Abstract
Histone methylation has diverse functions including transcriptional regulation via its lysine or arginine residue methylation. Studies indicate that deregulation of histone methylation is linked to human cancers including leukemia. Histone H3K27 methyltrnasferase response element II binding protein (RE-IIBP), as a transcriptional repressor to target gene IL-5, interacts with HDAC and is over-expressed in leukemia patient samples. In this study, we have identified that hematopoiesis-related genes GATA1 and HOXA9 are down-regulated by RE-IIBP in K562 and 293T cells. Transient reporter analysis revealed that GATA1 transcription was repressed by RE-IIBP. On the other hand, HOXA9 and PBX-related homeobox gene MEIS1 was up-regulated by RE-IIBP. These results suggest that RE-IIBP might have a role in hematopoiesis or leukemogenesis by regulating the transcription of target genes, possibly via its H3K27 methyltransferase activity.
Introduction
In eukaryote, chromatin structure plays a critical role in transcriptional regulation. Epigenetic modifications on either DNA or histone subunits are important factors for different transcriptional outcomes. It has been proposed that DNA methylation is involved in gene silencing and might play a role in carcinogenesis. As compared with normal cells, the cancer cells show major changes in their DNA methylation pattern (Bae and Kim Citation2008). The posttranslational modifications of histone N-terminus result in changes in chromatin structure and are related with transcription activation or repression (Li et al. Citation2007). Among the histone modifications, methylation of histone at specific residues, K4, K9, K27, and K36 on H3 and K20 on H4 is carried out by histone methyltransferases (HMTases) containing SET domain. Specificity of methylation sites on histone determines its biological effects including transcriptional regulation (Zhang and Reinberg Citation2001).
WHSC1/MMSET/NSD2 splice variants have been determined to be involved in certain human malignancies, including leukemia and multiple myeloma (MM). Among them, the response element II binding protein (RE-IIBP) has been reported to initiate translation at exon 15 of the transcript and possesses a SET domain within its C-terminal region (Garlisi et al. Citation2001). The expression of RE-IIBP appears to be universal and was shown to bind to response element II of the human interleukin-5 (IL-5) promoter and suppresses transcription (Garlisi et al. Citation2001; Keats et al. Citation2005). We have demonstrated that RE-IIBP represses IL-5 expression through histone H3K27 hypermethylation and H3 hypoacetylation around the promoter region (Kim et al. Citation2008). We further determined that the H3K27 HMTase RE-IIBP is up-regulated in the blood cells of various leukemic patients (Kim et al. Citation2008).
In this study, we have identified hematopoiesis-related genes regulated by RE-IIBP expression. GATA1 and HOXA9 gene expressions were down-regulated and MEIS1 gene expression was up-regulated by the RE-IIBP. Considering the importance of identified genes in regulation of blood cell differentiation and leukemia development, further research will identify a possible role of RE-IIBP in these processes.
Materials and methods
Cell culture and transient transfection
K562 cells were grown in RPMI-1640 and 293T cells were grown in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum and 0.05% penicillin–streptomycin at 37°C in a 5% CO2 atmosphere. K562 and 293T cells were transfected with the indicated DNA constructs for 48 hr using Lipofectamine 2000 (Invitrogen) or polyethylenimine (Sigma).
RNA isolation and RT-PCR
K562 and 293T cells were transfected with pcDNA6, pcDNA6–RE-IIBP, pSM2c-sh–RE-IIBP, and pcDNA6–RE-IIBP-N. Total RNA was isolated using RNAiso Plus (TaKaRa). RNA (5 µg) was reverse transcribed with oligo (dT) primer (Fermentas) and M-MLV Reverse Transcriptase (Enzynomics). The quantified cDNA was subjected to target gene mRNA expression pattern analysis. RT-PCR was performed using target gene specific primers. The sequences of primers used for PCR and reaction conditions are shown in .
Table 1. List of primer sets for RT-PCR.
