ABSTRACT
C-terminal binding protein 1 (CtBP1) is a critical transcriptional corepressor of many tumor suppressor genes and plays diverse roles in the progression of cancers. The transcriptional repression function of CtBP1 is mediated by recruiting histone-modifying enzymes, such as histone deacetylases and histone methyltransferases, to target genes by binding with DNA-interacting factors. Several post-translational modifications of CtBP1 have been identified, including ubiquitination, phosphorylation, and SUMOylation. This paper reports that CtBP1 is conjugated by ISG15. Endogenous CtBP1 was modified by ISG15 after interferon-α treatment in HeLa cells. The ISGylation process of CtBP1 was regulated by deISGylation enzyme USP18 and ISG15 E3 ligase EFP. Interestingly, CtBP1 ISGylation affected the binding affinity between CtBP1 and some components of CtBP1-associated transcriptional complexes. HDAC1 and LSD1 bound more efficiently to ISG15-conjugated CtBP1 than non-conjugated CtBP1. On the other hand, binding between CtBP1 and HDAC4 was unaffected by ISG15 modification. Furthermore, ISG15 modification enhanced the transcriptional repression activity of CtBP1 on several target genes related to EMT and apoptosis. These findings suggest that the ISG15 modification of CtBP1 modulates the function and activity of CtBP1 and that CtBP1 ISGylation may provide a new insight for CtBP1-mediated cancers.
KEYWORDS:
Introduction
C-terminal binding protein 1 (CtBP1) was initially recognized by its binding to the C-terminus of adenovirus E1A protein and is highly conserved among metazoans (Boyd et al. Citation1993). Humans have two CtBP family proteins, CtBP1 and CtBP2, which show high similarity in amino acid sequence and structure. Although CtBP1 and CtBP2 exhibit many overlapping functions, they perform distinct and unique functions in diverse cellular processes (Stankiewicz et al. Citation2014; Chen Citation2021). The role of CtBPs as an oncogene has been identified in various tumors, including melanoma, leukemia, breast, prostate, and colon cancers. The pro-tumorigenic function of CtBPs involves regulating a diverse range of genes involved in cell proliferation, apoptosis, EMT and invasion, and stem cell-like behaviors (Chinnadurai Citation2009; Chen Citation2021; Kwon et al. Citation2023). In CtBP-knockout mouse embryonic fibroblasts (MEFs), the expression of proapoptotic and epithelial genes like plakoglobin, keratin-8, PERP, p21, Bax, and E-cadherin, was increased. On the other hand, the effect was abrogated when the MEFs were rescued with CtBP1 (Grooteclaes et al. Citation2003). Furthermore, several tumor suppressors, including PTEN, p16INK4A, and GAS5, were negatively regulated by CtBP (Paliwal et al. Citation2007; Mroz et al. Citation2008; Blevins et al. Citation2017). Since CtBPs do not have a DNA binding motif, they are directed onto the promoters or enhancers of specific genes through interaction with DNA-binding transcription factors. All CtBP-binding transcription factors have a PXDLS sequence that can bind to the transcription factor-binding domain of CtBP1. Once bound to the transcription factors, CtBPs form a corepressor complex by recruiting several epigenetic modifiers, including histone methyl transferases (HMTases), histone deacetylases (HDACs), and lysine-specific demethylase (LSD1) (Shi et al. Citation2003; Chen Citation2021). Coordinated regulation of the chromatin structure by these CtBP-associated epigenetic regulators may contribute to the transcriptional repression of CtBPs.
