ABSTRACT
Circular RNA (circRNA) is a non-coding RNA with a covalently closed loop structure and usually more stable than messenger RNA (mRNA). However, coding sequences (CDSs) following an internal ribosome entry site (IRES) in circRNAs can be translated, and this property has been recently utilized to produce proteins as novel therapeutic tools. However, it is difficult to produce large proteins from circRNAs because of the low circularization efficiency of lengthy RNAs. In this study, we report that we successfully synthesized circRNAs with the splint DNA ligation method using RNA ligase 1 and the splint DNAs, which contain complementary sequences to both ends of precursor linear RNAs. This method results in more efficient circularization than the conventional enzymatic method that does not use the splint DNAs, easily generating circRNAs that express relatively large proteins, including IgG heavy and light chains. Longer splint DNA (42 nucleotide) is more effective in circularization. Also, the use of splint DNAs with an adenine analog, 2,6-diaminopurine (DAP), increase the circularization efficiency presumably by strengthening the interaction between the splint DNAs and the precursor RNAs. The splint DNA ligation method requires 5 times more splint DNA than the precursor RNA to efficiently produce circRNAs, but our modified splint DNA ligation method can produce circRNAs using the amount of splint DNA which is equal to that of the precursor RNA. Our modified splint DNA ligation method will help develop novel therapeutic tools using circRNAs, to treat various diseases and to develop human and veterinary vaccines.
Introduction
Circular RNA (circRNA) is a type of non-coding RNA with a covalently closed loop structure (Jeck et al. Citation2013). Single-stranded circRNA was first discovered in viroids in 1976 (Sanger et al. Citation1976) and then later in eukaryotes in 1979 (Hsu and Coca-Prados Citation1979). circRNAs have higher stability than messenger RNA (mRNA) due to a lack of free 5′- and 3′-ends (Enuka et al. Citation2016). They are involved in various cellular pathways, such as ribosome biogenesis, regulation of microRNA (miRNA) activity and mRNA translation, and control of β-cell activities (Hansen et al. Citation2013; Holdt et al. Citation2016; Abdelmohsen et al. Citation2017; Stoll et al. Citation2018). circRNAs are also implicated in various human diseases, such as cancer, neurological disorders, cardiovascular diseases, chronic inflammatory diseases, and diabetes (Holdt et al. Citation2016; Hanan et al. Citation2017; Fang et al. Citation2018; Kristensen et al. Citation2018; Li et al. Citation2018; Qin et al. Citation2023).
Initially, circRNAs were thought to be unable to produce proteins because they do not have a 5′-cap structure required for mRNA translation initiation (Kozak Citation1979). However, it was later revealed that circRNAs with an internal ribosome entry site (IRES) could produce protein products (Chen and Sarnow Citation1995). Currently, circRNA is explored as a novel tool for generating proteins more stably than linear mRNA (Wesselhoeft et al. Citation2018).
There are three different ways of in vitro synthesis of circRNA to produce antigens or proteins are divided into chemical, ribozymatic, and enzymatic methods (Obi and Chen Citation2021). The head and tail of linear RNA can be joined by using chemicals such as cyanogen bromide (BrCN) or 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) (Moore and Query Citation2000). Using the permuted intron-exon (PIE) method, a kind of ribozymatic method, circRNAs are generated from the self-splicing Group I introns in the thymidylate synthase (Td) gene of T4 phage or pre-tRNALeu gene of Anabaena (Puttaraju and Been Citation1992; Ford and Ares Citation1994). The enzymatic method synthesizes circRNA by using T4 DNA ligase or T4 RNA ligase with splint DNA or splint RNA, both of which have partial complementary sequences to the head and tail of the linear precursor RNA. Thus, it can enhance circularization (Bain and Switzer Citation1992; Moore and Query Citation2000; Kershaw and O'Keefe Citation2012; Stark and Rader Citation2014). It was reported that the enzymatic method produces circRNA with lower immunogenicity than the PIE method (Liu et al. Citation2022). However, all of these methods exhibit lower circularization efficiency when longer precursor RNAs are used (Wesselhoeft et al. Citation2018; Obi and Chen Citation2021).
To increase the circularization efficiency of the enzymatic method, we identified that the enzymatic method with splint DNA could efficiently produce various proteins compared to the enzymatic method that does not use the splint DNA. In particular, our splint DNA ligation method demonstrated that circRNA can produce IgG antibody in vivo for the first time. Next, we used the longer splint DNAs or replaced adenine with an adenine analog, 2,6-diaminopurine (DAP) in the splint DNA. These modifications significantly increased the circularization efficiency in generating long circRNAs.