Western blot analysis
K562 and 293T cells were transfected with pcDNA6, pcDNA6–RE-IIBP, pSM2c–sh-RE-IIBP, and pcDNA6–RE-IIBP-N. Total proteins were prepared from lysed cells using RIPA lysis buffer (50 mM Tris–HCl [pH8.0], 150 mM NaCl, 0.1% SDS, 0.5% SDC, 1% NP40, 1X protease inhibitor cocktail, and 1 mM EDTA), fractionated by 12% SDS–PAGE gel and transferred to nitrocellulose membranes. The membranes were probed overnight with α-GATA1, α-MEIS1 antibodies (Santa Cruz Biotechnology) at 4°C. The blots were incubated with HRP-conjugated α-goat antibody (Santa Cruz Biotechnology) and detected using an ECL system (Santa Cruz Biotechnology).
Luciferase assay
K562 and 293T cells were seeded at the density of 3.2×104 cells in 48 well plates and were transfected with pGL3-GATA1 promoter (100 ng), PMX-GATA1-FIG (200 ng), PMX-GATA1-FIG-ΔZF (100 ng, 200 ng) and pcDNA6-RE-IIBP (100 ng, 200 ng) using Lipofectamine 2000 (Invitrogen), or polyethylenimine (Sigma). After 48 hr of transfection, cells were collected and lysed in cell culture lysis reagent (Promega). Luciferase activities were measured by adding 15 µl luciferase assay substrate (Promega) into 80 µl of cell lysates using a Glomax luminometer (Promega). The results were confirmed by performing the experiment at least three times.
Statistical analysis
The data were presented as the means ± SD of three or more independent experiments. Statistical significance (P<0.05) was evaluated using Microsoft EXCEL software. Differences between groups were evaluated via one-way analysis of variance (ANOVA), followed by Student's t-tests.
Results and discussion
Identification of RE-IIBP target genes
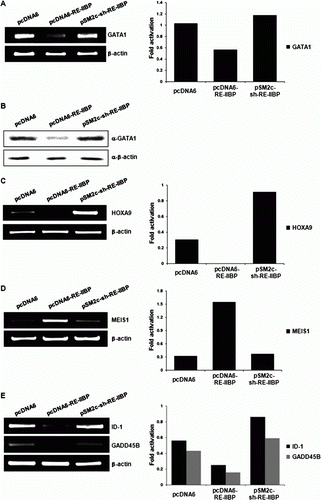
To investigate RE-IIBP target genes in hematopoiesis, we performed RT-PCR and Western blot for selected genes which have been known for their roles in blood cell differentiation. Transcription factor GATA1 has an important role in differentiation of erythroid cells. Over-expression of RE-IIBP significantly down-regulated GATA1 expression, in contrast knock down of RE-IIBP by sh-RE-IIBP RNA up-regulated the expression (A). Repression of GATA1 expression was further confirmed in the protein level by Western blot (B). Next, we looked at the expression level of one of the leukemogenesis-related gene, HOXA9. HOXA9 is a transcription factor and its translocation with the NUP98 gene has been associated with myeloid leukemogenesis. The expression of HOXA9 was almost completely repressed when RE-IIBP was over-expressed, however, knock down of RE-IIBP significantly increased HOXA9 expression (C). Furthermore, we tested the expression level of homeobox gene MEIS1. MEIS1 has diverse functions including early development and neoplasia. It has been known that MEIS1 forms triple complex with HOXA9 and PBX1 in myeloid cells (Shen et al. Citation1999). In contrast to GATA1 and HOXA9, RE-IIBP up-regulated MEIS1 expression (D). RE-IIBP knock down by sh-RE-IIBP RNA significantly down-regulated MEIS1 expression, which suggests positive regulatory role of RE-IIBP on MEIS1 expression (D). These results indicate that RE-IIBP might play both negative and positive regulatory roles in transcription depending on target genes. Recently, one of the RE-IIBP isoform, MMSET target genes were profiled using DNA microarray in erythroid leukemia cell line K562 (Hudlebusch et al. Citation2005). Among differently expressed genes, we screened helix-loop-helix protein inhibitor of differentiation (ID-1) and growth arrest and DNA-damage inducible beta (GADD45B) which are up-regulated by MMSET. In contrast to the result of differential expression profiling, we found that ID-1 and GADD45B expressions were repressed by RE-IIBP over-expression (E). Again, knock down of RE-IIBP restored both expressions (E). ID-1 can heterodimerize with members of the basic HLH family of transcription factors and can be used to mark endothelial progenitor cells which are critical to tumor growth and angiogenesis (Lyden et al. Citation1999). GADD45B has a function as stress sensor and is implicated in biological processes including DNA damage response, cell cycle arrest, apoptosis, and cell survival (Gupta et al. Citation2005).
