract. PLoS Pathog. 2019;15(3):e1007581. doi:10.1371/journal.ppat.1007581.
- Ridlon JM, Kang D-J, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47(2):241–259. doi:10.1194/jlr.R500013-JLR200.
- Sinha SR, Haileselassie Y, Nguyen LP, Tropini C, Wang M, Becker LS, Sim D, Jarr K, Spear ET, Singh G, et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host & Microbe. 2020;27(4):659–70.e5. doi:10.1016/j.chom.2020.01.021.
- Verstockt B, Parkes M, Lee JC. How do we predict a patient’s disease course and whether they will respond to specific treatments? Gastroenterology. 2022;162(5):1383–1395. doi:10.1053/j.gastro.2021.12.245.
- Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001;81(3):1031–1064. doi:10.1152/physrev.2001.81.3.1031.
- Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27(2):104–119. doi:10.1111/j.1365-2036.2007.03562.x.
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. doi:10.1038/nature12726.
- Inan MS, Rasoulpour RJ, Yin L, Hubbard AK, Rosenberg DW, Giardina C. The luminal short-chain fatty acid butyrate modulates NF-kappaB activity in a human colonic epithelial cell line. Gastroenterology. 2000;118:724–734. doi:10.1016/S0016-5085(00)70142-9.
- Klampfer L, Huang J, Sasazuki T, Shirasawa S, Augenlicht L. Inhibition of interferon gamma signaling by the short chain fatty acid butyrate. Mol Cancer Res. 2003;1:855–862.
- Rossi O, van Berkel LA, Chain F, Tanweer Khan M, Taverne N, Sokol H, Duncan SH, Flint HJ, Harmsen HJM, Langella P, et al. Faecalibacterium prausnitzii A2-165 has a high capacity to induce IL-10 in human and murine dendritic cells and modulates T cell responses. Sci Rep. 2016;6(1):18507. doi:10.1038/srep18507.
- Patterson AM, Mulder IE, Travis AJ, Lan A, Cerf-Bensussan N, Gaboriau-Routhiau V, Garden K, Logan E, Delday MI, Coutts AGP, et al. Human gut symbiont roseburia hominis promotes and regulates innate immunity. Front Immunol. 2017;8:1166. doi:10.3389/fimmu.2017.01166.
- O’Connell Motherway M, Houston A, O’Callaghan G, Reunanen J, O’Brien F, O’Driscoll T, Casey PG, de Vos WM, van Sinderen D, Shanahan F. A bifidobacterial pilus-associated protein promotes colonic epithelial proliferation. Molecular Microbiology. 2019;111:287–301. doi:10.1111/mmi.14155.
- Breuer RI, Soergel KH, Lashner BA, Christ ML, Hanauer SB, Vanagunas A, Harig JM, Keshavarzian A, Robinson M, Sellin JH, et al. Short chain fatty acid rectal irrigation for left-sided ulcerative colitis: a randomised, placebo controlled trial. Gut. 1997;40(4):485–491. doi:10.1136/gut.40.4.485.
- Scheppach W. Treatment of distal ulcerative colitis with short-chain fatty acid enemas. A placebo-controlled trial. German-Austrian SCFA study group. Dig Dis Sci. 1996;41:2254–2259. doi:10.1007/BF02071409.
- Steinhart AH, Hiruki T, Brzezinski A, Baker JP. Treatment of left-sided ulcerative colitis with butyrate enemas: a controlled trial. Aliment Pharmacol Ther. 1996;10(5):729–736. doi:10.1046/j.1365-2036.1996.d01-509.x.
- Ghouri YA, Richards DM, Rahimi EF, Krill JT, Jelinek KA, DuPont AW. Systematic review of randomized controlled trials of probiotics, prebiotics, and synbiotics in inflammatory bowel disease. Clin Exp Gastroenterol. 2014;7:473–487. doi:10.2147/CEG.S27530.
- Xiao F, Dong F, Li X, Yu G, Liu Z, Wang Y, Zhang T. Bifidobacterium longum CECT 7894 improves the efficacy of infliximab for DSS-Induced colitis via regulating the gut microbiota and bile acid metabolism. Front Pharmacol. 2022;13:902337. doi:10.3389/fphar.2022.902337.
- Matsuoka K, Uemura Y, Kanai T, Kunisaki R, Suzuki Y, Yokoyama K, Yoshimura N, Hibi T. Efficacy of Bifidobacterium breve fermented milk in maintaining remission of ulcerative colitis. Dig Dis Sci. 2018;63:1910–1919. doi:10.1007/s10620-018-4946-2.
- Pavlidis P, Powell N, Vincent RP, Ehrlich D, Bjarnason I, Hayee B. Systematic review: bile acids and intestinal inflammation-luminal aggressors or regulators of mucosal defence? Aliment Pharmacol Ther. 2015;42(7):802–817. doi:10.1111/apt.13333.
