ABSTRACT
Harmful substances like the cyanotoxin microcystin-leucine-arginine (MC-LR) are commonly found in eutrophic freshwater environments, posing risks to aquatic organisms. The water flea, Daphnia, is a well-established model organism for environmental toxicology research. Nevertheless, there is currently insufficient research on the genes that respond to MC-LR in Daphnia galeata. This study aimed to gain insights into the notable genes that react significantly to MC-LR. In this study, we generated an extensive RNA-Seq sequences isolated from the D. galeata HK strain, Han River in Korea. This strain was nourished with a diet of the green microalga Chlorella vulgaris and treated with pure MC-LR at a concentration of 36 ug/L. The transcriptome profile in response to the MC-LR treatment was obtained and 336 differentially expressed genes were subjected to Gene Ontology (GO) and euKaryotic Orthologous Groups of proteins analyses. GO enrichment analysis showed that chemical stimulus, amino sugar metabolic and catabolic process, oxidative stress, and detoxification were highly enriched, in reverse, proteolysis and fucosylation were underpresented. Detoxification process related genes such as peroxidase-like, chorion, and thyroid peroxidase-like were enriched for eliminating or neutralizing MC_LR from an organism's body. Furthermore, functional protein classification revealed an upregulation of lipid and inorganic ion transport processes, while amino acid and carbohydrate transport processes were found to be downregulated. These findings offer insights into how organisms respond to ecotoxic stimuli, providing valuable information for understanding adaptation or defense pathways.
Introduction
Daphnia are considered an indicator species of water toxicity (Wan et al. Citation2021), and their survival and reproductive success can reflect the health of aquatic ecosystems (Dao et al. Citation2010; Cao et al. Citation2014; Henning-Lucass et al. Citation2016). Studying the survival of Daphnia exposed to environmental toxins is of great importance in the field of ecotoxicology (Cui et al. Citation2018; Zimmermann et al. Citation2020; Wan et al. Citation2021). Researchers gain insights into the overall state of aquatic environments by understanding how environmental toxins affect the development, reproduction, growth, and life cycles of Daphnia (Lurling Citation2003; Herrera et al. Citation2015; Schwarzenberger and Martin-Creuzburg Citation2021). Excessive nutrient input to freshwater has increased the frequency of cyanobacterial blooms, which produce biological toxins referred to cyanotoxins that affect the feeding, growth, and life history parameters of the grazer Daphnia (Ma et al. Citation2014; Huisman et al. Citation2018; Feist and Lance Citation2021).
Previous studies have shown that when an organism is exposed to microcystin-leucine-arginine (MC-LR), a common cyanotoxin, its cells can attempt to detoxify the toxin by conjugating it with glutathione (GSH). This process involves the binding of MC-LR to the thiol (sulfhydryl) group of GSH, forming a covalent bond (Plugmacher et al. Citation1998). The resulting glutathione conjugate is generally less toxic than the original MC-LR molecule, which helps protect the organism from the harmful effects of the toxin. The glutathione conjugate of MC-LR can then be further processed and ultimately eliminated from the organism's body through various biochemical pathways, including transport and excretion mechanisms. This detoxification process is an essential defense mechanism that organisms employ to counteract the toxic effects of MC-LR and other xenobiotics (foreign substances). However, the efficiency of this detoxification process can vary among species, and some may be more susceptible to the harmful effects of MC-LR than others. Understanding the detoxification pathways of MC-LR and its interactions with cellular components, like glutathione, is of great interest in environmental and toxicological research, particularly in the context of aquatic ecosystems where cyanobacterial blooms and microcystin contamination can occur.
The water flea Daphnia is a well-established model organism for environmental toxicology. Nevertheless, there is currently insufficient research on the genes that respond to MC-LR in Daphnia galeata, a widespread Daphnia species found in Korea. Therefore, to gain insights into these genes, we generated an extensive RNA-Seq dataset and performed a comparative transcriptome analysis. The RNA-Seq method provides high-resolution data on gene expression and allows the identification of novel transcripts. This ecotoxicology experiment provides opportunities for developing predictive models and identifying biomarkers of stress response for freshwater assessments in Korea using Daphnia galeata.
Materials and methods
Field sample collection and laboratory culture
Zooplankton samples from Han River in Korea were collected in a layer 0–1 m deep using a plankton net with an opening diameter of 30 cm and mesh size of 50 µm. The sampling site was located at 37°34’50” N 127°10’42” E (). On 7 June 2023, the field environmental conditions included a temperature of 24°C, pH of 7.27, dissolved oxygen (DO) of 5.2 mg/L, electrical conductivity (EC) of 372 µs/cm, and total dissolved solids (TDS) of 186 ppm. Daphnia (Daphnia) galeata Sars, 1863 (Kim Citation1988; Tanaka Citation1992; Zuykova et al. Citation2010; Kirdiashcheva and Kotov Citation2013; Błędzki and Rybak Citation2016) is a dominant species in the Han River of Korea (NIER Citation2006, Citation2017). The D. galeata collected from Han River in Korea was named D. galeata HK clone for our laboratory culture. This HK clone was classified according to Daphnia (Daphnia) galeata Sars, 1863. We used diagnostic characters – such as the presence of an ocellus, a pointed head helmet, the rostrum shape, particular features of ephippial females, and the presence of postabdominal claw without fine external combs in which the spine size of combs 1, 2, and 3 are equal (). The classification was subsequently confirmed using molecular phylogenetic analysis (). Female individuals were isolated with the aid of a stereomicroscope SZ40 (Olympus, Japan) for parthenogenetic reproduction. In the laboratory, all clones were cultured at 21 ± 1°C and a light intensity ranging from 540 to 1000 lux with a 16:8 light:dark photoperiod. For the clonal culture, five to ten clones per 0.5 L of moderately hard water were maintained (NIER Citation2017). The culture medium was renewed twice a week and prepared a day before use, with DO concentrations ranging from 5.5 to 8.0 mg/L. Daphnia HK clones were fed every three days with a mixture of the pure green alga Chlorella vulgaris and YCT (Yeast, Alfalfa, and Tetramin) (EvergreenTOP, Korea).