Figure 1. RE-IIBP represses transcription of leukemogenesis-related genes. (A) mRNA level of GATA1 was regulated by RE-IIBP. K562 cells were transfected with pcDNA6, pcDNA6–RE-IIBP, and pSM2c-sh–RE-IIBP. Total mRNA was isolated from each cells and RT-PCR was performed using GATA1 primers. (B) GATA1 was regulated by RE-IIBP in protein level. Western blot was performed using α-GATA1 antibody in transfected K562 cells. β-Actin was used as a loading control. (C–E) 293T cells were transfected with pcDNA6, pcDNA6–RE-IIBP, and pSM2c-sh–RE-IIBP. RT-PCR was performed using HOXA9, MEIS1, ID-1, and GADD45B-specific primers. β-Actin was used as a loading control. The results are representative of at least two independent experiments. Each graph represents band intensity of target genes compared with β-Actin.
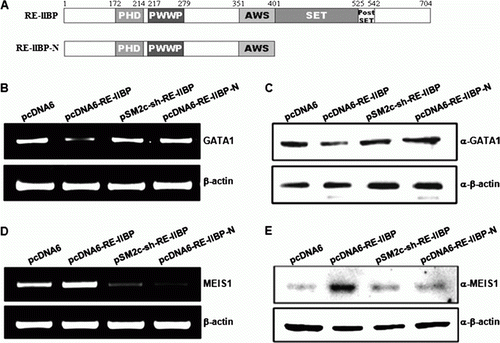
SET domain dependent regulation of target genes by RE-IIBP
The importance of the SET and post-SET domains in HMTase activity has been reported previously. Deletion of the post-SET domain drastically reduced the H3K27 HMTase activity of RE-IIBP, and deletion of the SET domain completely abolished enzyme activity (Kim et al. Citation2008). We further investigated whether RE-IIBP-mediated target gene regulation is SET domain dependent. We transfected wild type RE-IIBP and C-terminus deletion mutant excluding SET and post-SET domain to K562 and 293T cells and monitored the expression of target genes (A). GATA1 expression was down-regulated by RE-IIBP over-expression and knock down of RE-IIBP by sh-RE-IIBP RNA recovered the expression. When we transfected SET domain deletion mutant, expression level of GATA1 remained intact (B). Consistent results were obtained by Western blot suggesting the importance of HMTase activity in regulation of target gene GATA1 expression by RE-IIBP (C). When we looked at the expression of MEIS1, RE-IIBP up-regulated the MEIS1 expression and sh-RE-IIBP RNA and SET domain deletion mutant significantly down-regulated the MEIS1 expression (D). Again, similar patterns were observed in protein level by Western blot (E). Taken together, these results prove that the presence of SET domain-mediated intrinsic HMTase activity of RE-IIBP is important for the regulation of target gene expression.
Figure 2. Regulations of GATA1 and MEIS1 transcription by RE-IIBP are SET domain dependent. (A) Schematic view of wild type RE-IIBP and C-terminus deletion mutant. Full-length (amino acids 1-704) and RE-IIBP-N (amino acids 1-401). (B and C) GATA1 was down-regulated by RE-IIBP in SET domain-dependent manner. K562 cells were transfected with pcDNA6, pcDNA6–RE-IIBP, pSM2c-sh–RE-IIBP, and pcDNA6–RE-IIBP-N. RT-PCR and Western blot were performed with GATA1 specific primers and antibody. β-Actin was used as a loading control. (D and E) Up-regulation of MEIS1 by RE-IIBP was SET domain dependent. 293T cells were transfected with the indicated DNA constructs. RT-PCR and Western blot were performed with MEIS1 specific primers and antibody. β-Actin was used as a loading control. The results were confirmed by performing the experiment at least two times.