- Jena A, Dutta U, Shah J, Sharma V, Prasad KK, Shivaprakash RM, Mandavdhare HS, Samanta J, Sharma P, Popli P, et al. Oral fluconazole therapy in patients with active ulcerative colitis who have detectable Candida in the stool: a double-blind randomized placebo-controlled trial. J Clin Gastroenterol. 2022;56(8):705–711. doi:10.1097/MCG.0000000000001609.
- Imhann F, Vich Vila A, Bonder MJ, Fu J, Gevers D, Visschedijk MC, Spekhorst LM, Alberts R, Franke L, van Dullemen HM, et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut. 2018;67(1):108–119. doi:10.1136/gutjnl-2016-312135.
- Pittayanon R, Lau JT, Leontiadis GI, Tse F, Yuan Y, Surette M, Moayyedi P. Differences in gut microbiota in patients with vs without inflammatory bowel diseases: a systematic review. Gastroenterology. 2020;158:930–46.e1. doi:10.1053/j.gastro.2019.11.294.
- Carroll IM, Ringel-Kulka T, Keku TO, Chang Y-H, Packey CD, Sartor RB, Ringel Y. Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2011;301:G799–807. doi:10.1152/ajpgi.00154.2011.
- Meade S, Routledge E, Sharma E, Honap S, Zeki S, Ray S, Anderson SH, Sanderson J, Mawdsley J, Irving PM, Samaan MA. . How achievable are STRIDE-II treatment targets in real-world practice and do they predict long-term treatment outcomes? Frontline Gastroenterol. 2022. flgastro-2022-102309. doi:10.1136/flgastro-2022-102309.
- Rehman A, Rausch P, Wang J, Skieceviciene J, Kiudelis G, Bhagalia K, Amarapurkar D, Kupcinskas L, Schreiber S, Rosenstiel P, et al. Geographical patterns of the standing and active human gut microbiome in health and IBD. Gut. 2016;65(2):238–248. doi:10.1136/gutjnl-2014-308341.
- Misra R, Sarafian M, Pechlivanis A, Ding N, Miguens-Blanco J, McDonald J, Holmes E, Marchesi J., Arebi N Ethnicity associated microbial and metabonomic profiling in newly diagnosed ulcerative colitis. Clin Exp Gastroenterol. 2022;15:199–212. doi:10.2147/CEG.S371965.
- Estaki M, Pither J, Baumeister P, Little JP, Gill SK, Ghosh S, Ahmadi-Vand Z, Marsden KR, Gibson DL. Cardiorespiratory fitness as a predictor of intestinal microbial diversity and distinct metagenomic functions. Microbiome. 2016;4:42. doi:10.1186/s40168-016-0189-7.
- Biedermann L, Zeitz J, Mwinyi J, Sutter-Minder E, Rehman A, Ott SJ, Steurer-Stey C, Frei A, Frei P, Scharl M, et al. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS ONE. 2013;8(3):e59260. doi:10.1371/journal.pone.0059260.
- Hyams JS, Davis Thomas S, Gotman N, Haberman Y, Karns R, Schirmer M, Mo A, Mack DR, Boyle B, Griffiths AM, et al. Clinical and biological predictors of response to standardised paediatric colitis therapy (PROTECT): a multicentre inception cohort study. Lancet. 2019;393(10182):1708–1720. doi:10.1016/S0140-6736(18)32592-3.
- Seong G, Kim N, Joung JG, Kim ER, Chang DK, Chun J, Hong SN, Kim Y-H. Changes in the intestinal microbiota of patients with inflammatory bowel disease with clinical remission during an 8-week infliximab Infusion Cycle. Microorganisms. 2020;8:8. doi:10.3390/microorganisms8060874.
- Gorelik Y, Freilich S, Gerassy-Vainberg S, Pressman S, Friss C, Blatt A, Focht G, Weisband YL, Greenfeld S, Kariv R, et al. Antibiotic use differentially affects the risk of anti-drug antibody formation during anti-TNFα therapy in inflammatory bowel disease patients: a report from the epi-IIRN. Gut. 2022;71(2):287–295. doi:10.1136/gutjnl-2021-325185.
- Williams A-J, Paramsothy R, Wu N, Ghaly S, Leach S, Paramsothy S, Corte C, O’Brien C, Burke C, Wark G, et al. Australia IBD Microbiome (AIM) Study: protocol for a multicentre longitudinal prospective cohort study. BMJ Open. 2021;11(2):e042493. doi:10.1136/bmjopen-2020-042493.
- Zhao M, Bendtsen F, Petersen AM, Larsen L, Dige A, Hvas C, Seidelin JB, Burisch J. Predictors of response and disease course in patients with inflammatory bowel disease treated with biological therapy—the Danish IBD Biobank Project: protocol for a multicentre prospective cohort study. BMJ Open. 2020;10:e035756. doi:10.1136/bmjopen-2019-035756.