Abstract
In contrast to cell type-specific pre-mRNA alternative splicing, mechanisms controlling activity-dependent alternative splicing is under-studied and not well understood. In a recent study, we conducted a comprehensive analysis of calcium-mediated mechanism that regulates alternative exon skipping in mouse cardiomyocytes. Our results reveal a strong link between histone hyperacetylation and skipping of cassette exons, and provide support to the kinetic coupling model of the epigenetic regulation of alternative splicing at the chromatin level.
Alternative pre-mRNA splicing, a process that generates more than one mRNA transcripts from a single gene, is one of the major mechanisms that control gene expression. Alternative splicing is tightly regulated leading to differential expression patterns, often in temporal or spatial cell type-specific manners.Citation1 Alternative splicing can also be regulated in response to environmental stimuli.Citation2 A number of studies demonstrated that alternative splicing patterns can change responding to cell activation, UV irradiation, and neuronal depolarization.Citation3-6 However, compared to other patterns, this particular pattern of alternative splicing has been less studied.
Many years of investigations on cell type-specific alternative splicing have revealed a large number of complex regulatory mechanisms of alternative splicing. A great majority of these mechanisms involve RNA-binding proteins that regulate alternative splicing by interacting with RNA sequences on pre-mRNA molecules. The interaction either recruits the spliceosome to include the alternative exon associated with the sequences or blocks the spliceosome to skip the alternative exon depending on the location and context of the RNA regulatory sequences.Citation1
In recent years, a new theme of regulatory mechanisms, known as the epigenetic regulation of alternative splicing that occurs at the level of chromatin, has come to light. Given the tight coupling of transcription and splicing, it is conceivable that the factors influencing either one of the 2 individual processes would have an effect on the other one. Epigenetic changes that alter chromatin architecture and histone modifications affect the alternative splicing outcome in one of at least 2 modes.Citation7,8 In the first mode, epigenetic changes modulate the structure of a specific region on chromatin to be more or less open, which alters the transcriptional elongation rate of RNA polymerase II (RNAPII). Changes in RNAPII elongation rate is thought to affect alternative splicing through the kinetic coupling model. The model predicts that when the transcriptional elongation rate is high, an alternative exon, generally surrounded by suboptimal splicing signals, is likely to be outcompeted by the downstream constitutive exon, usually surrounded by strong splicing signals, and thus, is excluded from the transcript.Citation9 In the second mode, known as the chromatin-splicing adaptor model, it is thought that a splicing regulatory protein can be more efficiently recruited to an alternative exon through its interaction with a chromatin remodeler protein bound at specific histone marks.Citation7,8
Acetylation of core histone tails disrupts the condensed higher-order chromatin folding to enhance transcription.Citation10 In contrast to the well-established role of histone hyperacetylation in increasing transcription initiation and transcript levels, how histone hyperacetylation affects alternative splicing is poorly understood. In a recent publication, we reported our studies demonstrating a direct link between histone hyperacetylation and exon skipping of alternative exons. We also elucidated the underlying regulatory mechanism, revealing a cascade of molecular events that are mediated by the intracellular calcium levels.Citation11
Depolarization Induces a Robust and Reversible Increase of Exon Skipping