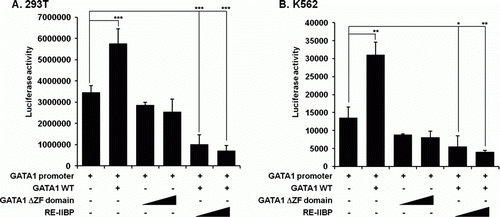
RE-IIBP negatively regulates GATA1 transcription
Erythroid transcription factor also known as GATA-binding factor 1 is a member of the GATA transcription factor family and is involved in cell growth and carcinogenesis (Ferreira et al. Citation2005). GATA1 is essential for erythroid and megakaryocytic development and knockout mice are embryonic lethal (Shivdasani et al. Citation1997). It contains three domains: the C-finger, the N-finger, and the activation domain. The C-finger, located in C-terminal, has a zinc finger DNA binding domain. Next, we investigated the effects of RE-IIBP on GATA1 promoter activity using reporter assay. Transient co-transfection experiments were conducted using pGL3-GATA1 luciferase reporter, PMX-GATA1-FIG, PMX-GATA1-FIG-ΔZF, and pcDNA6–RE-IIBP constructs in both 293T and K562 cells (). In this assay, luciferase activity was increased by GATA1 expression whereas the zinc finger-deleted GATA1 failed to elicit pGL3-GATA1 promoter-driven transactivation. This result confirmed the importance of the zinc finger domain of GATA1 (A and B). The elevated activity of the pGL3-GATA1 promoter in the presence of wild type GATA1 showed dramatic decrease in transcriptional activity when RE-IIBP was co-transfected in both cell lines (A and B). These results indicate that RE-IIBP functions as a negative transcriptional regulator of GATA1 expression.
Figure 3. RE-IIBP negatively regulates GATA1 transcription. (A and B) 293T and K562 cells were transfected with the pGL3-GATA1 promoter (100 ng), PMX-GATA1-FIG (200 ng), PMX-GATA1-FIG-ΔZF (100 ng, 200 ng) and pcDNA6–RE-IIBP (100 ng, 200 ng), and their cell extracts were assayed for luciferase activity. The pcDNA6 empty vector was used in all subsequent transfections as a negative control and was added to maintain equal amounts of total transfected DNA. Data in A and B are presented as mean ± SD; n=4. *P<0.05, **P<0.01, and ***P<0.001.
Previous reports suggested that MMSET has diverse lysine specificity including H3K4, 27, 36, and H4K20 (Kim et al. Citation2008; Marango et al. Citation2008; Pei et al. Citation2011). These reports suggest that MMSET regulates methylation level of diverse lysine residues directly or indirectly by interaction with other effectors. Another study suggests that these diversity or discrepancy on target lysine selection might be contributed by the nature of the substrates such as octamers versus nucleosomes, etc. (Li et al. Citation2009).
In this study, we selected list of genes involved in blood cell differentiation or leukemogenesis and tested gene expressions by H3K27 HMTase RE-IIBP. Among the tested genes, we found that zinc finger transcription factor GATA1 and homeobox containing transcription factor HOXA9 were down-regulated by RE-IIBP over-expression. These proteins have critical roles in hematopoietic cell development via transcriptional regulation of numerous target genes. We showed that RE-IIBP down-regulated GATA1 transcription, which indicated its possible role in GATA1-mediated hematopoiesis. We propose that the same mechanism is applied to transcriptional regulation of HOXA9. On the contrary, it is interesting that RE-IIBP up-regulated MEIS1 expression. MEIS1 expression is a strong inducer of caspase-dependent apoptosis via the interaction with cofactor PBX (Berthelsen et al. Citation1999). Further research such as the effects of RE-IIBP on MEIS1-mediated apoptosis, PBX dependence, and nuclear localization would reveal detailed role of HMTase RE-IIBP in leukemogenesis.