Diverse post-translational modifications (PTMs), such as ubiquitination, phosphorylation, and SUMOylation, influence the protein stability, transcriptional activity, and subcellular localization of CtBPs (Byun and Gardner Citation2013). CtBP1 is ubiquitinated by XIAP, and CtBP2 is ubiquitinated by CHIP, both of which result in degradation by proteasome (Lee et al. Citation2012; Lee and Yoo Citation2013). HIPK2 and JNK1 phosphorylate CtBP at Ser422 and trigger the ubiquitination and subsequent proteasomal degradation of CtBP (Zhang et al. Citation2003; Wang et al. Citation2006). AMPK and Pak1 phosphorylate CtBP1 at Ser158, leading to a decrease in the CtBP corepressor functions by triggering CtBP translocation to the cytoplasm (Barnes et al. Citation2003; Kim et al. Citation2013). SUMOylation at Lys428 in the consensus SUMOylation motif (ΨKxE) of CtBP1 increased the nuclear localization and corepressor activity. Several SUMO E3 ligases, including Pc3, PIAS1, and PIAS2, are involved in CtBP1 SUMOylation. Interestingly, CtBP2 has no SUMO consensus site (Lin et al. Citation2003). CtBP1 is retained in the cytoplasm and nucleus, whereas the localization of CtBP2 is limited to the nucleus. Although the sequence identity between CtBP1 and CtBP2 is high, their N-terminal regions differ. The N-terminus of CtBP2 is acetylated at three Lys residues (Lys6, Lys8, and Lys10) by the p300 nuclear acetyltransferase, and acetylation at Lys10 is critical for nuclear retention (Zhao et al. Citation2006). Recently, it was reported that KAT2A succinylated CtBP1 and regulated the pro-tumorigenic function of CtBP1 (Zhou et al. Citation2023).
Interferon (IFN)-stimulated gene 15 (ISG15) is a ubiquitin-like protein that is involved in diverse biological functions, such as immune modulation, DNA damage response, autophagy, and protein translation (Zhang and Zhang Citation2011). Like ubiquitination, E1 activating enzyme, E2 conjugating enzyme, and E3 ligase participate in the covalent conjugation of target proteins by ISG15. ISGylation processes are reversed by ISG15-deconjugating enzyme ubiquitin-specific protease 18 (USP18) (Zhang and Zhang Citation2011). Similar to other PTMs, ISGylation alters the stability, subcellular localization, activity, and interacting partners of the target proteins (Tecalco-Cruz Citation2021). Several molecules implicated in tumorigenesis have been identified as ISGylation targets. The ISGylation of PP2Cβ negatively regulates the activity of PP2Cβ against NF-κB activation (Takeuchi et al. Citation2006; Shao et al. Citation2023). The tumor suppressive function of p53 is promoted by ISGylation under DNA damage conditions (Park et al. Citation2016). The ubiquitination ligase activity of CHIP is enhanced by ISG15 conjugation, which leads to cell growth inhibition through c-Myc degradation (Yoo et al. Citation2018). ISG15 modification of YAP is essential for its stabilization and the malignant phenotypes of lung adenocarcinoma (Xue et al. Citation2022; Um et al. Citation2023).
Although the regulatory mechanisms of CtBP1 by several PTMs have been identified, it is unclear if CtBP1 is ISGylated and how ISGylation affects its corepressor activity. In this report, we found that CtBP1 is covalently modified by ISG15. In addition, the ISGylation of CtBP1 regulates the binding affinity between its interacting partners, including HDAC1 and LSD1. Furthermore, the ISGylation of CtBP1 enhances the corepressor activity on target genes related to EMT and apoptosis. In summary, CtBP1 ISGylation and other CtBP1 modifications may allow the more subtle and diverse regulation of the CtBP1 function and activity as an oncogene during tumorigenesis.
Materials and methods
Cells and antibodies
HeLa, HEK293T, and CtBP1−/− MEFs were maintained in DMEM containing 10% FBS and antibiotic–antimycotic solution. In the case of CtBP1−/− MEFs, 2 mM L-glutamine was added. All cells were grown in incubators with conditions at 37°C, 5% CO2, and 95% humidity. The manufacturers and catalog numbers of the antibodies used are as follows: CtBP1 (Santa Cruz, sc-17759), ISG15 (Santa Cruz, sc-166755), Myc (Santa Cruz, sc-40), HA (Cell signaling, #3724), Flag (Sigma, F3165), GFP (Proteintech, 66002), HDAC1 (Santa Cruz, sc-81598), HDAC4 (Proteintech 17449-1-AP), β-actin (Sigma–Aldrich, A1978).