Cells respond to environmental changes by altering their gene expression output. Although there are only a small number of examples, it has been shown that pre-mRNA alternative splicing can be regulated by environmental stimuli.Citation2 Several studies indicated that activity-dependent alternative splicing regulation occurs in neuronal cells.Citation12,13 To investigate activity-dependent alternative splicing in a different cell type, we depolarized mouse cardiomyocytes using potassium chloride (KCl) and examined alternative splicing of exon 23a of the Nf1 gene. We selected this particular alternative exon because its regulation had been extensively studied in our laboratory, and its expression plays a key role in regulating the active Ras activity in cells.Citation14,15 We prepared mouse cardiomyocytes in 2 different ways, either isolating primary cardiomyocytes from neonatal day 1-3 mice or differentiating cardiomyocytes from mouse embryonic stem (ES) cells using established methods.Citation16,17
The semi-quantitative reverse transcriptase-polymerase chain reactions (RT-PCR) indicated that when primary cardiomyocytes are treated with increasing concentrations of KCl (25–100 mM), the inclusion of Nf1 exon 23a was reduced from 70% to 15%.Citation11 Similar results were observed with ES-derived cardiomyocytes. No changes were observed when mouse ES cells were treated with KCl.Citation11 A time-course experiment indicated that the alternative splicing pattern change was first detected at 6 hours of treatment, which coincides with the half-life of Nf1 mRNA, and reached its maximum at 24 hours.Citation11 The splicing change is reversible as removal of KCl completely restored the splicing pattern, indicating the dynamic nature of this phenomenon.Citation11
It is known that KCl leads to calcium influx via L-type voltage-gated calcium channels.Citation18-20 To examine the role of calcium in the splicing regulation of Nf1 exon 23a, we used pharmacological drugs in conjunction with KCl to treat cardiomyocytes. Use of either the L-type calcium channel blocker nifedipine or the NMDA receptor agonist BAPTA-AM together with calcium chelator AP5 significantly reduced the effect of KCl on alternative splicing of Nf1 exon 23a.Citation11 The results support that changes of the intracellular calcium levels are responsible for KCl-induced splicing pattern change of Nf1 exon 23a.
The KCl-induced exon skipping is not unique to Nf1 exon 23a, as a similar dynamic change of exon inclusion/skipping was observed for several alternative exons including Ktn1 exon36, Ank2 exon 21, Enah exon 5 and Mef2A exon 9.Citation11 Thus, there appears to be a general mechanism that regulates the expression of a set of alternative exons. These exons, named calcium-responsive exons in our study, and Capzb exon 8, a calcium-nonresponsive exon, were used next to study the underlying mechanism.
Model
The demonstration of robust and reversible changes in exon skipping/inclusion for several alternative exons strongly suggested a calcium-mediated regulatory program that controls a network of alternative exons in cells in response to altered intracellular calcium levels. To study the underlying mechanism, we carried out a variety of experiments that combined genetic, biochemical, molecular, and pharmacological approaches. The results of these experiments reveal a cascade of events that are triggered by increased calcium levels and lead to skipping of alternative exons through a mechanism involving protein kinase signaling activity, HDAC protein transport, histone modification, chromatin structure and RNAPII transcriptional elongation rate. A working model developed from the results is depicted in . The following sections will describe the experiments that support this model.
Figure 1. A model indicating the cascade of molecular events triggered by increased intracellular calcium levels. Increased calcium levels in mouse cardiomyocytes induced by either depolarization or caffeine treatment (step 1) activates CaMK and PKD (step 2), which phosphorylate class II HDACs causing them to translocate from the nucleus to cytoplasm (step 3). The reduced association of HDAC and increased association of HAT on genes containing calcium-responsive alternative exons lead to histone hyperacetylation (step 4), which supports a more relaxed chromatin structure, allowing for RNAPII to elongate faster (step 5). The higher elongation rate leads to skipping of alternative exons (step 6).
