Abstract
Cell differentiation relies on tissue-specific transcription factors (TFs) that cooperate to establish unique transcriptomes and phenotypes. However, the role of ubiquitous TFs in these processes remains poorly defined. Recently, we have shown that the CCCTC-binding factor (CTCF) is required for adipocyte differentiation through epigenomic remodelling of adipose tissue-specific enhancers and transcriptional activation of Peroxisome proliferator-activated receptor gamma (PPARG), the main driver of the adipogenic program (PPARG), and its target genes. Here, we discuss how these findings, together with the recent literature, illuminate a functional role for ubiquitous TFs in lineage-determining transcriptional networks.
Abbreviations
| CTCF | = | CCCTC-binding factor |
| KLF | = | Krüppel-like factors |
| PPARG | = | Peroxisome proliferator-activated receptor gamma |
| CEBP | = | CCAAT/enhancer binding protein |
| H3K4me1 | = | monomethylation of histone H3 lysine 4 |
| H3K27ac | = | acetylation of histone H3 lysine 27 |
| TET | = | Ten-eleven translocation methylcytosine dioxygenase |
| TF | = | Transcription factor |
| 5mC | = | 5-methylcytosine |
| 5hmC | = | 5-hydroxymethylcytosine |
Transcriptional Control of Cell Differentiation
Cell differentiation is a highly dynamic process, which allows the establishment of defined phenotypes. This is accomplished through specification of unique transcriptomes, a process that heavily relies on usage of cell type-specific enhancers.Citation1,2 Enhancer activation can be achieved through sequential TF recruitment to the chromatin following initial binding of pioneer TFs or involve a concomitant and cooperative binding of multiple TFs.Citation3-5 Cooperative binding may involve assisted loading where TF binding to close or overlapping motifs may facilitate each other's binding because of the very dynamic nature of TF-chromatin interactions.Citation6,7 Either way, enhancer activation requires chromatin/epigenomic remodelling, which has now been widely used as a surrogate to monitor enhancer activities during cell differentiation, including adipocyte differentiation.Citation8-10 In this context, we have contributed to the identification of DNA methylation as a novel epigenetic mark actively controlled by TFs at enhancers.Citation11,12 Moreover, using adipocyte differentiation as a model system, we have shown that loss of DNA methylation at activated enhancers is reciprocally linked to a gain in DNA hydroxymethylation ().Citation13,14 The mechanistic connection between these observations is underlined by Ten-eleven translocation methylcytosine dioxygenase (TET)-mediated DNA hydroxymethylation being an intermediate in the DNA demethylation process.Citation15 Importantly, a recent study functionally linked DNA hydroxymethylation to DNA demethylation, enhancer activation and gene induction during cell differentiation.Citation16 However, DNA hydroxymethylation is relatively stableCitation17 and can also modulate TF or chromatin modifier DNA recognitionCitation18 suggesting that cytosine hydroxymethylation may represent a novel epigenetic mark per se.Citation19 Fujiki et al. subsequently ascribed to the adipocyte lineage-determining TF Peroxisome proliferator-activated receptor gamma (PPARG) the function of promoting DNA hydroxymethylation through chromatin recruitment of TET.Citation20
Figure 1. Involvement of ubiquitous TFs in the transcriptional regulation of cell differentiation. The main concepts discussed in the manuscript are summarized (see text for details). The absence or presence of histone modifications that characterize active enhancers (H3K4me1 and H3K27ac) is indicated together with DNA methylation (5mC) and DNA hydroxymethylation (5hmC). H3K4me1, monomethylation of histone H3 lysine 4; H3K27ac, acetylation of histone H3 lysine 27; 5mC, 5-methylcytosine; 5hmC, 5-hydroxymethylcytosine.