Figure 1. Sampling location of Daphnia galeata HK in Korea. The small orange box in the left panel corresponds to the large orange box (right panel). The white dot in the right panel indicates the sampling site (37°34'50"N, 127°10'42"E).
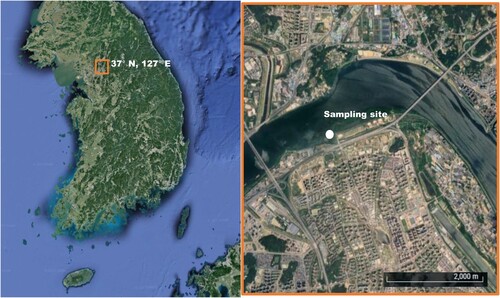
Figure 2. Morphological characteristics of female Daphnia galeata HK under a microscope. (a) A lateral view showing the broad oval carapace and bluntly pointed helmet. (b) A dorsal view. (c) A close-up view of the head showing the reduced antennules and only sensory setae protruding from under the pronounced rostrum. The white arrow indicates the large compound eye, the blue arrow indicates the ocellus, and the black arrow indicates sensory setae. (d) The postabdominal claw is shown with its fine, evenly sized combs. (e) A close-up view of an ephippium showing two ephippial eggs inside.

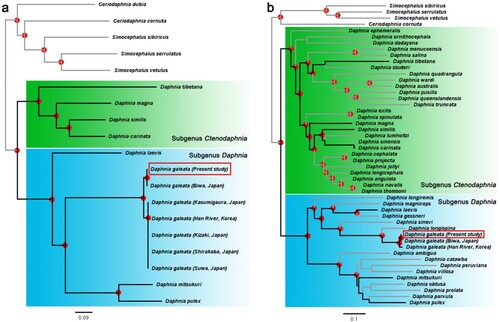
Figure 3. Maximum-likelihood phylograms based on Daphnia mitochondrial gene sequences. (a) A phylogenetic tree inferred based on an alignment of the nucleotide sequences of 13 mitochondrial genes. (b) A phylogenetic tree inferred based on an alignment of the partial nucleotide sequences of the mitochondrial 12S and 16S rRNA genes and the COX1 gene. In (b), the individuals used in the phylogeny of (a) are indicated by black bold lines. Nodes with the bootstrap support values greater than 85% are indicated with a red dot. The D. galeata HK clone used in this study is indicated with a red box. Species from the genera Simocephalus and Ceriodaphnia were used as the outgroups.
The growth observations of Daphnia HK strain
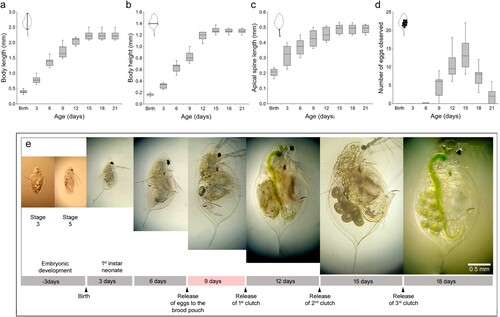
To record growth stages, we measured the body length, height, and apical spine length on days 3, 6, 9, 12, 15, 18 and 21. Also, we monitored the reproduction success of Daphnia by counting the number of observed eggs. For these experiments, five Daphnia possessing 8 or 10 eggs, isolated using a microscope, were cultured in 500 ml of the culture medium. The next day, neonates were collected to perform a semi-static test. Ten neonates cultured in 500 ml of culture medium were fed every 3 days, and the medium was renewed every 3 days to measure morphological sizes and count the number of eggs (). After 9 days, the first instar and subsequent neonates were separated from their mothers and cultured separately.
Figure 4. Boxplots of the growth and reproduction characteristics of Daphnia galeata HK and photomicrographs showing growth stages. (a) Body length measured from the point of the helmet to the base of the apical spin. (b) Body height measured from the dorsal to the ventral side of the body at its widest point. (c) Apical spine length measured from the base to the tip. (d) The number of eggs produced by D. galeata HK. (e) Embryonic development stages 3 and 5 and 3-, 6-, 9-, 12-, and 18-day-old adult daphnids. Below the photos, there is a timeline annotating the life stages based on studies of D. magna (Campos et al. Citation2018). To measure the sizes, we used 10 Daphnia. The relevant morphometric measurement ranges are illustrated in the upper left of each boxplot.
Short term (24 h) exposure to MC-LR
For this experiment, we used 9-day-old mature D. galeata HK (). The mRNAs of Daphnia HK clones exposed to MC-LR were sequenced to obtain transcript expression. For the MC-LR exposure experiment, a concentration of 36 µg/L MC-LR (MC-LR) was used, and a positive control (EtOH; 36 µl/L EtOH) and a negative control (Ctrl; no treatment) were included. After treatment, cultures were incubated for 24 h in 50 ml of the test medium with 15 Daphnia HK clones that had reached maturity (8–9 days of age). The MC-LR (> 95% purity, ALX-350-012) was purchased from Enzo Biochem (USA) and dissolved in an organic solvent, absolute ethanol (99.9%) at a ratio of 1:1 (w/v). The dissolved MC-LR was stored at −20°C for 6 months.