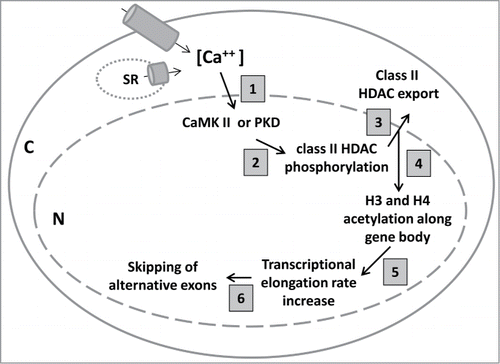
The signaling pathways: CaMKII and PKD
In several previous studies, depolarization has been found to induce exon skipping in neurons.Citation12,13 Interestingly, mixed results were reported regarding the signaling pathway that is responsible for increased exon skipping. For example, while protein CaMKIV, the neuronal member of the Ca/calmodulin-dependent protein kinase (CaMK) family, was shown to be responsible for skipping of the stress-axis regulated exon of the Slo1 gene in depolarized neurons,Citation21 pharmacological inhibition of CaMK had no effect on depolarization-induced skipping of exon 18 of the neural cell adhesion molecule (NCAM) gene.Citation21
To investigate the responsible signaling pathway(s), we also started with pharmacological approaches. Interestingly, we found that blocking the function of CaMK in cardiomyocytes had no effect on depolarization-induced skipping of Nf1 exon 23a.Citation11 Further investigations using small molecule inhibitors or siRNAs revealed that CaMK II, the CaMK protein family member expressed in cardiomyocytes, and protein kinase D (PKD) have a redundant role in regulating Nf1 exon 23a skipping. Only when both CaMK II and PKD are blocked simultaneously, the effect of depolarization on skipping of Nf1 exon 23a, Ktn1 exon 36 and Ank2 exon 21 is abolished.Citation11
Histone hyperacetylation
It is known that the calcium-mediated up-regulated activity of CaMK II or PKD leads to phosphorylation of class II histone deacetylases (HDAC), which include HDACs 4,5,7,9, resulting in nuclear export and cytoplasmic accumulation of these proteins.Citation22,23 Our immunostaining assay and western blot analysis using fractionated proteins indicated a nuclear to cytoplasmic transport of class II HDAC proteins, consistent with the previous studies.Citation11
A consequence of the HDAC nuclear export is disruption of the normal histone acetyltransferase (HAT) to HDAC ratio in the nucleus. It was predicted that, as a result of the HDAC export, HAT association with chromatin would increase and histones would be hyperacetylated. A ChIP analysis using antibodies specific for the HAT CBP or p300 indicated a moderate increase of their association with Nf1 and Ktn1.Citation11 In addition, a western blot analysis using antibodies against total histone H3 or H4, or acetylated H3 or H4, showed that depolarization of cardiomyocytes led to hyperacetylation of both H3 and H4, with a 2.5-3-fold increase of histone acetylation levels.Citation11 Importantly, histone acetylation levels returned to the basal level when cells recovered from depolarization.Citation11 The histone hyperacetylation occurs downstream of the CaMK II and PKD activation, as inhibition of both pathways simultaneously abolished the depolarization-induced stronger HAT association and histone hyperacetylation.Citation11 As with the splicing assay, blocking either kinase by itself did not have any effect.Citation11
It appears that hyperacetylation occurs in a pan-lysine manner. All of the lysines we examined, including H3K9, H3K14, H3K27, and H4K16, showed increased acetylation.Citation11 In contrast, no changes on lysine methylation, e.g., H3K4me3 and H3K36me3, were observed.Citation11 Also consistent with the pan-lysine hyperacetylation observation, our chromatin-immunoprecipitation (ChIP) experiment indicated that hyperacetylation occurs throughout the gene body, and was not just limited to the promoter region of a given gene.Citation11 However, specificity does exist, as hyperacetylation only occurred at genes that contain calcium-responsive alternative exons, such as Nf1 and Ktn1, but not at genes that contain nonresponsive alternative exons, such as Capzb.Citation11
To provide further connection between histone hyperacetylation and alternative exon skipping, we carried out a siRNA knockdown of HAT experiment in depolarized cardiomyocytes. We found that knockdown of either CBP or p300 singly partially rescued skipping of the Nf1 exon 23a, and knockdown of both HAT had an additive effect.Citation11