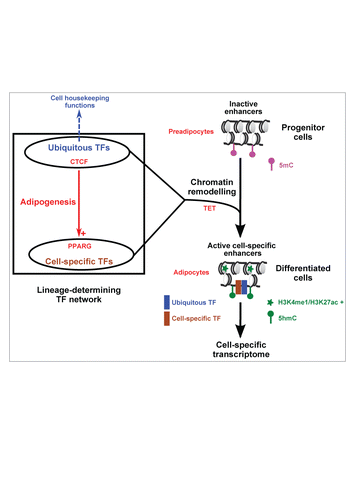
In addition to lineage-determining factors such as PPARG, which are expressed in specific tissues, the TF repertoire comprises ubiquitous TFs with similar expression in almost all tissues.Citation21 However, with the exception of general TFs involved in the pre-initiation complex (PIC), the role of ubiquitous TFs in cell differentiation has long remained elusive. Here, we discuss the general implications for our most recent findings showing a requirement for ubiquitous CCCTC-binding factor (CTCF) in defining the adipocyte-specific epigenome and transcriptome ().Citation22
CTCF As a Driver of Cell Type-Specific Enhancer Activities
CTCF, initially identified as a TF binding to insulators,Citation23 was latter revealed to harbour pleiotropic activities including transcriptional activator/repressor, nucleosome positioning and chromatin 3-dimensional organizer activities.Citation2,23,24 Based on this latter function and on the identification of conserved CTCF binding events across various cell-types and tissues,Citation25-27 this TF was proposed to fulfil a general chromatin organization role.Citation28 At the same time, specific roles for CTCF in neuronal and haematopoietic cell differentiation had been defined.Citation29 In this context, detailed analyses of CTCF cistromic studies revealed the existence of cell type-specific chromatin binding sites.Citation30 Indeed, thorough comparison of the CTCF binding landscape in 19 human cell-types revealed that 64% of identified CTCF binding sites are not conserved.Citation30 This may have been previously overlooked owing to a more limited number of analyzed cell types or to the use of stringent criteria to call bound regions. Indeed, conserved binding sites show stronger CTCF recruitment and show less degenerate CTCF binding motifs compared to cell type-specific CTCF bound regions.Citation22,30,31 We have now shown that the CTCF cistrome is highly dynamic during the course of adipogenesis since we found that more than half of the CTCF binding sites (>30,000 sites) identified across 4 stages of the differentiation process were not constitutively bound.Citation22 Moreover, our study provides evidence that this dynamic CTCF chromatin binding occurs at lineage-specific enhancers, which promote adipocyte differentiation ().Citation22 CTCF binding at these enhancers is not detected in preadipocytes and occurs upon differentiation when these regulatory sites are activated. As discussed hereafter, CTCF is necessary for DNA hydroxymethylationCitation22 of enhancers, a process required for cell differentiation.Citation16,20 Interestingly, another recent study has uncovered a similar role for another ubiquitous TF, the NF-Y complex, in activation of cell type-specific enhancers.Citation32 How these ubiquitous TFs are targeted to cell type-specific regulatory sites remains to be defined. Regarding CTCF, post-translational modifications and the existence of different modes of DNA recognition might help to modulate target sequence recognition.Citation33,34 Alternatively, combinatorial TF binding to enhancers, which now arises as a prevalent mechanism, could shape the CTCF cistrome. Direct CTCF interaction with TFs may, for instance, be involved in this process.Citation23,35 Of note, differentially active enhancers in mast cell and multipotent haematopoietic progenitors require TFs which are both specific and shared between the 2 cell-types to regulate transcription, further indicating that commonly expressed TFs are instrumental in regulating cell type-specific enhancer activities.Citation27,36
Involvement of CTCF in Chromatin Remodelling at Cell Type-Specific Enhancers
Multiple mechanisms may explain the requirement for combinatorial TF binding to enhancers. For instance, combinatorial TF binding to enhancers during adipocyte differentiation allows for cooperative coactivator recruitment and induction of histone acetylation.Citation5 In this context, in addition to PPARG, we have found that CTCF promotes TET-mediated DNA hydroxymethylation of enhancers driving adipocyte differentiation.Citation22 In addition to such ”quantitative” effects where multiple TFs cooperate to reinforce or amplify a given chromatin modification, individual contributions of different TFs to different enhancer functionalities may occur. TF binding to enhancers is not always simultaneous and can rather involve different kinetics implying a sequential or even a mutually