Transcriptome sequencing, data filtering and de novo assembly
After the toxicant exposure tests, surviving daphnids in each condition were retrieved and immediately preserved in 1.8 ml of RNAlater™ Stabilization Solution (Thermo Fisher Scientific, USA). Short-read mRNA sequencing was performed on multiple individuals of D. galeata: 15 individuals each from the Et-OH treatment and Ctrl groups, and 14 from the MC-LR treatment group. For each experiment group, a paired-end library was generated using the TruSeq Stranded mRNA library kit (Illumina, USA) with an insert size of approximately 155 bp (Jung et al. Citation2020; Kim et al. Citation2022; Jeon et al. Citation2023). The mRNA sequencing was conducted on the Novaseq6000 platform (Illumina, USA), generating 101 bp reads from both ends. Sequencing data filtering was performed using fastp (ver. 0.23.4) (Chen et al. Citation2018). Adapter sequences (–detect_adapter_for_pe) and bases with a mean quality lower than Q20 (–cut_mean_quality 20, –cut_window_size 4) were removed. In addition, reads shorter than 75 bp (–length_required 75) and/or that contained an undetermined base ‘N’ (–n_base_limit 0) were also filtered out. De novo transcriptome assembly was carried out using SPAdes (ver. 3.15.5) (Bushmanova et al. Citation2019) in RNA-seq mode (–rna) with two auto-determined k-mer parameters (-k 33,49). Quality of the transcriptome assembly was assessed using BUSCO (ver. 5.3.0) (Manni et al. Citation2021) with the Arthropoda lineage database (arthropoda_odb10, as of September 2020) containing a total 1,013 BUSCO marker genes.
Molecular phylogenetic analysis
Phylogenetic trees of the genus Daphnia were constructed based on the nucleotide sequences of the mitochondrial genes. For this analysis, GetOrganelle (ver. 1.7.7.0) (Jin et al. Citation2020) was used to assemble the mitochondrial genome of D. galeata HK using their transcriptome sequencing data with a previous mitochondrial genome of D. galeata available on GenBank (LC177071.1; from Biwa Lake, Japan) as a reference. In addition, both publicly available mitochondrial genomes and partial sequences of the mitochondrial 12S, 16S rRNA and cytochrome c oxidase subunit 1 (COX1) genes from other daphnids were retrieved from the database. The information of the sequences used are summarized in Table S1. Multiple sequence alignments were initially generated using MAFFT (ver. 7.520) (Katoh and Standley Citation2013) with the L-INS-i algorithm, and then the alignments were concatenated into 13 mitochondrial genes or three partial genes, using a custom-written Perl script conCat.pl (Eyun Citation2017). The concatenated sequence alignments used for phylogenetic inferences are available in Supplemental File 1. The maximum-likelihood (ML) analyses were performed using RAxML-NG (ver. 1.2.0) (Kozlov et al. Citation2019), applying the substitution model GTR + F + I + G4 for both alignment datasets. The best tree for each ML analysis was selected among the initial 15 random and 15 parsimony trees. Bootstrap support values were estimated with the option ‘–bs-trees autoMRE’ and presented as the transfer bootstrap expectation values (Lemoine et al. Citation2018). Tree visualization was performed using FigTree (ver. 1.4.4; available at http://tree.bio.ed.ac.uk/software/figtree).
Differentially expressed gene analysis and functional annotation
Open reading frames (ORFs) of assembled transcriptome contigs were predicted to generate unigene sequences with a minimum protein length of 100 amino acids using TransDecoder (ver. 5.7.1) (https://github.com/TransDecoder/TransDecoder). The ORF redundancies with less than 95% sequence similarity were eliminated (-c 0.95) using cd-hit-est (ver. 4.8.1) (Li and Godzik Citation2006). Contig and unigene completeness were assessed and compared with BUSCO (ver. 5.3.0) based on the single-copy orthologs represented in the arthropoda_odb10 database. The filtered reads of each sample were mapped to the unigene set using Bowtie2 (ver. 2.5.1) (Langmead and Salzberg Citation2012) with ‘–very-sensitive’ for read count calculations. The mapping results were normalized using the Cufflinks package (ver. 2.1.1) (Trapnell et al. Citation2010). Fragments per kilobase of transcript per million mapped reads (FPKM) values were used to estimate gene expression levels. The criteria for identifying significantly differentially expressed genes (DEGs) was set at a p-value < 0.05 and | log2(fold-change) | between samples ≥ 1. Gene annotation and functional DEG enrichment analyses were performed on the KOBAS website (http://bioinfo.org/kobas) with a Benjamini–Hochberg adjusted p-value < 0.05 as the criteria for significance based on the genome of Daphnia pulex (Bu et al. Citation2021). Gene ontology enrichment and graphical visualization of the results and gene characteristics were performed with ShinyGO (ver. 0.77) with an FDR cutoff of 0.05 (http://bioinformatics.sdstate.edu/go). The number of euKaryotic Orthologous Groups of proteins (KOGs) in each regulated group was counted and assigned to their specific categories using eggNOG-mapper (ver. 2.1.11) (Cantalapiedra et al. Citation2021). The genes in the significantly affected KOG categories were identified and annotated by BLAST against the NCBI nr database.