Material and methods
Reagents
DH5α chemically competent Escherichia coli cells (CP010), XbaI (R013L), 10X Tris/glycine/SDS (EBT005), 10X Tris/glycine (EBT006), 10X TBS (RBT004), TOPscript™ RT–PCR Kit (RT410S), and EZ™ MEGA T7 Transcription Kit (EZ039S) were purchased from Enzynomics. QIAfilter Plasmid Midi Kit (12243) and QIAquick PCR Purification Kit (28106) were purchased from Qiagen. T4 RNA Ligase 1 (M0204S) and Quick CIP (M0525L) were purchased from NEB. RNase R (ab286929) was purchased from Abcam. RiboRuler High Range RNA Ladder (SM1821), SuperSignal™ West Pico PLUS Chemiluminescent Substrate (34579), Gaussia Luciferase Glow Assay Kit (16161), Lipofectamine 3000 (L3000015), E-Gel™ EX 2% agarose gels (G401002), and HRP-conjugated goat anti-human IgG Fc cross-adsorbed secondary antibody (31314) were purchased from Thermo Scientific. GMP (G8377), formamide (F9037), and hexylamine (219703) were purchased from Sigma. Nitrocellulose (NC) membrane (1215471) was purchased from GVS. Splint DNA was synthesized by Macrogen (Seoul, Korea). pUC-GW-Kan-Gluc vector and IgG heavy chain (HC)−light chain (LC) vector were synthesized by Genewiz (South Plainfield, NJ, USA). DMEM (11995), FBS (16140089), Opti-MEM™ (31985-070), and penicillin/streptomycin (15140) were purchased from Gibco. Acetonitrile (9017-88) and glacial acetic acid (9522) were purchased from J.T. Baker.
In vitro-transcribed (IVT) template preparation
Template-transformed DH5α chemically competent cells were plated on LB plates supplemented with kanamycin (50 μg/ml) and were incubated at 37°C overnight. The colonies were inoculated into LB media containing kanamycin at 50 μg/ml and were cultured in a shaking incubator at 200 rpm, 37°C overnight. The template was purified using the QIAfilter Plasmid Midi Kit. In order to linearize the template, 15 μg of the template was reacted with 80 U of XbaI restriction enzyme at 37°C overnight. The linearized template was purified with the QIAquick PCR Purification Kit.
Precursor synthesis
IVT mixture (2 μg of linearization template, 2 μl of MEGA T7 enzyme mix, 2 μl of 10X MEGA T7 reaction buffer, 2 μl of 100 mM rATP, 2 μl of 100 mM rCTP, 2 μl of 100 mM rGTP, 2 μl of 100 mM rUTP, and 1.5 μl of 400 mM GMP, total volume to 20 μl) was prepared. After incubation at 37°C for 2 h, 2 μl of DNase I was added, followed by heating at 37°C for 15 min. Synthesized precursors were purified using the RNeasy Mini Kit.
circRNA synthesis
Precursor and splint DNA mixture was heated at 70°C for 5 min and then cooled down at room temperature for 5 min. Five microliters of T4 RNA ligase 1, 12 μl of 10X T4 RNA ligase reaction buffer, and 12 μl of 10 mM ATP were added, and the mixture was incubated at 37°C for 30 min. Two microliters of DNase I was added to the ligation reaction and incubated at 37°C for 15 min. After adding rCutSmart™ buffer (final 1X) and Quick CIP at a final concentration of 1 U/μg, it was incubated at 37°C for 15 min. Synthesized circRNAs were purified using the RNeasy Mini Kit.
RNase R treatment
Twenty micrograms of circRNA, RNase R buffer (final 1X), and RNase R at a final concentration of 0.25 U/μg were mixed, and the total volume was adjusted to 100 μl, followed by incubation at 37°C for 15 min. RNase R-treated circRNAs were purified using the RNeasy Mini Kit.
HPLC
circRNA was purified using an Agilent 1260 Infinity II LC system equipped with Agilent PLRP-S 4000 Å (10 × 250 mm) and Sepax SRT 2000 Å (10 × 3000 mm) columns. Mobile phase A buffer (100 mM HAA, pH 7) and mobile phase B (50% acetonitrile in mobile phase A buffer, pH 7) were prepared and filtered using a bottle top vacuum filter. The temperature of the thermostat attached to the Agilent PLRP-S 4000 Å (10 × 250 mm) column was set to 60°C. circRNA was purified by gradient elution (0 min, A 20%, B 80%; 30 min, A 0%, B 100%) with the mobile phases at a 2 ml/min flow rate. Fractions of the circRNA peak were collected and purified using the ethanol precipitation method. TE buffer (pH 6) for SEC column purification was filtered using a bottle-top vacuum filter. The temperature of the thermostat attached to the Sepax SEC 2000 Å (10 × 300 mm) column was set to 60°C. circRNA was purified under the mobile phase condition of A 100% at a 1.42 ml/min flow rate. Purified RNA was concentrated in an Amicon tube (10 kDa MWCO).
Electrophoresis assay for circRNA validation
After mixing 500 ng of precursor and circRNA in 10 μl of RNase-free water, the solution was then mixed with 10 μl of formamide. For the size marker, 2 μl of RiboRuler High Range RNA Ladder, 8 μl of RNase-free water, and 10 μl of formamide were combined and used. After reacting at 70°C for 5 min using a SimpliAmp thermal cycler, it was cooled down in ice for 5 min. The ladder and sample were loaded onto an E-Gel™ EX Agarose Gel 2% mounted on an E-Gel™ Power Snap Electrophoresis Device. Following electrophoresis, the gel was cooled for 15 min and imaged using the ChemiDoc™ XRS + System. The circularization efficiency was quantified by measuring the relative amounts of circRNA and precursor linear RNA.