exclusive recruitment.Citation5,37 This may in part be driven by the cyclic nature of transcriptional regulatory events that require instructed and sequential TF/cofactor chromatin binding and dissmisal.Citation38,39 In this context, it will be of great interest to better define whether and how the different functionalities of CTCF on the chromatin structure are involved at enhancers and how they relate to those of collaborating TFs. Indeed, CTCF is able to control local epigenetic modifications of DNA and histones.Citation22,40 It is also able to position surrounding nucleosomesCitation24 and to act on chromatin 3-dimentional foldingCitation41. Regarding NF-Y, it was hypothesized that this factor promotes chromatin opening at enhancers (potentially through nucleosome displacement) to favor recruitment of master lineage-determining TFs.Citation32
An important aspect of this research area will be to better define the relative influence of DNA methylation/hydroxymethylation on chromatin binding of ubiquitous and cell-specific TF. For instance, CTCF DNA binding can be inhibited by DNA methylationCitation36 while, reciprocally, CTCF is able to promote DNA demethylation suggesting a mutual influence.Citation12,35,42 Importantly, this relationship may be strongly linked to the genomic environnment as, for example, convertion of methylated DNA into hydroxymethylated DNA correlates with CTCF binding mostly outside of CpG islands (CGIs).Citation43
Finally, cell-specific CTCF binding has been shown to influence chromatin looping to promote interaction between Ubx enhancers and promoter to trigger gene transcriptional activation in Drosophila.Citation41
Therefore, CTCF emerges as a crucial chromatin remodeler at cell-type specific enhancers.
Ubiquitous TFs and Lineage-Determining TF Networks
We identified the adipose lineage-determining factor PPARG as one main target gene dynamically bound and transcriptionally regulated by CTCF during adipocyte differentiation, ().Citation22 Reminiscent of the establishment of specialized transcriptomes in other cell types such as hepatocytes,Citation44 adipocyte differentiation involves the build-up of a TF network, which includes, in addition to PPARG, members of the Krüppel-like factor (KLF) and CCAAT/enhancer binding protein (CEBP) families. These TFs show cross-regulation of their expression and cooperate to regulate important adipose-specific target genes.Citation45,46 Our study therefore shows that CTCF is part of this network through the regulation of both PPARG expression and activities (). These findings are reminiscent of those of Delgado-Olguín et al. regarding myogenesis during which CTCF both regulates the expression of, and cooperates with MyoD Citation47. These studies functionally validate predictions made from the analyses of DNAseI footprints from 41 cell and tissue-types indicating that CTCF was surprisingly involved in most cell type-specific TF networks. Interestingly, a similar prediction was made for additional ubiquitous TFs including SP1, NFYA, and MAX suggesting that this could be a general organization scheme of lineage-determining transcriptional networks.Citation48
Concluding Remarks
Our recent study provides an important new contribution to the recent litterature describing how ubiquitously expressed TFs functionally contribute to establishing cell-specific transcriptomes and phenotypes. However, important questions still remain to be answered. For instance, CTCF is characterized by an uncommonly large number of potential activities when bound to chromatin, making it central to the regulation of the functional genomic landscape. However, a major challenge arising is to better understand how CTCF's multiple functionalities are specified in space and time. With this aim, it will be instrumental to better understand how cues provided by the local chromatin environment and collaborating TF/cofactors allow for region and cell-specific CTCF binding and functions. This will undoubtly significantly improve our understanding of the functional connection between ubiquitous and cell type-specific TFs in lineage-determining transcriptional networks.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Work in our laboratory is supported by grants from “European Genomic Institute for Diabetes” (E.G.I.D., ANR-10-LABX-46), OSEO-ANVAR (IT-DIAB) and the Région Nord-Pas de Calais (contrat de plan Etat-Région). B.S. is a member of the Institut Universitaire de France.