Results
Morphological identification keys of a female Daphnia galeata HK
A typical planktonic species, D. galeata is widely distributed and very frequently reported from Korea, commonly in lakes, reservoirs, fishponds, and rivers, but not from temporary waters (Kim Citation1988; Błędzki and Rybak Citation2016). The most recent morphological description of D. galeata and discussion of its taxonomic status was released in 2013 (Kirdiashcheva and Kotov Citation2013). To identify D. galeata daphnids to species, we used the pointed shape of the head helmet (a), the medial crest located high on the head (b), the presence of a single ocellus (c, blue arrow), the large compound eye (c, white arrow), pointed rostrum shape (c), the presence of postabdominal claws without fine external combs (d), and the particular features of ephippium (e). Distinctively, in this taxon, the first antennae of females are located noticeably nearer the end of the rostrum (c, black arrow) and the bases of the first antennae are uniquely positioned along the sides of the medial crest when compared to those of its closest relatives, such as D. cucullata and D. hyalina (Kirdiashcheva and Kotov Citation2013). In this study, 9-day-old D. galeata HK clones showed one to four eggs under the carapace. On average, a body length with an apical spine was 2.143 mm and a body heigh was 0.9 mm at this age (). The identification of D. galeata HK was consistent with all diagnostic characters of this taxon. Despite this morphological concordance, we applied molecular phylogenetics to corroborate the species classification ().
Phylogenetic analysis
To perform genetic classification of the Daphnia galeata HK clone, two phylogenetic trees of Daphnia were constructed using an ML method. The first tree used a dataset consisting of 13 concatenated mitochondrial (mt) genes and included 19 species, with a total of 11,201 positions in the final dataset (a) (Plessis et al. Citation2023). The second ML analysis used a dataset consisting of three concatenated partial mt genes (12S, 16S rRNAs, and COX1), comparing 49 species using a total of 3,864 positions in the final dataset (b). The nucleotide sequences were aligned with MAFFT (Katoh and Standley Citation2013), and when aligned sequences contained gaps, the sequences corresponding to the gaps were included from all other sequences. Sequences from the genera Simocephalus and Ceriodaphnia made up the outgroups in both trees. The genus Daphnia is divided into two subgenera: Ctenodaphnia and Daphnia. Using molecular methods, Kotov and Taylor (Citation2011) showed that this splits dates back to the Mesozoic. Both of our trees showed a genetical separation between subgenus Ctenodaphnia – including Daphnia (C.) magna Straus, 1820 (green boxes in ) – and subgenus Daphnia – including Daphnia (D.) pulex Leydig, 1860 and Daphnia (D.) galeata G.O. Sars, 1863 (blue boxes in ) – which is consistent with the split seen in a previous study (Błędzki and Rybak Citation2016). Also, the tree based on three partial mt genes showed high compatibility with the tree based on the full lengths of 13 mt gene sequences. Therefore, mitochondrial 12S rRNA, 16S rRNA, and COX1 appear to be sufficient for species identification within the genus Daphnia. In summary, the species identification of D. galeata HK was verified by comparative phenotypic () and genetic () analyses, and the two results confirmed each other.
The life cycles of Daphnia galeata HK in the culture conditions
To investigate the developmental phenomena in D. galeata HK, the body length, body height, and the size of an apical spine were measured and the number of eggs under the carapace were counted at three-day intervals for 21 days (). In this experiment, from 3 to 21 days, the median total body length (not including the apical spine) grew from 0.597 to 2.744 mm, body heigh from 0.168 to 1.280 mm, and apical spine size from 0.205 to 0.503 mm. At day 21, total body lengths with the apical spine were 2.600 to 2.950 mm and body heights were 1.250 to 1.320 mm (a–c). Adult female Daphnia with mature ovaries release eggs 13 min after molting (Hiruta and Tochinai Citation2014; Campos et al. Citation2018). Eight out of ten mature females formed subitaneous eggs by day 9, with 3–9 eggs observed (d). The genus Daphnia molts four to six times before reaching maturity, but continues to molt and grow in regular intervals throughout their life (Sumiya et al. Citation2016; Zuykova et al. Citation2017). During the subculture, embryos were forced out of the carapace to observe embryo developmental traits from the egg stage to stage 5. Daphnia galeata HK clones showed two red eyes at stage 3, two brown eyes at stage 4, and one black eye at stage 5 (e). This experiment showed the escape of a juvenile stage at day 9 under our laboratory culture conditions (), revealing the maturation time of the Korean strain D. galeata HK.
Gene expression profiling
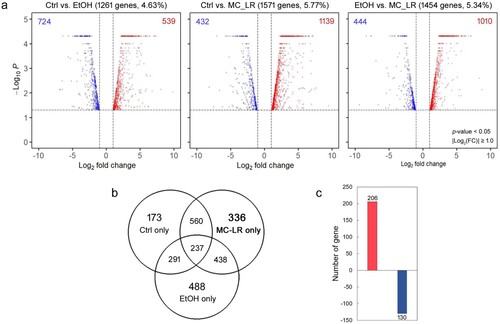
Comparative transcriptome studies provide high-resolution data on transcript levels and allow the identification of novel transcripts expressed under experimental conditions in the laboratory or in response to environmental changes in nature (Orsini et al. Citation2016; Toyota et al. Citation2016; Cui et al. Citation2018). We performed RNA sequencing (RNA-Seq) to obtain comprehensive data on gene expression changes in response to cyanotoxin MC-LR exposure. Paired-end Illumina RNA sequencing generated a total of 116.4 million reads under the MC-LR treatment, and negative (Ctrl) and positive (EtOH) controls produced 151.8 and 116.0 million reads, respectively (Table S2). Low-quality reads and adapter sequences were filtered, leaving 11.3–14.4 Gbp per treatment containing 112–142.7 million high-quality reads (Table S2). The filtered reads were assembled de novo into full-length transcripts, and predicted ORFs were extracted, resulting in a total of 29,163 unigenes (Table S2). Under cyanotoxin MC-LR exposure, we identified significant up- or down-regulated transcripts based on two-fold or greater changes with a p-value ≥ 0.05. A total of 1,263 (Ctrl vs. EtOH), 1,571 (Ctrl vs. MC-LR), and 1,454 (EtOH vs. MC-LR) genes were differentially expressed across the three comparisons (a). In the daphnids treated with MC-LR, 336 unique DEGs were identified (b). The number of up-regulated genes was more than 1.5 times that of the down-regulated genes (c).