Acknowledgements
This work was supported by grants from Chung-Ang University.
References
- Bae , JB and Kim , YJ. 2008 . Cancer and epigenetics . Anim Cells Syst (Seoul) , 12 : 117 – 125 .
- Berthelsen , J , Kilstrup-Nielsen , C , Blasi , F , Mavilio , F and Zappavigna , V. 1999 . The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH . Genes Dev , 13 : 946 – 953 .
- Ferreira , R , Ohneda , K , Yamamoto , M and Philipsen , S. 2005 . GATA1 function, a paradigm for transcription factors in hematopoiesis . Mol Cell Biol , 25 : 1215 – 1227 .
- Garlisi , CG , Uss , AS , Xiao , H , Tian , F , Sheridan , KE , Wang , L , Motasim Billah , M , Egan , RW , Stranick , KS and Umland , SP. 2001 . A unique mRNA initiated within a middle intron of WHSC1/MMSET encodes a DNA binding protein that suppresses human IL-5 transcription . Am J Respir Cell Mol Biol , 24 : 90 – 98 .
- Gupta , M , Gupta , SK , Balliet , AG , Hollander , MC , Fornace , AJ , Hoffman , B and Liebermann , DA. 2005 . Hematopoietic cells from Gadd45a- and Gadd45b-deficient mice are sensitized to genotoxic-stress-induced apoptosis . Oncogene , 24 : 7170 – 7179 .
- Hudlebusch , HR , Theilgaard-Monch , K , Lodahl , M , Johnsen , HE and Rasmussen , T. 2005 . Identification of ID-1 as a potential target gene of MMSET in multiple myeloma . Br J Haematol , 130 : 700 – 708 .
- Keats , JJ , Maxwell , CA , Taylor , BJ , Hendzel , MJ , Chesi , M , Bergsagel , PL , Larratt , LM , Mant , MJ , Reiman , T Belch , AR . 2005 . Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients . Blood , 105 : 4060 – 4069 .
- Kim , JY , Kee , HJ , Choe , NW , Kim , SM , Eom , GH , Baek , HJ , Kook , H and Seo , SB. 2008 . Multiple-myeloma-related WHSC1/MMSET isoform RE-IIBP is a histone methyltransferase with transcriptional repression activity . Mol Cell Biol , 28 : 2023 – 2034 .
- Li , B , Carey , M and Workman , JL. 2007 . The role of chromatin during transcription . Cell , 128 : 707 – 719 .
- Li , Y , Trojer , P , Xu , CF , Cheung , P , Kuo , A , Drury , WJ 3rd , Qiao , Q , Neubert , TA , Xu , RM Gozani , O . 2009 . The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate . J Biol Chem , 284 : 34283 – 34295 .
- Lyden , D , Young , AZ , Zagzag , D , Yan , W , Gerald , W , O'Reilly , R , Bader , BL , Hynes , RO , Zhuang , Y Manova , K . 1999 . Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts . Nature , 401 : 670 – 677 .
- Marango , J , Shimoyama , M , Nishio , H , Meyer , JA , Min , DJ , Sirulnik , A , Martinez-Martinez , Y , Chesi , M , Bergsagel , PL Zhou , MM . 2008 . The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor . Blood , 111 : 3145 – 3154 .
- Pei , H , Zhang , L , Luo , K , Qin , Y , Chesi , M , Fei , F , Bergsagel , PL , Wang , L , You , Z and Lou , Z. 2011 . MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites . Nature , 470 : 124 – 128 .
- Shen , WF , Rozenfeld , S , Kwong , A , Kom ves , LG , Lawrence , HJ and Largman , C. 1999 . HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells . Mol Cell Biol , 19 : 3051 – 3061 .
- Shivdasani , RA , Fujiwara , Y , McDevitt , MA and Orkin , SH. 1997 . A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development . EMBO J , 16 : 3965 – 3973 .
- Zhang , Y and Reinberg , D. 2001 . Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails . Genes Dev , 15 : 2343 – 2360 .