Reverse transcription PCR
The PCR reaction mixture (5 μl of TOPscript™ One-step RT PCR Kit, 10 ng circRNA, 10 pmol of gene-specific primer, RNase-free sterile water) was prepared. The PCR conditions were as follows: reverse transcription at 50°C for 30 min; initial denaturation at 95°C for 10 min; annealing at 55°C for 30 s; elongation at 72°C for 30 s; number of cycles, 30; final elongation at 72°C for 5 min.
Cell culture and transfection
293T cells were maintained in DMEM supplemented with 10% heat-inactivated FBS. Cells were cultured overnight in a 24-well plate (2.5 × 105 cells/well). The next day, P3000 reagent solution (1 μl P3000, 500 ng circRNA in 25 μl of Opti-MEM™) and Lipofectamine 3000 solution (1.5 μl of Lipofectamine 3000 in 25 μl of Opti-MEM™) were mixed, followed by incubation for 10 min. The lipofectamine mixture was added to each well, and then the plate was incubated for 18 h (Chen et al. Citation2021).
Dot blot
IgG secreted from transfected cells into media was measured using a Bio-Dot microfiltration apparatus (Bio-Rad). First, the NC membrane was pre-wetted in 1X TBS buffer for 15 min. Then, the Bio-Dot microfiltration apparatus and membrane were assembled. Samples were loaded and filtered at 10 kPa pressure under vacuum. The blotted membrane was blocked with 5% skim milk in TBST for 1 h. HRP-conjugated goat anti-human IgG Fc cross-adsorbed secondary antibody diluted in 5% skim milk in TBST at 1:5000 was bound for 1 h. After washing five times with TBST, the experimental results were imaged using ECL.
Western blot
IgG secreted from transfected cells into media was measured using a Mini-Protein Tetra Cell (Bio-Rad). Samples were loaded on the prepared 12% acrylamide gel. The sample-loaded acrylamide gel was run at 100 V for 20 min and then at 150 V for 40 min. Afterward, the NC membrane was pre-wet in transfer buffer (1X Tris/glycine and 10% methanol) for 15 min. The protein in the acrylamide gel was transferred to the NC membrane using a Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad). The blotted membrane was blocked with 5% skim milk in TBST for 1 h. HRP-conjugated goat anti-human IgG Fc cross-adsorbed secondary antibody (1:5000 diluted in TBST containing 5% skim milk) was bound for 1 h. After washing five times with TBST, the experimental results were imaged using ECL (Jung et al. Citation2022).
Luciferase assay
Gaussia luciferase secreted from transfected cells into media was measured using the Gaussia Luciferase Glow Assay Kit. Luciferase assay working solution was prepared by diluting 100X coelenterazine to 1X in Gaussia Glow Assay Buffer. In a 96-well white plate, 10 μl of media sample was put into each well, followed by the addition of 50 μl of working solution. After incubation for 10 min, the light was measured with a microplate reader.
IgG ELISA assay
White 96-Well Immuno Plate (Thermo, 436110) was coated with human recombinant PD-1 (1 ug/mL in PBS) (R&D systems, 8986-PD-100), then incubated overnight at 4°C. The next day, the plate was washed 3 times with 200 ul of washing buffer (PBS containing 0.05% Tween 20) and blocked using 200 ul of blocking buffer (PBS containing 1% BSA) for 1 h at 37°C. Blocking buffer was removed from the plate and washed 3 times with 200 ul of washing buffer. Then, 100ul of 1:1000 dilution of each mouse plasma sample in blocking buffer, and the plate was incubated for 1 h at 37°C. After washing the plate 3 times, 100 ul of 1:5000 dilution of HRP-conjugated goat-anti-human IgG Fc antibody (Invitrogen, A1883) in blocking buffer were added to each well incubated for 1 h at 37°C and the plate was washed 5 times. HRP reaction was measured using ELISA femto substrate (Thermo, 37074).
In vivo expression validation in mice
The 7-week-old female ICR mice used in pharmacokinetic evaluation were obtained of SAMTAKOBIOKOREA, Inc (Osans-si, Republic of Korea) and raised in the animal facility of Nonclinical Research Center, QuBEST BIO Co., Ltd (Yongin-si, Republic of Korea) with the condition of specific pathogen free (SPF). LNP-IgG ringRNA at the doses of 1 mg/kg were intravenously (I.V.) treated to the mice via tail veins. The mouse plasma was collected from the orbit into heparin-coated capillaries and then further centrifuged at 12,000 rpm for approximately 2 min at 4 °C. The amount of IgG in the blood was measured through ELISA analysis and corrected by subtracting the day 0 sample value.