RNAPII transcriptional elongation rate
Histone hyperacetylation is generally associated with more permissive chromatin structure for faster elongation of RNAPII.Citation10 Thus, the positive correlation between histone hyperacetylation and increased exon skipping prompted us to consider the kinetics model that connects transcriptional elongation rate and alternative splicing outcome. The kinetics model posits that faster transcriptional elongation rate leads to more skipping of alternative exons because these exons are often surrounded by sub-optimal splicing signals and easily outcompeted by the fast coming downstream constitutive exons for recognition by the splicing machinery.Citation7
To test the hypothesis that histone hyperacetylation induced by depolarization leads to higher transcriptional elongation rate of RNAPII, we conducted 3 sets of experiments. First, we carried out ChIP to examine the RNAPII occupancy on the genes of interest. Using an antibody that recognizes the elongating RNAPII in this assay, we showed that RNAPII accumulation along the gene body of Nf1 and Ktn1 is approximately 3-fold lower in depolarized than in control cardiomyocytes, consistent with faster RNAPII elongation rate in depolarized cells.Citation11 Second, using an established procedure,Citation24,25 we directly measured RNAPII transcriptional elongation rate on several genes comparing control and depolarized cardiomyocytes. Our results indicate an expected significant increase of transcriptional elongation rate on Nf1 and Ktn1, but not on Capzb, in cardiomyocytes after depolarization.Citation11 Specifically, the rate increased from 3.2 to 6.8 kb/min for Nf1 and from 2.4 to 4.1 kb/min for Ktn1. However, importantly, the rate did not change for Capzb. Third, we took advantage of a drug called camptothecin, which is used to slow down transcriptional elongation,Citation24,26 to manipulate the transcriptional elongation rate of RNAPII. We found that depolarization of cardiomyocytes could no longer induce skipping of Nf1 exon 23a when camptothecin was added to the cells at the same time with KCl.Citation11 These results provide strong support to the model that depolarization-induced transcriptional elongation rate increase is responsible for skipping of alternative exons.
Effect of caffeine on exon skipping and histone acetylation
Many environmental factors affect intracellular calcium levels. One example is caffeine, which functions as an agonist of the ryanodine receptor located on the sarcoplasmic reticulum (SR) in cardiomyocytes to promote calcium release from inside of the SR to the cytoplasm.Citation27 We predicted that caffeine treatment of cardiomyocytes would lead to an exon skipping phenotype similar to depolarization. When we treated primary or ES-derived cardiomyocytes with increasing concentrations of caffeine, we did observe a similarly significant increase of exon skipping of Nf1 exon 23a, as in depolarized cells.Citation11
However, interestingly, there is a clear difference between caffeine- and KCl-treated cells in the changes of alternative splicing of Nf1 exon 23a. Unlike KCl treatment, the caffeine-induced skipping cannot be restored completely to the level in untreated cells after caffeine removal. Specifically, 24 hours after caffeine was removed, exon 23a inclusion increased to 48% but not 69% (). We do not know the reason for the partial recovery. It is possible that caffeine was not completely removed during recovery. Nonetheless, we took advantage of the opportunity to examine the histone acetylation levels. Significantly, as shown in , although histone acetylation level increased more than 4-fold after caffeine treatment, it did not go back to the basal level after caffeine removal, which correlates well with the partial reversion of the alternative splicing levels (). Thus, these results provide a stronger correlation between skipping of calcium-responsive alternative exons and histone hyperacetylation.