References
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen C, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 2013; 503: 290-4; PMID:24141950
- Heidari N, Phanstiel DH, He C, Grubert F, Jahanbanian F, Kasowski M, Zhang MQ, Snyder MP. Genome-wide map of regulatory interactions in the human genome. Genome Res 2014; PMID:25228660
- Magnani L, Eeckhoute J, Lupien M. Pioneer factors: directing transcriptional regulators within the chromatin environment. Trends Genet 2011; 27: 465-74; PMID:21885149; http://dx.doi.org/10.1016/j.tig.2011.07.002
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 2011; 25: 2227-41; PMID:22056668; http://dx.doi.org/10.1101/gad.176826.111
- Siersbæk R, Rabiee A, Nielsen R, Sidoli S, Traynor S, Loft A, La Cour Poulsen L, Rogowska-Wrzesinska A, Jensen ON, Mandrup S. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep 2014; 7: 1443-55; PMID:24857652; http://dx.doi.org/10.1016/j.celrep.2014.04.042
- Madsen MS, Siersbæk R, Boergesen M, Nielsen R, Mandrup S. Peroxisome proliferator-activated receptor γ and C/EBPα synergistically activate key metabolic adipocyte genes by assisted loading. Mol Cell Biol 2014; 34: 939-54; PMID:24379442; http://dx.doi.org/10.1128/MCB.01344-13
- Voss TC, Schiltz RL, Sung M, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell 2011; 146: 544-54; PMID:21835447; http://dx.doi.org/10.1016/j.cell.2011.07.006
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. Comparative epigenomic analysis of murine and human adipogenesis. Cell 2010; 143: 156-69; PMID:20887899; http://dx.doi.org/10.1016/j.cell.2010.09.006
- Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J, Manduchi E, Stoeckert CJJ, Lazar MA. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev 2010; 24: 1035-44; PMID:20478996; http://dx.doi.org/10.1101/gad.1907110
- Siersbæk R, Nielsen R, John S, Sung M, Baek S, Loft A, Hager GL, Mandrup S. Extensive chromatin remodelling and establishment of transcription factor 'hotspots' during early adipogenesis. EMBO J 2011; 30: 1459-72; PMID:21427703; http://dx.doi.org/10.1038/emboj.2011.65
- Sérandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Métivier R, et al. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res 2011; 21: 555-65; PMID:21233399; http://dx.doi.org/10.1101/gr.111534.110
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 2011; 480: 490-5; PMID:22170606
- Oger F, Dubois-Chevalier J, Gheeraert C, Avner S, Durand E, Froguel P, Salbert G, Staels B, Lefebvre P, Eeckhoute J. Peroxisome proliferator-activated receptor γ (PPARγ) regulates genes involved in insulin/IGF signalling and lipid metabolism during adipogenesis through functionally distinct enhancer classes. J Biol Chem 2014; 289: 708-722; PMID:24288131; http://dx.doi.org/10.1074/jbc.M113.526996
- Sérandour AA, Avner S, Oger F, Bizot M, Percevault F, Lucchetti-Miganeh C, Palierne G, Gheeraert C, Barloy-Hubler F, Péron CL, et al. Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. Nucleic Acids Res 2012; 40: 8255-65; PMID:22730288; http://dx.doi.org/10.1093/nar/gks595
- Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol 2013; 14: 341-56; PMID:23698584; http://dx.doi.org/10.1038/nrm3589
- Hon GC, Song C, Du T, Jin F, Selvaraj S, Lee AY, Yen C, Ye Z, Mao S, Wang B, et al. 5mC Oxidation by Tet2 Modulates Enhancer Activity and Timing of Transcriptome Reprogramming during Differentiation. Mol Cell 2014; 56: 286-97; PMID:25263596; http://dx.doi.org/10.1016/j.molcel.2014.08.026