Results
Synthesis of circRNA using splint DNA and T4 RNA ligase 1
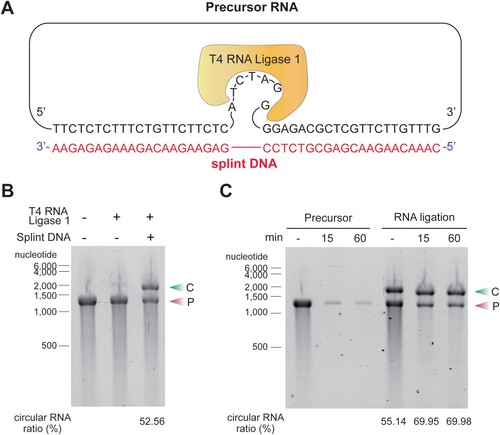
circRNA was generated by the splint DNA ligation method, which utilizes a ligation reaction using in vitro transcribed linear RNA, splint DNA, and T4 RNA ligase 1. The synthesized splint DNA has complementary sequences to both ends of the precursor linear RNA ((A)). The splint DNA with T4 RNA ligase 1 significantly improved the circularization efficiency compared to T4 RNA ligase 1 alone. Slower migrating circRNA was distinguished from faster migrating linear RNA by agarose gel electrophoresis ((B)). To confirm that the slow migrating band on the agarose gel were circular RNA, the RNA sample was treated with RNase R enzyme, an exonuclease that degrades linear RNA without affecting circRNA. RNase R treatment eliminated the fast-migrating band but did not degrade the slow-migrating band. This result confirms that most of the slow-migrating bands are circRNAs. RNase R treatment also increased the purity of circRNA from 55.14% to 69.98% ((C)).
Figure 1. Ligation of circular RNA (circRNA) using the splint DNA ligation method. (A) Splint DNAs that bind complementarily to the precursor RNAs were synthesized. When the splint DNA hybridizes with the precursor RNA, there are 6 nucleotide (nt) of single-stranded RNA at the 3′-end of the precursor and a single unbound RNA nucleotide at the 5′-end of the precursor RNA. T4 RNA ligase recognizes the free single-stranded RNA at the 3′-end of the precursor and the single unbound nucleotide at the 5′-end of the precursor and joins the ends. (B) Addition of splint DNA facilitates the production of circRNA. A circRNA synthesis reaction was performed using the T4 RNA ligase 1 enzyme with or without splint DNA. The reaction solutions were analyzed on an E-Gel EX 2% agarose gel. Circularization efficiency was quantified by measuring densities of circRNAs (upper band) and linear precursor RNAs (lower band). (C) circRNA formation was confirmed by RNase R treatment. Precursor RNA, with or without ligation reaction, was incubated with an RNase R, a 3′-to-5′ exonuclease, for the indicated minutes and then analyzed on an E-Gel EX 2% agarose gel. The experiments in B and C were performed in triplicate and a representative gel image for each experiment is presented. C, circular RNA; P, precursor RNA.
To ensure that the ligation was performed accurately at the desired location, we performed the sanger sequencing method and determined that cytidine residue (C) was added to the ligation site (Supplementary Data 1). This addition has been considered to occur during the IVT process rather than ligation step (Gholamalipour et al. Citation2018).
Protein production from circRNA generated by splint DNA ligation method
To determine whether the circRNA generated by our splint DNA ligation method can be used for protein production in cells, the precursor RNA containing the IRES of the CVB3 virus and the Gaussia luciferase (G. luciferase) gene was circularized by our splint DNA ligation method. The circularization efficiency was 52.40% ((A)). circRNA was transfected into 293T cells, and the luciferase protein production was determined by luciferase assay. The circRNA produced about 170 times more luciferase protein than the precursor RNA ((B)). These results demonstrate that our splint DNA ligation method generates circRNAs that can efficiently produce proteins.
Figure 2. Recombinant protein expression from circRNA synthesized by splint DNA ligation method. (A) A circRNA containing CVB3 IRES and Gaussia luciferase gene was formed from linear precursor RNA. Both precursor RNA and circRNA were visualized and quantified on an E-Gel EX 2% agarose gel. (B) Protein products from precursor RNA and circRNA were measured by luciferase assay. The relative luciferase values on the y-axis are a logarithmic scale. (C) circRNA containing an IgG heavy chain (HC) gene was generated by the splint DNA ligation method and transfected with 293T cells. (D) IgG HC protein was measured by western blot. (E) The circ RNAs containing CVB3 IRES and IgG HC gene as well as CVB3 IRES and IgG light chain (LC) gene, were generated from their respective linear precursor RNAs. Both precursor RNAs and circRNAs were visualized and quantified on an E-Gel EX 2% agarose gel. (F) Secreted IgG product amounts from circRNA. circRNAs expressing IgG HC and LC genes were co-transfected with 293T cells, and the cell culture media was collected at 48 h post-transfection. Secreted IgG was measured using dot-blot analysis. The left image is the dot blot result of the indicated amounts of IgG as a standard protein. The right image is the dot blot results showing the IgG produced from the circRNA in 293T-grown media. A total of 1.43 μg/ml of IgG in the right image was produced compared to the standard protein (left). The experiments were performed in duplicate. Data shown as mean ± s.d; indicated P value by two-tailed t-test. C, circular RNA; P, precursor RNA; W, whole cell; S, soluble fraction.