Figure 2. Effects of caffeine on exon skipping and histone acetylation. (A) Effect of caffeine on alternative splicing of Nf1 exon 23a. Semi-quantitative RT-PCR analysis using total RNA isolated from mouse cardiomyocytes treated for 24 hours with EtOH, 10 mM caffeine (dissolved in EtOH), or caffeine for 24 hours followed by recovery for 24 hours. Primers annealing to exons 23 and 24 were used.Citation11 Products representing inclusion (open circle) or skipping (solid circle) of an alternative exon are indicated. The graph shows the percentage of exon 23a inclusion. n = 5. (B) Effect of caffeine on histone acetylation. Cardiomyocytes were treated as described above. Total histones were acid-extracted from the treated cells. Western blot analysis was carried out using antibodies specific for total or pan-acetylated histone H3. The graph shows the fold differences between caffeine- and EtOH-treated cells. n = 1. (C) Effect of caffeine on alternative splicing of additional alternative exons. Cardiomyocytes were treated with EtOH or 10 mM caffeine for 24 hours. Semi-quantitative RT-PCR analysis was carried out using primers annealing to exons surrounding the indicated alternative exons, as described.Citation11 Products representing inclusion (open circle) or skipping (solid circle) of an alternative exon are indicated. n = 2.
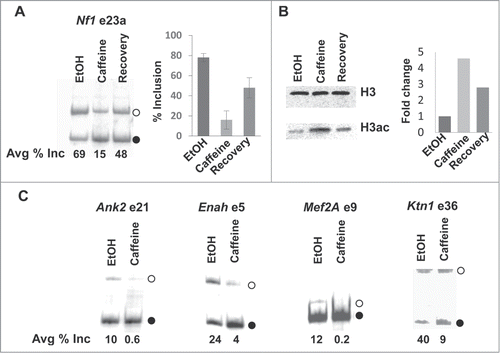
We also examined several additional alternative splicing events that respond to depolarization in caffeine-treated cardiomyocytes and found similar changes (). These results strongly support that increases of intracellular calcium levels triggered by different factors lead to similar downstream events.
Discussion
In this study, we carried out a comprehensive analysis of calcium-mediated regulation of alternative splicing in cardiomyocytes. We first demonstrated that calcium regulates the inclusion of a set of alternative exons in a dynamic fashion. Specifically, increased intracellular calcium leads to reversible skipping of at least 5 alternative exons. We then discovered a general mechanism that explains the observed splicing changes for these alternative exons. We found that an increase in calcium level activates either one of the 2 signaling pathways, CaMKII and PKD, which have redundant roles in this regulation. The activation of either kinase leads to an epigenetic change, histone hyperacetylation, along the body of the genes that contain calcium-responsive exons, which relaxes the chromatin structure to allow for increased RNAPII transcriptional elongation rate. Consequently, alternative exons are skipped, as predicted by the kinetic coupling model ().
A limited number of previous studies have investigated the role of changes of histone acetylation in alternative splicing regulation. In several cases, pan HDAC inhibitors are used to induce a global histone hyperacetylation. For example, when HeLa cells were treated with sodium butyrate, a pan-HDAC inhibitor, close to 700 alternative exons changed their splicing patterns.Citation28 This study further implicated, using ChIP analysis, that the treatment induced histone H4 hyperacetylation, and increased RNAPII processivity, inferred from reduced RNAPII association, across chromatin regions containing alternative exons, although a direct measurement of transcriptional elongation rate was not conducted.Citation28 In another study using depolarized neuronal cells, it was suggested that increased skipping of exon 18 in the NCAM pre-mRNA was correlated with a localized increase of H3K9 acetylation and H3K36 trimethylation, as well as increased local RNAPII movement.Citation21 A previous study from our own laboratory also provided a link between localized histone hyperacetylation and exon skipping, although in this case, the histone hyperacetylation depends on a splicing regulator.Citation25 Taken together, histone hyperacetylation induced through different mechanisms, and occurring either locally or globally throughout the gene body, can lead to exon skipping.