Figure 5. Comparison of differentially expressed genes in Daphnia galeata HK following treatment with microcystin-LR (MC-LR) or ethanol (EtOH) or in an untreated control (Ctrl). (a) Volcano plots of the three pairwise comparisons. The number and percentage of genes is indicated above each plot. Red and blue circles represent highly upregulated or downregulated genes in female D. galeata HK, respectively. (b) A Venn diagram showing the number of shared or unique differentially expressed genes for each treatment. (c) Unique genes expressed after exposure to MC-LR.
Functional enrichment and frequency analysis
To determine the function of specific unigenes expressed in response to MC-LR, Gene Ontology (GO) enrichment and euKaryotic Orthologous Groups of proteins (KOGs) analysis were performed. GO-enrichment analysis is a powerful bioinformatics approach to understand the biological significance of a set of genes (Mi et al. Citation2013) and KOG analysis is a useful for functional and evolutionary studies to classify the database of clusters of orthologous groups of proteins encoded in Eukaryotes (Tatusov et al. Citation2000).
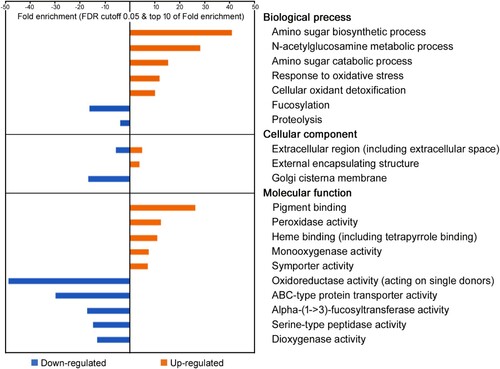
Firstly, in our GO-enrichment analysis, 248 (73.8%) unigenes were annotated to the D. pulex genome and divided into 166 up-regulated and 82 down-regulated unigenes (Table S3 and S4). Only 176 unigenes were assigned to GO terms, including 73 to biological processes, 43 to cellular components, and 60 to molecular functions. In biological processes (GO:0008150), metabolic processes (parent GO:008152) and responses to stimulus (parent GO:0050896) were highly enriched (). Amino sugar metabolic process (GO:0006040) genes were over-represented, including amino sugar biosynthetic process (GO:0046349), N-acetylglucosamine metabolic process (GO:0006044), and amino sugar catabolic process (GO:0046348) genes. Also, responses to stimulus were over-represented, including response to oxidative stress (GO:0006979) and cellular oxidant detoxification (GO:0098869) genes. Proteolysis (GO:0006508) and fucosylation (GO:0036065) genes were under-represented. In general, proteolysis is the hydrolysis of proteins into smaller polypeptides and/or amino acids by cleavage of their peptide bonds, and fucosylation is a subclass of glycosylation (GO:0070085). In Cellular Components (GO:0005575), extracellular region (GO:0005576) genes, including extracellular space and matrix (GO:0031012) genes were both over- and under-represented, but none of the assigned unigenes overlapped with each other (). Golgi cisterna membrane genes, which are polarized between receiving cargo from the endoplasmic reticulum and sending cargo forward to post-Golgi organelles, were down-represented (Ito et al. Citation2014). In molecular functions (GO:0003674), the GO terms of DEGs coincided those assigned under the biological process and cellular component categories. For instance, peroxidase activity (GO:00004601) and monooxygenase activity (GO:0004497) are associated with the response to oxidative stress and oxidant detoxification, categories in biological processes, and both were over-represented in this study (). In addition, α-(1->3)-fucosyltransferase activity (GO:0046920) is related to fucosylation (a biological process) and was down-represented. Transporter activity (GO:0005215), the parent GO term of symporter activity (GO:0015293) and ABC-type protein transporter activity (GO:0008320), were enriched under molecular functions, but not under biological processes. Among the subclasses of oxidoreductase activity pathway (GO:0016491), monooxygenase (GO:0004497), oxidoreductase (GO:0016684), oxidoreductase (GO:0016702), and peroxidase (GO:0004601) activity genes were over-represented. However, dioxygenase (GO:0051213) and oxidoreductase (GO:0016702) activity genes were under-represented.
Figure 6. Gene ontology enrichment analysis with 336 differentially expressed genes specific to MC-LR-treated Daphnia galeata HK daphnids. Over- and under-represented genes in each GO-term are shown as orange and blue bars, respectively. The individual GO terms are categorized as belonging to cellular components, biological processes, and molecular functions.