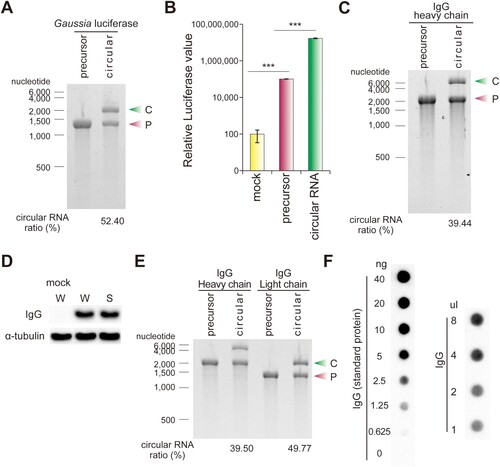
Monoclonal antibodies are extensively employed as therapeutic agents. Therefore, we sought to investigate whether our circRNA could also efficiently express antibodies in both cell culture and animal models. A circRNA containing the IgG heavy chain (HC, Pembrolizumab) gene and an IRES were synthesized by our splint DNA ligation method ((C)), which produced the antibody HC in a soluble form ((D)). Both HC and light chain (LC, Pembrolizumab) must be present together to express and secrete functional antibodies, thus we also generated a circRNA expressing the IgG LC by our splint DNA ligation method ((E)). We next performed dot blot analysis to determine the antibody concentration secreted in the media. Based on a comparison with a dilution series of recombinant IgG (the left image in (F)), the concentration of secreted IgG in the media was estimated to be approximately 1.43 μg/ml IgG (the right image in (F)).
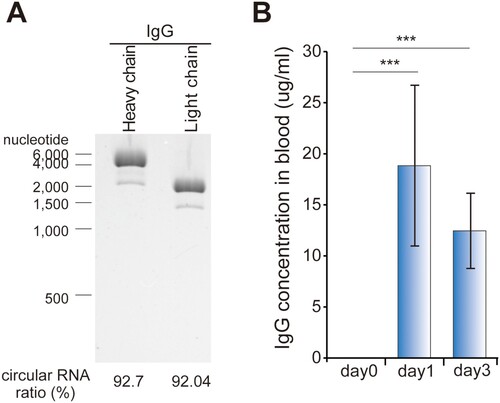
To determine that our circRNA could produce antibodies in animals besides cell culture, each circRNA expressing IgG HC and LC was purified by HPLC ((A)), encapsulated in lipid nanoparticles (LNPs), and then injected intravenously into mice. On the first day post-injection, 18.84 μg/ml of IgG protein was detected in the blood of mice, and the IgG protein concentration was maintained at 12.45 μg/ml on the third-day post-injection ((B)).
Figure 3. In vivo expression of IgG using circRNA. (A) The circRNA shown in (E) was purified using HPLC to a purity of ≥90%. (B) The circRNA was encapsulated in lipid nanoparticles (LNPs) and intravenously injected into mice. Blood IgG production was quantified by ELISA. The IgG concentration at each day is represented by subtracting the concentration measured at day 0 from the measured concentration at each indicated day. The experiment in A and B were performed in duplicate and triplicate, respectively. Data shown as mean ± s.d; indicated P value by two-tailed t-test.
circRNA synthesis containing genes with various length
In most of the circularization protocols, the efficiency of RNA circularization decreases as the length of the RNA precursor increases (Wesselhoeft et al. Citation2018). To determine the effect of precursor length on the efficiency of splint DNA-mediated circularization, the precursors with various lengths were evaluated. The circularization efficiency using splint DNA decreased from 42.06% to 15.08% as the length of the precursor increased from 1300 nucleotides to 4500 nucleotides ().
Figure 4. Ligation efficiencies of precursors containing coding sequences (CDS) of different sizes. Precursors with the indicated lengths were ligated using T4 RNA ligase 1 and 26-nt splint DNA (splint 2-1 in ). Precursors and corresponding circularization reaction products were visualized and quantified by E-Gel EX 2% agarose gel analysis. Ligation efficiency was calculated as the ratio of the band intensity of circRNA to precursor RNA. The experiment was performed in duplicate. C, circular RNA; P, precursor RNA.
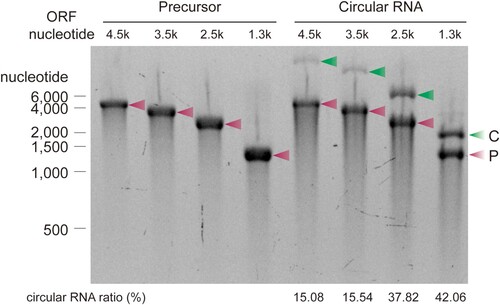
Table 1. The list of splint DNA.
circRNA synthesis using splint DNA containing modified nucleotides
Circularization efficiency decreases as the length of the precursor RNA increases (). This characteristic can be problematic when attempting to express relatively long genes using the circRNA. To overcome this problem, we tested both longer splint DNA and the splint DNA with modified nucleotides, such as an adenine analog, DAP ((A)). DAP can increase the binding force between complementary sequences of splint DNA and precursor RNA because it forms three hydrogen bonds with thymine (Cristofalo et al. Citation2019). As the splint DNA length increased from 26 nt to 42 nt, the circularization efficiency was improved by approximately 1.67-fold ((B)). The 26-nt modified splint DNA containing DAP also resulted in an approximate 1.67-fold increase in circularization efficiency compared to the same length of splint DNA ((B)).