Western blotting
PBS-washed cells were pipetted with lysis buffer (50 mM Tris-HCl (pH 8.0), 0.5% NP-40, 120 mM NaCl, and complex protease inhibitors (containing aprotinin, leupeptin, and NEM)) for suspension and rotated for 30 min at 4°C. The cell extract was cleared by centrifugation, and the lysate was mixed with sampling buffer and boiled for 10 min. The protein samples were subjected to SDS-PAGE and subsequent transfer to PVDF membranes (Millipore). After membrane blocking with 5% non-fat milk, the primary antibodies were treated for one hour or O/N. The HRP-conjugated secondary antibodies (CELLNEST; CNG004-0005, CNG005-0005) were treated for one hour. And the protein signals were visualized by an enhanced chemiluminescence system.
In vivo ISGylation assay
In HEK293T cells, MycHis-CtBP1, HA-UBE1L, HA-UbcH8, and Flag-ISG15 were expressed as indicated for 48 h. After PBS washing, the cells were lysed in lysis buffer (25 mM Tris-HCl (pH 8.0), 0.1% NP-40, 150 mM NaCl, and 10 mM Imidazole) supplemented with complex protease inhibitors. After centrifugation, 30 µl of Ni-NTA agarose beads (Clontech) was added to the clarified supernatants and rotated O/N. After washing with wash buffer (20 mM Tris-HCl, 900 mM NaCl, 0.5% NP-40, and 10 mM Imidazole), the slurry was suspended with 150 mM Imidazole containing lysis buffer. The elutes were boiled, processed by SDS-PAGE, and immunoblotted.
Realtime RT–PCR and primers
Invitrogen Trizol reagent was used for total RNA extraction from CtBP1−/− MEFs. The first cDNA was synthesized using RevertAid reverse transcriptase (Thermo ScientificTM; EP0441) and oligo (dT) primers. Semiquantitative realtime RT–PCR was performed with qPCR PreMIX (Enzynomics; RT501M). To calculate the relative mRNA transcript of the specific genes, we used the ΔΔCt method normalized to the GAPDH gene. The sequences of primer pairs for the mouse genes used in this study are as follows: Bax (5′-CACTAAAGTGCCCGAGCTGA-3′ and 5′-GGGTCCCGAAGTAGGAGAGG-3′), Noxa (5′-AGTGCACCGGACATAACTGTG-3′ and 5′-GAGTTGAGCACACTCGTCCTT-3′), E-cadherin (5′-GGTCATCAGTGTGCTCACCTCT-3′ and 5′-GCTGTTGTGCTCAAGCCTTCAC-3′) (Zhang et al. Citation2022), and GAPDH (5′-TGCGACTTCAACAGCAACTC-3′ and 5′-GCCTCTCTTGCTCAGTGTCC-3′) (Kim et al. Citation2017). All measurements were taken in triplicate.
Results
CtBP1 is a target for ISG15 conjugation
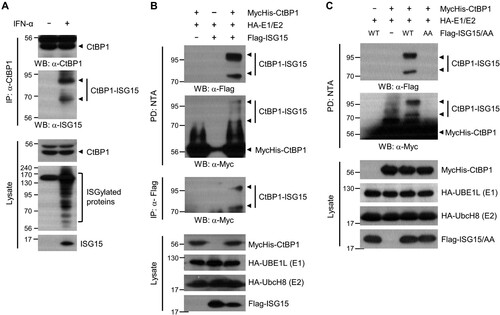
To identify the novel regulatory mechanism of CtBP1, we investigated whether CtBP1 is ISGylated. Type Ι IFNs induce the ISG15-conjugation system, including ISG15 (Zhang and Zhang Citation2011). Immunoprecipitation analysis revealed two CtBP1 bands conjugated with ISG15 in HeLa cells after IFN-α treatment (A). The MycHis-tagged CtBP1, ISG15, and enzymes for ISG15 conjugation (UBE1L(E1), UbcH8(E2)) were overexpressed in HEK293T cells to determine if the overexpressed CtBP1 could also be modified with ISG15. Two ISGylated CtBP1 bands were detected in the pulldown of MycHis-CtBP1 or the immunoprecipitation of Flag-ISG15 (B). The C-terminal diglycine residue of mature ISG15 is critical for conjugation to target proteins. When ISG15 mutant, in which the C-terminal diglycine was replaced with Ala–Ala (ISG15 AA), was overexpressed instead of the ISG15 wild-type, no ISGylated CtBP1 band was detected (C). These results suggest that CtBP1 is modified by ISG15.