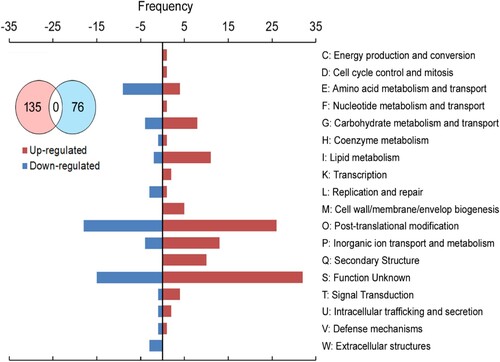
Secondly, KOGs analysis showed that 211 unigenes were assigned to KOG database and divided into 135 up-regulated and 76 down-regulated genes (). The M and Q class only involved in the upregulation. In the M class, C–C motif chemokine ligand 2 is one of several cytokine genes, which are a superfamily of secreted proteins involved in immunoregulatory and inflammatory processes in human (Zhang et al. Citation2010b). The hexosyl transferases [chs1] is responsible for chitin synthesis in cuticle of fungi, nematodes, and arthropods (Zhang et al. Citation2010a). The Q class includes the secondary membrane or lipid transporters. The sulfonylurea receptor [Sur], the removal or sequestration of unwanted toxic compounds, forms a large tetrameric complex with an inward-complex functions in regulations of the activity of the channel by sensing cellular ATP levels (Wilkens Citation2015). The lipid transporter, ATP binding cassette, class A3 [ABCA3] plays a critical role in the regulation of pulmonary surfactant homeostasis (Beers and Mulugeta Citation2017). In reverse, the W class only involved in the upregulation, C1q and tumor necrosis factor related protein 3 [C1QTNF3] in human involved in several processes, including cellular triglyceride homeostasis, regulation of cytokine production, and negative regulation of gluconeogenesis (Wölfing et al. Citation2008). None of the assigned unigenes overlapped between up- and down-regulated genes (Table S5). Excluding unknown function as the S class, the high frequency was post-translational modification as the O class. Interestingly, some of unigenes involved in the same pathway, but having unique genes each (marked in dark grey on Table S5). For instance, in the O class, triacylglycerol [PNLIP], the peptidase S1 family [Tpsg1], and serine-type endopeptidase activity involved in proteolysis appeared in both up- and down-regulated, but the genes were unique. In the same way to the P class, the sodium ion transport [Nach] was represented in both up- and down-regulated, but showing unique genes associated with each.
Discussion
Researchers and environmental agencies often study the effects of microcystin on aquatic organisms to better understand the potential ecological impacts of harmful algal blooms and to develop strategies for managing and mitigating the risks associated with these toxins. Cyanobacteria can be regarded as a key taxon in many aquatic ecosystems. Especially given that increased nutrient inputs into lakes has led to more frequent cyanobacterial blooms (Dokulil and Teubner Citation2000) and those high cyanobacterial abundances negatively affect herbivorous zooplankton such as Daphnia. The strong impact of cyanobacteria on freshwater ecosystem services is in part due to the very low food quality of cyanobacteria for herbivorous zooplankton (von Elert et al. Citation2003), but it can at least partly also be assigned to cyanobacterial secondary metabolites, many of which are known to have detrimental effects on zooplankton (Rohrlack et al. Citation1999).
Bioinformatics tools were used to analyze the biological functions, pathways, and processes associated with the genes differentially expressed in response to MC-LR. This can provide insights into the molecular mechanisms of toxin response. In this study, we discussed the implications of the gene expression changes and their relevance to toxin response mechanisms, and our RNA-seq analysis provides a foundation for further research into molecular and cellular responses to cyanotoxin exposure using gene expression profiling. This may include investigating specific pathways, regulatory elements, or potential therapeutic targets. By pursuing these avenues, we can profile the expression of significant genes responding to cyanotoxin exposure in Daphnia or other organisms, contributing to a better understanding of how these organisms adapt to and cope with environmental stressors. Specifically, a novel biomarker may be developed in Daphnia galeata to detect organic pollution in groundwater.
Supplemental Material
Download MS Word (74.6 KB)Acknowledgements
Conceptualization, E.K. and D.J.; data curation, E.K., D.J. and S.E.; writing–original draft preparation, E.K.; writing–reviewing and editing, E.K. and S.E.; sequence data and phylogenetic tree analysis, D.J.; technical support Y.P.; Funding acquisition, S.E. and D.J.; supervision and project administration, S.E. All authors have read and agreed to the published version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Beers MF, Mulugeta S. 2017. The biology of the ABCA3 lipid transporter in lung health and disease. Cell Tissue Res. 367(3):481–493. doi:10.1007/s00441-016-2554-z.
- Błędzki LA, Rybak Jl. 2016. Freshwater Crustacean Zooplankton of Europe: Cladocera & Copepoda (Calanoida, Cyclopoida) Key to species identification, with notes on ecology, distribution, methods and introduction to data analysis. Springer. doi:10.1007/978-3-319-29871-9.
- Bu D, Luo H, Huo P, Wang Z, Zhang S, He Z, Wu Y, Zhao L, Liu J, Guo J, et al. 2021. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 49(W1):W317–W325. doi:10.1093/nar/gkab447.
- Bushmanova E, Antipov D, Lapidus A, Prjibelski AD. 2019. rnaSPAdes: a de novo transcriptome assembler and its application to RNA-Seq data. GigaScience. 8(9):giz100. doi:10.1093/gigascience/giz100.
- Campos B, Fletcher D, Pina B, Tauler R, Barata C. 2018. Differential gene transcription across the life cycle in Daphnia magna using a new all genome custom-made microarray. BMC Genomics. 19(1):370. doi:10.1186/s12864-018-4725-7.
- Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. 2021. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol. 38(12):5825–5829. doi:10.1093/molbev/msab293.
- Cao H, Lyu K, Xiang F, Yang Z. 2014. Life history responses of Daphnia similoides simultaneously exposed to microcystin-LR and ammonia and their postexposure recovery. Environ Toxicol Chem. 33(11):2497–2505. doi:10.1002/etc.2701.
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890. doi:10.1093/bioinformatics/bty560.