Figure 5. Diaminopurine splint DNA improves circularization efficiency. (A) Hydrogen bondings in the adenine−thymine and the diaminopurine (DAP)−thymine base pairs. (B) circRNA ligation efficiencies in the ligation methods using the splint DNA or DAP splint DNA. A precursor having a 3.5-kb CDS was ligated by T4 RNA ligase 1 using splint DNA and DAP-splint DNA of the indicated lengths, respectively. Circularization reaction products were quantified, and the relative ligation efficiencies were calculated, as shown in the graph. The characteristics and sequences of each splint DNA are described in . (C) Ligation efficiency using various splint DNA and precursor ratios. Circularization reactions were performed using splint DNA and precursors at the indicated ratios. The efficiency of synthesis of circRNA was measured through E-Gel EX 2% agarose gel analysis. 26-nt splint DNA and DAP-splint DNA are 2-5 and 3-5 splint DNA in , respectively. DAP splint DNA, DAP-contained modified splint DNA. The experiment was performed in triplicate. Data shown as mean ± s.d; indicated P value by two-tailed t-test (B, C). For gel source data, see Supplementary .
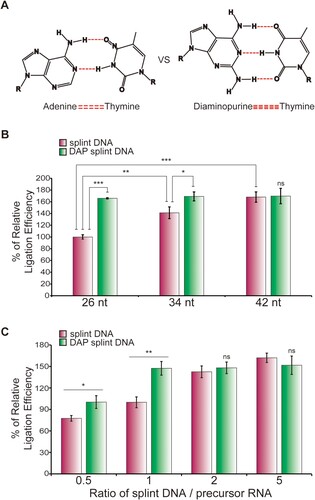
Alteration in the splint DNA to precursor RNA ratio also caused significant changes in the circularization efficiency ((C)). Increasing the ratio of the splint DNA to the precursor RNA from 0.5 to 1 resulted in a 1.29-fold increase in circularization efficiency. Further increasing the ratio from 1 to 2 augmented the circularization efficiency by 1.47-fold. Notably, even at the lower ratio of 1, there was still a significant production of circRNA when the splint DNA contained DAP.
Taken together, these data indicate that if these modified splint DNA ligation methods are used for circRNA synthesis, the circularization efficiency of long precursors can be increased, even when a relatively small amount of short splint DNA is used.
Discussion
During the coronavirus disease 2019 (COVID-19) pandemic, the advantages of mRNA vaccines have become apparent, leading to the exploration of various mRNA-based drug development strategies, such as protein replacement therapies, CAR-T cell therapies, therapeutic antibodies, and cancer vaccines (Sun et al. Citation2023). However, mRNA vaccines have several limitations. First, due to the instability of mRNA and its high susceptibility to degradation by RNase, mRNA vaccines require ultra-low temperatures for storage and transportation (Uddin and Roni Citation2021; Korosec et al. Citation2022). Second, the delivery of exogenous mRNA into cells could be recognized by intracellular pattern recognition receptors, such as RIG-I, leading to unwanted inflammation, suppression of its translation, and accelerated decay of the mRNA as an antiviral defense mechanism (Akira Citation2012; Frederickson and Herzog Citation2021). Third, additional nucleotide modification is required to improve the stability of the mRNA produced by in vitro transcription (Crommelin et al. Citation2021).
The high stability of circRNA presents a promising approach to solving stability concerns in mRNA-based vaccine development (Chaudhary et al. Citation2021). An extensive repertoire of innate immune receptors has evolved to detect and respond to foreign substances (Hawash et al. Citation2021; Buckley and Yoder Citation2022). Notably, nucleic acid receptors have undergone significant evolutionary adaptations to recognize the DNA or RNA of intracellular intruders, such as viruses. The stable closed-loop structure of circRNA could shield it from recognition of RNA 5′- or 3′-end by innate immune receptors as well as impede degradation (Wesselhoeft et al. Citation2019; He et al. Citation2021). As a result, circRNA could exhibit slower decay rates without induction of strong inflammation compared to the linear form of mRNA. In this regard, Qu et al. recently reported a circRNA-based vaccine that significantly increased the amount and duration of the receptor-binding domain (RBD) of SARS-CoV-2 spike protein, resulting in the generation of potent neutralizing antibodies against RBD. Furthermore, there was no significant reduction in RBD expression, even when circRNA was stored at room temperature for 2 weeks (Qu et al. Citation2022).
However, the technology for generating circRNA from linear RNA is still in the developmental stage, and various techniques, such as the PIE method and the RNA ligase method, are being developed to enhance its efficiency (Wesselhoeft et al. Citation2018).
In this study, we demonstrated that splint DNA and T4 RNA ligase 1 significantly improved the circularization efficiency compared to T4 RNA ligase 1 alone. This splint DNA ligation method can produce various proteins from the circRNA and presents efficient protein production in both cell cultures and animal models. The circRNA containing the G. luciferase gene showed a significant increase in luciferase protein expression compared to the precursor RNA alone ((B)). Furthermore, circRNA containing the IgG HC and LC genes successfully expressed soluble antibodies in cell culture, and the simultaneous in vivo expression of IgG HC and LC using LNPs resulted in the secretion of functional antibodies in the blood ((B)). These findings highlight the potential of circRNA as a platform for protein expression, including therapeutic antibodies.