A genome-wide exon-array analysis indicated that depolarization in neurons induces changes of many alternative splicing events.Citation29 We propose that some of the events may be explained by the mechanism we uncovered. Interestingly, manipulations of RNAPII elongation rate globally have generated mixed results of alternative splicing pattern changes. On the one hand, reduction of RNAPII elongation rate with CPT or DRB led to the expected increase of alternative exon inclusion.Citation30 On the other hand, use of RNAPII mutants that have either faster or slower elongation rates yielded alternative splicing changes in both directions, i.e., increase and decrease of inclusion of alternative exons.Citation31 It also be noted that a recent study demonstrated that slower transcriptional elongation led to increased skipping of an alternative exon instead of the expected increased inclusion, through recruiting a splicing silencer protein to its target sequence on the pre-mRNA.Citation32 Clearly, regulation of alternative splicing by the transcriptional elongation rate is more complicated that may involve different mechanisms for specific exons.
Why do only a set of alternative exons respond to increased calcium levels to change splicing pattern? How is specificity achieved? We started to address this question using ChIP analyses. We used antibodies specific to HAT and HDAC enzymes to examine association of these enzymes to the genes of interests in unstimulated or stimulated cells. In unstimulated cells, we found no differential association of any HAT or HDAC enzyme between genes containing calcium-responsive exons and those containing calcium-nonresponsive exons, indicating that these histone modifying enzymes do not distinguish these 2 groups of genes, associating with them similarly at the basal levels.Citation11 However, in depolarized cardiomyocytes, association of class II HDAC is reduced and association of HAT is increased on the genes containing calcium-responsive alternative exons, while no difference in association of HDAC and HAT was observed on genes containing calcium-nonresponsive exons.Citation11 These results suggest that the key in this regulation is whether class II HDAC dissociates from the genes after stimulation. It is possible that additional chromatin remodeler proteins are involved in this regulation. Future experiments will be aimed to reveal the mechanism by which HDACs dissociate from the set of genes containing calcium-responsive exons.
Lastly, even though we speculate that the mechanism we described here do not involve changes of RNA binding proteins (expression level or cellular localization), we cannot rule out such possibility. Several previous studies have demonstrated that depolarization of neuronal cells induced changes of splicing regulators, which in turn led to alternative splicing changes.Citation33-38 It is possible that the calcium-induced export of class II HDACs changes of post-translational modification of some splicing regulators, e.g., making some proteins more acetylated, which alters their function and then leads to downstream alternative splicing changes. It is very likely that multiple mechanisms exist that control calcium-mediated alternative splicing changes.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Institutes of Health (NS049103 to HL, S10RR021228 and S10RR024536 National Center for Research Resources Shared Instrumentation Grants for the Leica DM6000 wide-field microscope and the GE Healthcare Typhoon Trio Variable Mode Imager). LC was supported by National Natural Science Foundation of China (61271448). HN was supported by a fellowship from the Vietnam Education Foundation.
References
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet 2010; 11:75-87; http://dx.doi.org/10.1038/nrg2673