- Cui R, Kwak JI, An YJ. 2018. Comparative study of the sensitivity of Daphnia galeata and Daphnia magna to heavy metals. Ecotoxicol Environ Saf. 162:63–70. doi:10.1016/j.ecoenv.2018.06.054.
- Dao TS, Do-Hong LC, Wiegand C. 2010. Chronic effects of cyanobacterial toxins on Daphnia magna and their offspring. Toxicon. 55(7):1244–1254. doi:10.1016/j.toxicon.2010.01.014.
- Dokulil MT, Teubner K. 2000. Cyanobacterial dominance in lakes. Hydrobiologia. 438(1):1–12. doi:10.1023/A:1004155810302.
- Eyun S. 2017. Phylogenomic analysis of Copepoda (Arthropda, Crustacea) reveals unexpected similarities with earlier proposed morphological phylogenies. BMC Evol Biol. 17:23. doi:10.1186/s12862-017-0883-5.
- Feist SM, Lance RF. 2021. Genetic detection of freshwater harmful algal blooms: A review focused on the use of environmental DNA (eDNA) in Microcystis aeruginosa and Prymnesium parvum. Harmful Algae. 110:102124. doi:10.1016/j.hal.2021.102124.
- Henning-Lucass N, Cordellier M, Streit B, Schwenk K. 2016. Phenotypic plasticity in life-history traits of Daphnia galeata in response to temperature - a comparison across clonal lineages separated in time. Ecol Evol. 6(4):881–891. doi:10.1002/ece3.1924.
- Herrera NA, Echeverri LF, Ferrao-Filho AS. 2015. Effects of phytoplankton extracts containing the toxin microcystin-LR on the survival and reproduction of cladocerans. Toxicon. 95:38–45. doi:10.1016/j.toxicon.2014.12.016.
- Hiruta C, Tochinai S. 2014. Formation and structure of the ephippium (resting egg case) in relation to molting and egg laying in the water flea Daphnia pulex De Geer (Cladocera: Daphniidae). J Morphol. 275(7):760–767. doi:10.1002/jmor.20255.
- Huisman J, Codd GA, Paerl HW, Ibelings BW, Verspagen JMH, Visser PM. 2018. Cyanobacterial blooms. Nat Rev Microbiol. 16(8):471–483. doi:10.1038/s41579-018-0040-1.
- Ito Y, Uemura T, Nakano A. 2014. Formation and maintenance of the Golgi apparatus in plant cells. Int Rev Cell Mol Biol. 310:221–287. doi:10.1016/B978-0-12-800180-6.00006-2.
- Jeon M-S, Jeong DM, Doh H, Kang HA, Jung H, Eyun S. 2023. A practical comparison of the next-generation sequencing platform and assemblers using yeast genome. Life Science Alliance. 6(4):e202201744. doi:10.26508/lsa.202201744.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21:241. doi:10.1186/s13059-020-02154-5.
- Jung H, Ventura T, Chung JS, Kim W-J, Nam B-H, Kong HJ, Kim Y-O, Jeon M-S, Eyun S. 2020. Twelve quick steps for genome assembly and annotation in the classroom. PLoS Comput Biol. 16(11):e1008325. doi:10.1371/journal.pcbi.1008325.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kim H, Choo H, Cha J, Jang M, Son J, Jeong T, Choi B-H, Lim Y, Chai H-H, Lee J, et al. 2022. Blood transcriptome comparison between sexes and their function in 4-week Rhode Island red chickens. Animal Cells and Systems. 26(6):358–368. doi:10.1080/19768354.2022.2146187.
- Kim IH. 1988. Key to the Korean Freshwater Cladocera. Korean J Syst Zool. 2:43–65.
- Kirdiashcheva AG, Kotov AA. 2013. Morphology and age variation of Daphnia galeata Sars (Cladocera: Daphniidae) in two adjacent water bodies of the Kola Peninsula. Izv Akad Nauk Ser Biol. 2:174–185. doi:10.1134/S1062359013010093.
- Kotov AA, Taylor DJ. 2011. Mesozoic fossils (>145 Mya) suggest the antiquity of the subgenera of Daphniaand their coevolution with chaoborid predators. BMC Evol Biol. 129:1–9. doi:10.1186/1471-2148-11-129.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455. doi:10.1093/bioinformatics/btz305.
- Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie2. Nat Methods. 9:357–359. doi:10.1038/nmeth.1923.
- Lemoine F, Domelevo Entfellner J-B, Wilkinson E, Correia D, Dávila Felipe M, De Oliveira T, Gascuel O. 2018. Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature. 556:452–456. doi:10.1038/s41586-018-0043-0.
- Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 22(13):1658–1659. doi:10.1093/bioinformatics/btl158.
- Lurling M. 2003. Effects of microcystin-free and microcystin-containing strains of the cyanobacterium Microcystis aeruginosa on growth of the grazer Daphnia magna. Environ Toxicol. 18(3):202–210. doi:10.1002/tox.10115.
- Ma J, Brookes JD, Qin B, Paerl HW, Gao G, Wu P, Zhang W, Deng J, Zhu G, Zhang Y, et al. 2014. Environmental factors controlling colony formation in blooms of the cyanobacteria Microcystis spp. in Lake Taihu, China. Harmful Algae. 31:136–142. doi:10.1016/j.hal.2013.10.016.
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol. 38(10):4647–4654. doi:10.1093/molbev/msab199.
- Mi H, Muruganujan A, Casagrande JT, Thomas PD. 2013. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 8(8):1551–1566. doi:10.1038/nprot.2013.092.
- NIER. 2006. Investigation of ecosystem and structure of Southern Han River’s Basin and Lake Chungju. Han River Basin Environmental Office. National Institute of Environmental Research.