The synthesis of circRNA becomes more challenging as the length of the precursor RNA increases. Previous studies reported decreased circularization efficiency with longer precursors (Wesselhoeft et al. Citation2018). Our results also demonstrated reduction in circularization efficiency as the precursor length increased (). To overcome this limitation, we explored the use of longer splint DNA and modified nucleotides, specifically DAP, in the splint DNA sequences. The inclusion of DAP in the splint DNA resulted in thermodynamically more stable binding with the precursor RNA (Cristofalo et al. Citation2019). As a result, both longer splint DNA and DAP-modified splint DNA showed increased circularization efficiency compared to shorter splint DNA ((B)). Moreover, variations in the ratio of splint DNA to precursor RNA also influenced the circularization efficiency, with higher ratios leading to improved efficiency ((C)). These findings suggest that optimizing the length and composition of the splint DNA can enhance the circularization of longer RNA precursors.
In conclusion, this study demonstrated the successful synthesis of circRNA using the splint DNA ligation method with T4 RNA ligase 1 and showed efficient protein production capability using the synthesized circRNA. Additionally, modifications in the splint DNA ligation method, such as longer length and the incorporation of DAP, improved the circularization efficiency, especially for longer RNA precursors. These findings can contribute to the development of circRNA as a robust tool for protein expression and highlight the potential for its application in various research and therapeutic contexts in humans and animals.
Supplemental Material
Download Zip (9.9 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, et al. 2017. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 14(3):361–369. doi:10.1080/15476286.2017.1279788.
- Akira S. 2012. The role of mRNA degradation in immunity and inflammation. Arthritis Res Ther. 14(1):O7. doi:10.1186/ar3562.
- Bain JD, Switzer C. 1992. Regioselective ligation of oligoribonucleotides using DNA splints. Nucleic Acids Res. 20(16):4372–4372. doi:10.1093/nar/20.16.4372.
- Buckley KM, Yoder JA. 2022. The evolution of innate immune receptors: investigating the diversity, distribution, and phylogeny of immune recognition across eukaryotes. Immunogenetics. 74(1):1–4. doi:10.1007/s00251-021-01243-4.
- Chaudhary N, Weissman D, Whitehead KA. 2021. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat Rev Drug Discov. 20(11):817–838. doi:10.1038/s41573-021-00283-5.
- Chen C-Y, Sarnow P. 1995. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 268(5209):415–417. doi:10.1126/science.7536344.
- Chen J, Sun Y, Huang S, Shen H, Chen Y. 2021. Grub polypeptide extracts protect against oxidative stress through the NRF2-ARE signaling pathway. Anim Cells Syst. 25(6):405–415. doi:10.1080/19768354.2021.2018043.
- Cristofalo M, Kovari D, Corti R, Salerno D, Cassina V, Dunlap D, Mantegazza F. 2019. Nanomechanics of diaminopurine-substituted DNA. Biophys J. 116(5):760–771. doi:10.1016/j.bpj.2019.01.027.
- Crommelin DJA, Anchordoquy TJ, Volkin DB, Jiskoot W, Mastrobattista E. 2021. Addressing the cold reality of mRNA vaccine stability. J Pharm Sci. 110(3):997–1001. doi:10.1016/j.xphs.2020.12.006.
- Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. 2016. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 44(3):1370–1383. doi:10.1093/nar/gkv1367.
- Fang Y, Wang X, Li W, Han J, Jin J, Su F, Zhang J, Huang W, Xiao F, Pan Q, et al. 2018. Screening of circular RNAs and validation of circANKRD36 associated with inflammation in patients with type 2 diabetes mellitus. Int J Mol Med. 42(4):1865–1874. Eng.
- Ford E, Ares M Jr. 1994. Synthesis of circular RNA in bacteria and yeast using RNA cyclase ribozymes derived from a group I intron of phage T4. Proc Natl Acad Sci USA. 91(8):3117–3121. Eng. doi:10.1073/pnas.91.8.3117.
- Frederickson R, Herzog RW. 2021. RNA-based vaccines and innate immune activation: not too hot and not too cold. Mol Ther. 29(4):1365–1366. doi:10.1016/j.ymthe.2021.03.005.
- Gholamalipour Y, Karunanayake Mudiyanselage A, Martin CT. 2018. 3’ end additions by T7 RNA polymerase are RNA self-templated, distributive and diverse in character-RNA-seq analyses. Nucleic Acids Res. 46(18):9253–9263. doi:10.1093/nar/gky796.
- Hanan M, Soreq H, Kadener S. 2017. CircRNAs in the brain. RNA Biol. 14(8):1028–1034. doi:10.1080/15476286.2016.1255398.
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. 2013. Natural RNA circles function as efficient microRNA sponges. Nature. 495(7441):384–388. doi:10.1038/nature11993.