Figure 1. CtBP1 is a target for ISG15 modification. (A) HeLa cells were treated for 48 h with or without IFN-α (2000 U/ml). The cell lysates were immunoprecipitated with anti-CtBP1 antibody, followed by immunoblotting with anti-ISG15 antibody to detect ISG15-conjugated CtBP1. (B) MycHis-CtBP1, HA-UBE1L (E1), HA-UbcH8 (E2), and Flag-ISG15 were expressed in HEK293T cells as indicated. After 48 h, the cell lysates were subjected to NTA pulldown under denaturing conditions, followed by immunoblotting with anti-Flag or anti-Myc antibodies. The same cell lysates were also immunoprecipitated with anti-Flag antibody, followed by immunoblotting with anti-Myc antibody. (C) MycHis-CtBP1, HA-E1 and E2, Flag-ISG15 WT or ISG15 AA (conjugation-defective mutant) were expressed in HEK293T as indicated. The cell lysates were subjected to NTA pulldown under denaturing conditions and immunoblotting with anti-Flag or anti-Myc antibodies to detect ISG15-conjugated CtBP1.
USP18 and EFP regulate the ISGylation process of CtBP1
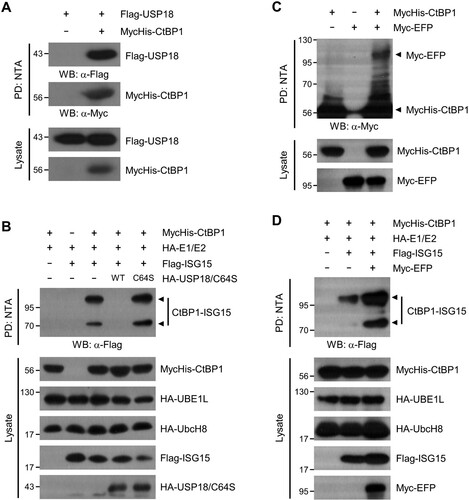
Like ubiquitination, the ISGylation process is dynamically regulated by a protein that removes ISG15 from the targets and E3 ligases that increase the interaction between ISG15 and the substrates. USP18 belongs to the ubiquitin-specific protease (USP) family and cleaves ISG15 from its conjugates (Dao and Zhang Citation2005). One HECT domain E3 (HERC5) and two RING finger E3s (EFP and HHARI) are known for ISGylation (Zou and Zhang Citation2006). To further determine the molecular constituents for CtBP1 ISGylation process, we explored whether USP18 and EFP are involved in the CtBP1 ISGylation process. The interaction between CtBP1 and USP18 was verified by Ni-NTA pulldown assay (A). ISG15 was efficiently removed from CtBP1 by wild-type USP18 but not by the catalytically inactive mutant USP18 (C64S) (B). EFP interacts with CtBP1 and strongly increases the ISGylation of CtBP1 (C and D). These data suggest that USP18 and EFP dynamically regulate the ISGylation status of CtBP1.
Figure 2. USP18 and EFP regulate the CtBP1 ISGylation process. (A) As indicated, HEK293T cells were transfected with plasmids that encode Flag-USP18 and MycHis-CtBP1. The cell lysates were subjected to NTA pulldown under denaturing conditions, followed by immunoblotting with anti-Flag or anti-Myc antibodies. (B) MycHis-CtBP1, HA-E1 and E2, Flag-ISG15, and HA-USP18 or USP18 C64S (active-site mutant) were expressed in HEK293T cells as indicated for 48 h. The cell lysates were subjected to NTA pulldown under denaturing conditions, followed by immunoblotting with anti-Flag antibody to detect ISG15-conjugated CtBP1. (C) As indicated, HEK293T cells were transfected with plasmids that encode MycHis-CtBP1 and Myc-EFP. The cell lysates were subjected to NTA pulldown, and immunoblotted with anti-Myc antibody. (D) MycHis-CtBP1, HA-E1 and E2, Flag-ISG15, and Myc-EFP were expressed in HEK293T cells as indicated for 48 h. The cell lysates were subjected to NTA pulldown under denaturing conditions and immunoblotting with anti-Flag antibody to detect ISG15-conjugated CtBP1.