- NIER. 2017. Water pollution process test standards in Korea. National Institute of Environmental research: Ministry of Environment.
- Orsini L, Gilbert D, Podicheti R, Jansen M, Brown JB, Solari OS, Spanier KI, Colbourne JK, Rusch DB, Decaestecker E, et al. 2016. Daphnia magna transcriptome by RNA-Seq across 12 environmental stressors. Sci Data. 3:160030. doi:10.1038/sdata.2016.30.
- Plessis SJ, Hong S, Lee B, Koepfli K-P, Chadwick EA, Hailer F. 2023. Mitochondrial genome-based synthesis and timeline of Eurasian otter (Lutra lutra) phylogeography. Animal Cells and Systems. 27(1):366–377. doi:10.1080/19768354.2023.2283763.
- Plugmacher S, Wiegand C, Oberemm A, Beattie KA, Krause E, Codd GA, Steinberg CEW. 1998. Identifcation of an enzymatically formed glutathione conjugate of the cyanobacterial hepatotoxin microcystin-LR: the first step of detoxication. Biochimica et Biophysica. 1425:527–533. doi:10.1016/S0304-4165(98)00107-X.
- Rohrlack T, Henning M, Kohl J-G. 1999. Mechanisms of the inhibitory effect of the cyanobacterium Microcystis aeruginosa on Daphnia galeata's ingestion rate. J Plankton Res. 21(8):1489–1500. doi:10.1093/plankt/21.8.1489.
- Schwarzenberger A, Martin-Creuzburg D. 2021. Daphnia's Adaptive Molecular Responses to the Cyanobacterial Neurotoxin Anatoxin-alpha Are Maternally Transferred. Toxins (Basel). 13(5):326. doi:10.3390/toxins13050326.
- Sumiya E, Ogino Y, Toyota K, Miyakawa H, Miyagawa S, Iguchi T. 2016. Neverland regulates embryonic moltings through the regulation of ecdysteroid synthesis in the water flea Daphnia magna, and may thus act as a target for chemical disruption of molting. J Appl Toxicol. 36(11):1476–1485. doi:10.1002/jat.3306.
- Tanaka S. 1992. Morphology and Variation of Daphnia galeata SARS from Lake Biwa and Lake Kizaki in Japan. Jpn J Limnol. 53(1):47–54. doi:10.3739/rikusui.53.47.
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28(1):33–36. doi:10.1093/nar/28.1.33.
- Toyota K, Hiruta C, Ogino Y, Miyagawa S, Okamura T, Onishi Y, Tatarazako N, Iguchi T. 2016. Comparative Developmental Staging of Female and Male Water Fleas Daphnia pulex and Daphnia magna During Embryogenesis. Zoolog Sci. 33(1):31–37. doi:10.2108/zs150116.
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 28:511–515. doi:10.1038/nbt.1621.
- von Elert E, Martin-Creuzburg D, Le Coz JR. 2003. Absence of sterols constrains carbon transfer between cyanobacteria and a freshwater herbivore (Daphnia galeata). Proc Biol Sci. 270:1209–1214. doi: 10.1098/rspb.2003.2357.
- Wan X, Cheng C, Gu Y, Shu X, Xie L, Zhao Y. 2021. Acute and chronic toxicity of microcystin-LR and phenanthrene alone or in combination to the cladoceran (Daphnia magna). Ecotoxicol Environ Saf. 220:112405. doi:10.1016/j.ecoenv.2021.112405.
- Wilkens S. 2015. Structure and mechanism of ABC transporters. F1000Prime Rep. 7:14. doi:10.12703/P7-14.
- Wölfing B, Buechler C, Weigert J, Neumeier M, Aslanidis C, Schöelmerich J, Schäffler A. 2008. Effects of the new C1q/TNF-related protein (CTRP-3) “cartonectin” on the adipocytic secretion of adipokines. Obesity (Silver Spring). 16(7):1481–1486. doi:10.1038/oby.2008.206.
- Zhang J, Liu X, Zhang J, Li D, Sun Y, Guo Y, Ma E, Zhu KY. 2010a. Silencing of two alternative splicing-derived mRNA variants of chitin synthase 1 gene by RNAi is lethal to the oriental migratory locust, Locusta migratoria manilensis (Meyen). Insect Biochem Mol Biol. 40(11):824–833. doi:10.1016/j.ibmb.2010.08.001.
- Zhang J, Patel L, Pienta KJ. 2010b. Targeting chemokine (C-C motif) ligand 2 (CCL2) as an example of translation of cancer molecular biology to the clinic. Prog Mol Biol Transl Sci. 95:31–53.
- Zimmermann L, Gottlich S, Oehlmann J, Wagner M, Volker C. 2020. What are the drivers of microplastic toxicity? Comparing the toxicity of plastic chemicals and particles to Daphnia magna. Environ Pollut. 267:115392. doi:10.1016/j.envpol.2020.115392.
- Zuykova EI, Bochkareva NA, Semenova AS, Katokhinc AV. 2010. Morphological differentiation, mitochondrial and nuclear DNA variability between geographically distant populations of Daphnia galeata and Daphnia cucullata (Anomopoda, Daphniidae). Biology.: Journal Of Siberian Federal University.
- Zuykova EI, Simonov EP, Bochkarev NA. 2017. Comparative morphological and genetic analysis of populations and species of the genus Daphnia O.F. Müller, 1785 (Crustacea; Daphniidae) from Lake Glubokoe and Lake Chany. Biology Bulletin. 44(3):277–289. doi:10.1134/S106235901703013X.