- Hawash MBF, Sanz-Remon J, Grenier JC, Kohn J, Yotova V, Johnson Z, Lanford RE, Brinkworth JF, Barreiro LB. 2021. Primate innate immune responses to bacterial and viral pathogens reveals an evolutionary trade-off between strength and specificity. Proc Natl Acad Sci USA. 118(13):e2015855118. doi:10.1073/pnas.2015855118.
- He AT, Liu J, Li F, Yang BB. 2021. Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduct Target Ther. 6(1):185. doi:10.1038/s41392-021-00569-5.
- Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, et al. 2016. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 7:12429. doi:10.1038/ncomms12429.
- Hsu M-T, Coca-Prados M. 1979. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 280(5720):339–340. doi:10.1038/280339a0.
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. 2013. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 19(2):141–157. doi:10.1261/rna.035667.112.
- Jung H, Kim YS, Jung DM, Lee KS, Lee JM, Kim KK. 2022. Melittin-derived peptides exhibit variations in cytotoxicity and antioxidant, anti-inflammatory and allergenic activities. Anim Cells Syst. 26(4):158–165. doi:10.1080/19768354.2022.2099971.
- Kershaw CJ, O'Keefe RT. 2012. Splint ligation of RNA with T4 DNA ligase. Methods Mol Biol. 941:257–269. doi:10.1007/978-1-62703-113-4_19.
- Korosec CS, Farhang-Sardroodi S, Dick DW, Gholami S, Ghaemi MS, Moyles IR, Craig M, Ooi HK, Heffernan JM. 2022. Long-term durability of immune responses to the BNT162b2 and mRNA-1273 vaccines based on dosage, age and sex. Sci Rep. 12(1):21232. doi:10.1038/s41598-022-25134-0.
- Kozak M. 1979. Inability of circular mRNA to attach to eukaryotic ribosomes. Nature. 280(5717):82–85. doi:10.1038/280082a0.
- Kristensen LS, Hansen TB, Veno MT, Kjems J. 2018. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 37(5):555–565. doi:10.1038/onc.2017.361.
- Li H, Li K, Lai W, Li X, Wang H, Yang J, Chu S, Wang H, Kang C, Qiu Y. 2018. Comprehensive circular RNA profiles in plasma reveals that circular RNAs can be used as novel biomarkers for systemic lupus erythematosus. Clin Chim Acta. 480:17–25. doi:10.1016/j.cca.2018.01.026.
- Liu CX, Guo SK, Nan F, Xu YF, Yang L, Chen LL. 2022. RNA circles with minimized immunogenicity as potent PKR inhibitors. Mol Cell. 82(2):420–434 e426. doi:10.1016/j.molcel.2021.11.019.
- Moore MJ, Query CC. 2000. [7] Joining of RNAs by splinted ligation. Methods Enzymol. 317:109–123. doi:10.1016/S0076-6879(00)17009-0.
- Obi P, Chen YG. 2021. The design and synthesis of circular RNAs. Methods. 196:85–103. doi:10.1016/j.ymeth.2021.02.020.
- Puttaraju M, Been MD. 1992. Group I permuted intron-exon (PIE) sequences self-splice to produce circular exons. Nucleic Acids Res. 20(20):5357–5364. Eng. doi:10.1093/nar/20.20.5357.
- Qin K, Zhang F, Wang H, Wang N, Qiu H, Jia X, Gong S, Zhang Z. 2023. circRNA circSnx12 confers cisplatin chemoresistance to ovarian cancer by inhibiting ferroptosis through a miR-194-5p/SLC7A11 axis. BMB Rep. 56(2):184–189.
- Qu L, Yi Z, Shen Y, Lin L, Chen F, Xu Y, Wu Z, Tang H, Zhang X, Tian F, et al. 2022. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell. 185(10):1728–1744 e1716. doi:10.1016/j.cell.2022.03.044.
- Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. 1976. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 73(11):3852–3856. doi:10.1073/pnas.73.11.3852.
- Stark MR, Rader SD. 2014. Efficient splinted ligation of synthetic RNA using RNA ligase. Methods Mol Biol. 1126:137–149. doi:10.1007/978-1-62703-980-2_10.
- Stoll L, Sobel J, Rodriguez-Trejo A, Guay C, Lee K, Veno MT, Kjems J, Laybutt DR, Regazzi R. 2018. Circular RNAs as novel regulators of beta-cell functions in normal and disease conditions. Mol Metab. 9:69–83. doi:10.1016/j.molmet.2018.01.010.
- Sun H, Zhang Y, Wang G, Yang W, Xu Y. 2023. mRNA-based therapeutics in cancer treatment. Pharmaceutics. 15(2):622. doi:10.3390/pharmaceutics15020622.
- Uddin MN, Roni MA. 2021. Challenges of storage and stability of mRNA-based COVID-19 vaccines. Vaccines. 9(9):1033. doi:10.3390/vaccines9091033.
- Wesselhoeft RA, Kowalski PS, Anderson DG. 2018. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 9(1):2629. doi:10.1038/s41467-018-05096-6.
- Wesselhoeft RA, Kowalski PS, Parker-Hale FC, Huang Y, Bisaria N, Anderson DG. 2019. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol Cell. 74(3):508–520 e504. doi:10.1016/j.molcel.2019.02.015.