ISGylation of CtBP1 enhances interaction with its binding partners
The molecular consequences of ISGylation vary according to the substrates. For some target proteins, ISG15 conjugation modulates the interaction between the target protein and its binding partners. 4EHP inhibits translation by binding to the 5′ cap of mRNA. ISG15 modification of 4EHP enhanced its binding affinity to the 5′ cap structure (Okumura et al. Citation2007). Filiamin B is a scaffold protein for RAC1 and JNK signaling components. Interaction between filamin B and JNK signaling components, including RAC1, MEKK1, and MKK4, was inhibited by filamin B ISGylation (Jeon et al. Citation2009). p53 ISGylation induced by DNA damage promotes the occupancy of p53 to its target genes and leads to the positive regulation of p53 tumor suppressive function (Park et al. Citation2016).
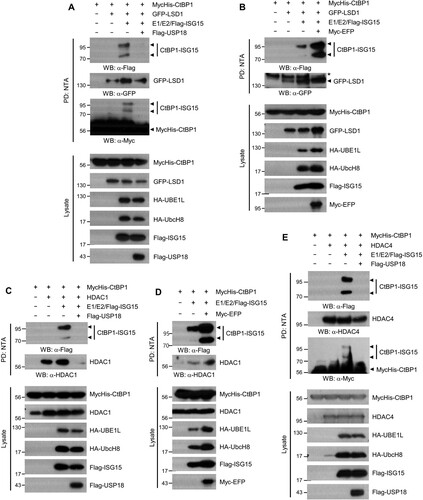
The CtBP proteins are targeted onto the promoters of specific genes through interaction with DNA-interacting transcription factors, such as KLF4, ZEB1/2, and E2F7 (Stankiewicz et al. Citation2014). Once bound to the transcription factors, CtBPs form a corepressor complex by recruiting many epigenetic modifiers, including HDACs, G9a, and LSD1 (Shi et al. Citation2003; Dcona et al. Citation2017). We examined whether the interaction of CtBP1 with its binding partners are regulated by CtBP1 ISGylation. The binding between LSD1 and CtBP1 increased when CtBP1 ISGylation was induced, whereas the removal of ISG15 from CtBP1 by USP18 returned the binding affinity to that when CtBP1 was not ISGylated (A). LSD1 binding to CtBP1 increased as CtBP1 ISGylation was enhanced by EFP (B). Similarly, in the case of HDAC1, the binding of the two proteins increased when CtBP1 was ISGylated, and the binding decreased when ISGylation was abrogated by USP18 (C). Furthermore, the binding affinity between CtBP1 and endogenous HDAC1 was enhanced when CtBP1 ISGylation was increased by EFP (D). On the other hand, the binding between CtBP1 and HDAC4 was not changed depending on the ISGylaton status of CtBP1 (E). These results indicate that binding affinities of several epigenetic modifiers recruited to the CtBP1 corepressor complex are regulated by the ISGylation status of CtBP1.
Figure 3. ISGylation of CtBP1 increases its interaction with LSD1 and HDAC1. HEK293T cells were transfected with plasmids that encode MycHis-CtBP1, ISG15 conjugation system (HA-E1 and E2, Flag-ISG15), Flag-USP18, Myc-EFP, and GFP-LSD1 (A and B) or HDAC1 (C and D) or HDAC4 (E) as indicated. After 48 h, Cell lysates were subjected to NTA pulldown under denaturing conditions, followed by immunoblotting. *: nonspecific bands.