- Lynch KW. Regulation of alternative splicing by signal transduction pathways. Adv Exp Med Biol 2007; 623:161-74; PMID:18380346; http://dx.doi.org/10.1007/978-0-387-77374-2_10
- Munoz MJ, Pérez Santangelo MS, Paronetto MP, de la Mata M, Pelisch F, Boireau S, Glover-Cutter K, Ben-Dov C, Blaustein M, Lozano JJ, et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell 2009; 137:708-20; PMID:19450518; http://dx.doi.org/10.1016/j.cell.2009.03.010
- Batsche, E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol 2006; 13:22-9; http://dx.doi.org/10.1038/nsmb1030
- Xie J, Black DL. A CaMK IV responsive RNA element mediates depolarization-induced alternative splicing of ion channels. Nature 2001; 410:936-9; PMID:11309619; http://dx.doi.org/10.1038/35073593
- Heyd F, Lynch KW. Phosphorylation-dependent regulation of PSF by GSK3 controls CD45 alternative splicing. Mol Cell 2010; 40:126-37; PMID:20932480; http://dx.doi.org/10.1016/j.molcel.2010.09.013
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell 2012; 144:16-26; http://dx.doi.org/10.1016/j.cell.2010.11.056
- Kornblihtt AR. et al.. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol 2013 14:153-65; PMID:23385723; http://dx.doi.org/10.1038/nrm3525
- de la Mata, M, Lafaille C, Kornblihtt AR. First come, first served revisited: factors affecting the same alternative splicing event have different effects on the relative rates of intron removal. RNA 2010; 16:904-12; PMID:20357345; http://dx.doi.org/10.1261/rna.1993510
- Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol 2015; 16:258-264; PMID:25549891
- Sharma A, Nguyen H, Geng C, Hinman MN, Luo G, Lou H. Calcium-mediated histone modifications regulate alternative splicing in cardiomyocytes. Proc Natl Acad Sci USA 2014; 111:E4920-4928; PMID:25368158; http://dx.doi.org/10.1073/pnas.1408964111
- Sharma A, Lou H. Depolarization-mediated regulation of alternative splicing. Front Neurosci 2011; 5:141; PMID:22207834; http://dx.doi.org/10.3389/fnins.2011.00141
- Razanau A, Xie J. Emerging mechanisms and consequences of calcium regulation of alternative splicing in neurons and endocrine cells. Cell Mol Life Sci 2013; 70:4527-36; http://dx.doi.org/10.1007/s00018-013-1390-5
- Fleming VA, Geng C, Ladd AN, Lou H. Alternative splicing of the neurofibromatosis type 1 pre-mRNA is regulated by the muscleblind-like proteins and the CUG-BP and ELAV-like factors. BMC Mol Biol 2012; 13:35; PMID:23227900; http://dx.doi.org/10.1186/1471-2199-13-35
- Hinman MN, Sharma A, Luo G, Lou H. Neurofibromatosis Type 1 Alternative Splicing is a Key Regulator of Ras Signaling in Neurons. Mol Cell Biol 2014; 34(12):2188-97; PMID:24710274
- Sreejit P, Kumar S, Verma RS. An improved protocol for primary culture of cardiomyocyte from neonatal mice. In Vitro Cell Dev Biol Anim 2008; 44:45-50; PMID:18297366; http://dx.doi.org/10.1007/s11626-007-9079-4
- Wobus AM, Guan K, Yang HT, Boheler KR. Embryonic stem cells as a model to study cardiac, skeletal muscle, and vascular smooth muscle cell differentiation. Methods Mol Biol 2002; 185:127-56; PMID:11768985
- Narang D. et al. Triton X-100 inhibits L-type voltage-operated calcium channels. Can J Physiol Pharmacol 2013; 91:316-24; http://dx.doi.org/10.1139/cjpp-2012-0257
- Rathi SS, Saini HK, Xu YJ, Dhalla NS. Mechanisms of low Na+-induced increase in intracellular calcium in KCl-depolarized rat cardiomyocytes. Mol Cell Biochem 2004; 263:151-62; PMID:15524176; http://dx.doi.org/10.1023/B:MCBI.0000041857.86178.f6
- Viola HM, Arthur PG, Hool LC. Evidence for regulation of mitochondrial function by the L-type Ca2+ channel in ventricular myocytes. J Mol Cell Cardiol 2009; 46:1016-26; http://dx.doi.org/10.1016/j.yjmcc.2008.12.015