- Bachman M, Uribe-Lewis S, Yang X, Williams M, Murrel A, Balasubramanian S. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat Chem 2014; PMID:25411882; http://dx.doi.org/10.1038/nchem.2064
- Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PWTC, Bauer C, Münzel M, Wagner M, Müller M, Khan F, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 2013; 152: 1146-59; PMID:23434322; http://dx.doi.org/10.1016/j.cell.2013.02.004
- Lu F, Liu Y, Jiang L, Yamaguchi S, Zhang Y. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev 2014; 28:2103-19; PMID:25223896; http://dx.doi.org/10.1101/gad.248005.114
- Fujiki K, Shinoda A, Kano F, Sato R, Shirahige K, Murata M. PPARγ-induced PARylation promotes local DNA demethylation by production of 5-hydroxymethylcytosine. Nat Commun 2013; 4: 2262; PMID:23912449; http://dx.doi.org/10.1038/ncomms3262
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009; 10: 252-63; PMID:19274049; http://dx.doi.org/10.1038/nrg2538
- Dubois-Chevalier J, Oger F, Dehondt H, Firmin FF, Gheeraert C, Staels B, Lefebvre P, Eeckhoute J. A dynamic CTCF chromatin binding landscape promotes DNA hydroxymethylation and transcriptional induction of adipocyte differentiation. Nucleic Acids Res. 2014; 42: 10943-59; PMID:25183525; http://dx.doi.org/10.1093/nar/gku780
- Zlatanova J, Caiafa P. CTCF and its protein partners: divide and rule?. J. Cell. Sci. 2009; 122: 1275-84; PMID:19386894; http://dx.doi.org/10.1242/jcs.039990
- Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008; 4: e1000138; PMID:18654629; http://dx.doi.org/10.1371/journal.pgen.1000138
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature 2012; 488: 116-20; PMID:22763441; http://dx.doi.org/10.1038/nature11243
- Lee B, Bhinge AA, Battenhouse A, McDaniell RM, Liu Z, Song L, Ni Y, Birney E, Lieb JD, Furey TS, et al. Cell-type specific and combinatorial usage of diverse transcription factors revealed by genome-wide binding studies in multiple human cells. Genome Res. 2012; 22: 9-24; PMID:22090374; http://dx.doi.org/10.1101/gr.127597.111
- Calero-Nieto FJ, Ng FS, Wilson NK, Hannah R, Moignard V, Leal-Cervantes AI, Jimenez-Madrid I, Diamanti E, Wernisch L, Göttgens B. Key regulators control distinct transcriptional programmes in blood progenitor and mast cells. EMBO J. 2014; 33: 1212-26; PMID:24760698
- Sheffield NC, Thurman RE, Song L, Safi A, Stamatoyannopoulos JA, Lenhard B, Crawford GE, Furey TS. Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and long-range interactions. Genome Res. 2013; 23: 777-88; PMID:23482648; http://dx.doi.org/10.1101/gr.152140.112
- Herold M, Bartkuhn M, Renkawitz R. CTCF: insights into insulator function during development. Development 2012; 139: 1045-57; PMID:22354838; http://dx.doi.org/10.1242/dev.065268
- Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012; 22: 1680-8; PMID:22955980; http://dx.doi.org/10.1101/gr.136101.111
- Essien K, Vigneau S, Apreleva S, Singh LN, Bartolomei MS, Hannenhalli S. CTCF binding site classes exhibit distinct evolutionary, genomic, epigenomic and transcriptomic features. Genome Biol. 2009; 10: R131; PMID:19922652
- Oldfield AJ, Yang P, Conway AE, Cinghu S, Freudenberg JM, Yellaboina S, Jothi R. Histone-Fold Domain Protein NF-Y Promotes Chromatin Accessibility for Cell Type-Specific Master Transcription Factors. Mol Cell 2014; 55: 708-22; PMID:25132174; http://dx.doi.org/10.1016/j.molcel.2014.07.005
- Nakahashi H, Kwon KK, Resch W, Vian L, Dose M, Stavreva D, Hakim O, Pruett N, Nelson S, Yamane A, et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep 2013; 3: 1678-89; PMID:23707059; http://dx.doi.org/10.1016/j.celrep.2013.04.024