- Schor IE, Rascovan N, Pelisch F, Allo M, Kornblihtt AR. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci USA 2009; 106, 4325-30; PMID:19251664; http://dx.doi.org/10.1073/pnas.0810666106
- Vega RB, Harrison BC, Meadows E, Roberts CR, Papst PJ, Olson EN, McKinsey TA. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol Cell Biol 2004; 24:8374-85; PMID:15367659; http://dx.doi.org/10.1128/MCB.24.19.8374-8385.2004
- Backs J, Song K, Bezprozvannaya S, Chang S, Olson EN. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J Clin Invest 2006; 116:1853-64; PMID:16767219; http://dx.doi.org/10.1172/JCI27438
- Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol 2009; 16:1128-33; PMID:19820712; http://dx.doi.org/10.1038/nsmb.1666
- Zhou HL, Hinman MN, Barron VA, Geng C, Zhou G, Luo G, Siegel RE, Lou H. Hu proteins regulate alternative splicing by inducing localized histone hyperacetylation in an RNA-dependent manner. Proc Natl Acad Sci U S A 2011; 108:E627-635; PMID:21808035; http://dx.doi.org/10.1073/pnas.1103344108
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol 2007; 14:796-806; http://dx.doi.org/10.1038/nsmb1280
- Zucchi R, Ronca-Testoni S. The sarcoplasmic reticulum Ca2+ channel/ryanodine receptor: modulation by endogenous effectors, drugs and disease states. Pharmacol Rev1997; 49:1-51; PMID:9085308
- Hnilicova J, Hozeifi S, Dušková E, Icha J, Tománková T, Staněk D. Histone deacetylase activity modulates alternative splicing. PloS one 2011; 6:e16727; PMID:21311748; http://dx.doi.org/10.1371/journal.pone.0016727
- McKee AE, Neretti N, Carvalho LE, Meyer CA, Fox EA, Brodsky AS, Silver PA. Exon expression profiling reveals stimulus-mediated exon use in neural cells. Genome Biol 2007; 8:R159; http://dx.doi.org/10.1186/gb-2007-8-8-r159
- Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, Blencowe BJ. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res 2011; 21:390-401; PMID:21163941; http://dx.doi.org/10.1101/gr.111070.110
- Fong N, Kim H, Zhou Y, Ji X, Qiu J, Saldi T, Diener K, Jones K, Fu XD, Bentley DL. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev 2014; 28:2663-76; PMID:25452276; http://dx.doi.org/10.1101/gad.252106.114
- Dujardin G, Lafaille C, de la Mata M, Marasco LE, Muñoz MJ, Le Jossic-Corcos C, Corcos L, Kornblihtt AR. How slow RNA polymerase II elongation favors alternative exon skipping. Mol Cell 2014; 54:683-90; PMID:24793692; http://dx.doi.org/10.1016/j.molcel.2014.03.044
- Dembowski JA, An P, Scoulos-Hanson M, Yeo G, Han J, Fu XD, Grabowski PJ. Alternative Splicing of a Novel Inducible Exon Diversifies the CASK Guanylate Kinase Domain. J Nucleic Acids 2012; 2012:816237; PMID:23008758; http://dx.doi.org/10.1155/2012/816237
- Rozic G, Lupowitz Z, Zisapel N. Exonal elements and factors involved in the depolarization-induced alternative splicing of neurexin 2. J Mol Neurosci 2013; 50:221-33; http://dx.doi.org/10.1007/s12031-012-9919-x
- Lee JA, Tang ZZ, Black DL. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev 2009; 23:2284-93; PMID:19762510; http://dx.doi.org/10.1101/gad.1837009
- Yu J, Hai Y, Liu G, Fang T, Kung SK, Xie J. The heterogeneous nuclear ribonucleoprotein L is an essential component in the Ca2+/calmodulin-dependent protein kinase IV-regulated alternative splicing through cytidine-adenosine repeats. J Biol Chem 2009; 284:1505-13; ; http://dx.doi.org/10.1074/jbc.M805113200
- Lee JA, Xing Y, Nguyen D, Xie J, Lee CJ, Black DL. Depolarization and CaM kinase IV modulate NMDA receptor splicing through two essential RNA elements. PLoS Biol 2007; 5:e40; PMID:17298178; http://dx.doi.org/10.1371/journal.pbio.0050040
- An P, Grabowski PJ. Exon silencing by UAGG motifs in response to neuronal excitation. PLoS Biol 2007; 5:e36; PMID:17298175; http://dx.doi.org/10.1371/journal.pbio.0050036