- Schmidt D, Schwalie PC, Wilson MD, Ballester B, Gonçalves A, Kutter C, Brown GD, Marshall A, Flicek P, Odom DT. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 2012; 148: 335-48; PMID:22244452; http://dx.doi.org/10.1016/j.cell.2011.11.058
- Weth O, Renkawitz R. CTCF function is modulated by neighboring DNA binding factors. Biochem. Cell Biol. 2011; 89: 459-68; PMID:21895576; http://dx.doi.org/10.1139/o11-033
- Davidson EH. The uncommon roles of common gene regulatory factors in the genomes of differentiating cells. EMBO J 2014; 33: 1193-4; PMID:24788410; http://dx.doi.org/10.1002/embj.201488693
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38: 576-89
- Métivier R, Penot G, Hübner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 2003; 115: 751-63; PMID:14675539; http://dx.doi.org/10.1016/S0092-8674(03)00934-6
- Quintin J, Le Péron C, Palierne G, Bizot M, Cunha S, Sérandour AA, Avner S, Henry C, Percevault F, Belaud-Rotureau M, et al. Dynamic estrogen receptor interactomes control estrogen-responsive trefoil Factor (TFF) locus cell-specific activities. Mol Cell Biol 2014; 34: 2418-36; PMID:24752895; http://dx.doi.org/10.1128/MCB.00918-13
- Weth O, Paprotka C, Günther K, Schulte A, Baierl M, Leers J, Galjart N, Renkawitz R. CTCF induces histone variant incorporation, erases the H3K27me3 histone mark and opens chromatin. Nucleic Acids Res 2014; 42: 11941-51; PMID:25294833; http://dx.doi.org/10.1093/nar/gku937
- Magbanua JP, Runneburger E, Russell S, White R. A variably occupied CTCF binding site in the Ultrabithorax gene in the Drosophila Bithorax Complex. Mol Cell Biol 2014; PMID:25368383
- Feldmann A, Ivanek R, Murr R, Gaidatzis D, Burger L, Schübeler D. Transcription factor occupancy can mediate active turnover of DNA methylation at regulatory regions. PLoS Genet. 2013; 9: e1003994; PMID:24367273
- Teif VB, Beshnova DA, Vainshtein Y, Marth C, Mallm J, Höfer T, Rippe K. Nucleosome repositioning links DNA (de)methylation and differential CTCF binding during stem cell development. Genome Res. 2014; 24: 1285-95; PMID:24812327; http://dx.doi.org/10.1101/gr.164418.113
- Odom DT, Dowell RD, Jacobsen ES, Nekludova L, Rolfe PA, Danford TW, Gifford DK, Fraenkel E, Bell GI, Young RA. Core transcriptional regulatory circuitry in human hepatocytes. Mol. Syst. Biol. 2006; 2: 2006.0017; PMID:16738562; http://dx.doi.org/10.1038/msb4100059
- Eeckhoute J, Oger F, Staels B, Lefebvre P. Coordinated regulation of PPARγ expression and activity through control of chromatin structure in adipogenesis and obesity. PPAR Res 2012; 2012: 164140; PMID:22991504; http://dx.doi.org/10.1155/2012/164140
- Siersbæk R, Nielsen R, Mandrup S. Transcriptional networks and chromatin remodeling controlling adipogenesis. Trends Endocrinol. Metab. 2011; 23: 56-64; PMID:22079269; http://dx.doi.org/10.1016/j.tem.2011.10.001
- Delgado-Olguín P, Brand-Arzamendi K, Scott IC, Jungblut B, Stainier DY, Bruneau BG, Recillas-Targa F. CTCF promotes muscle differentiation by modulating the activity of myogenic regulatory factors. J. Biol. Chem. 2011; 286: 12483-94; PMID:21288905; http://dx.doi.org/10.1074/jbc.M110.164574
- Neph S, Stergachis AB, Reynolds A, Sandstrom R, Borenstein E, Stamatoyannopoulos JA. Circuitry and dynamics of human transcription factor regulatory networks. Cell 2012; 150: 1274-86; PMID:22959076; http://dx.doi.org/10.1016/j.cell.2012.04.040