CtBP1 ISGylation promotes the transcriptional inhibition of its target genes
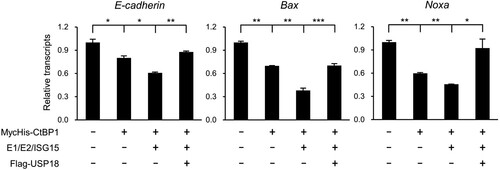
CtBP proteins repress a wide range of genes that play a role in proliferation and apoptosis, EMT, and stem cell-like behaviors (Chinnadurai Citation2009; Chen Citation2021). CtBP1-associated proteins, including LSD1, HDAC1, and HDAC4, belong to histone-modifying enzymes that facilitate the epigenetic silencing of target promoters by coordinating the demethylation of histone H3K4 or the deacetylation of histone H3K9 (Chinnadurai Citation2007). Thus, regulating the binding between CtBP1 and its associated epigenetic modifiers would affect the corepressor function of CtBP1. As CtBP1 ISGylation increases the association between CtBP1 and its binding partners, such as LSD1 or HDAC1, we examined whether the CtBP1 ISGylation status modulates the transcriptional corepression activity of CtBP1 on its target genes. It has been reported that CtBP1 corepresses epithelial and proapoptotic genes, including E-cadherin, Noxa, and Bax (Grooteclaes et al. Citation2003; Chen Citation2021). CtBP1 overexpression in CtBP1−/− MEFs suppressed the expression of E-cadherin, Bax, and Noxa. Coexpression of the ISGylation system enhances the transcriptional suppression of these genes. USP18, which removes ISG15 from CtBP1, alleviates the transcriptional repression-enhancing effects of CtBP1 ISGylation (). Overall, ISGylation of CtBP1 augments the transcriptional corepression activity of CtBP1 by increasing the affinity between CtBP1 and its binding partners.
Figure 4. ISGylation of CtBP1 augments the repression of its target genes. MycHis-CtBP1, ISG15 conjugation system (HA-E1 and E2, Flag-ISG15), or Flag-USP18 were overexpressed in CtBP1−/− MEFs as indicated for 48 h. The total RNA was isolated using the Trizol-Chloroform extraction method. Briefly, 2 µg of RNA was used to synthesize cDNA, and realtime quantitative RT-PCR analysis of E-cadherin, Bax, and Noxa was performed. The analysis was done in triplicate, and the results are presented as mean ± SD. The p-value was calculated using a Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
CtBP1 is overexpressed in a wide range of cancers and is implicated in the pro-tumorigenic features, including cell proliferation and survival, EMT, and stem cell-like behaviors (Chinnadurai Citation2009; Blevins et al. Citation2017). Diverse PTMs, including ubiquitination, phosphorylation, SUMOylation, acetylation, and succinylation, are involved in the dynamic regulation of CtBPs, but the link between CtBP1 and ISGylation is unclear. In this study, we identified that CtBP1 was modified by ISG15. USP18 removed ISG15 from CtBP1, and EFP acted as an ISG15 E3 ligase for CtBP1. The ISG15 conjugation of CtBP1 modulated the binding affinity between its interacting partners, including HDAC1 and LSD1. Furthermore, ISGylation of CtBP1 enhanced the corepressor activity on repressing the target genes related to EMT and apoptosis.
The oncogenic role of CtBP1 is executed by the transcriptional regulation of target genes as a corepressor. CtBP1 assembles on the specific target gene promoters by binding with several transcriptional factors and facilitates the recruitment of epigenetic modifiers to regulate gene expression. The association of CtBP1 with epigenetic modifiers in the CtBP1-associated complexes occurs directly or indirectly. The direct interactions with CtBP1 are mediated through the PXDLS-containing epigenetic modifiers, such as HDAC3/4, p300, and CBP. It appears that non-PXDLS-containing epigenetic modifiers, such as HDAC1/2, LSD1, and G9 are recruited to CtBP1 at other regions or indirectly mediated through other cofactors (Turner and Crossley Citation2001; Chinnadurai Citation2007; Byun and Gardner Citation2013; Chen Citation2021). Interestingly, LSD1 and HDAC1 bind to CtBP1 more efficiently when CtBP1 is ISGylated, whereas the association between CtBP1 and HDAC4 is unaffected by CtBP1 ISGylation. Further studies will be needed to elucidate the mechanism of CtBP1 ISGylation and histone-modifying enzyme recruitment by identifying the ISG15 modification site of CtBP1.
The CtBP1-associated transcriptional corepressor complexes repress epithelial and proapoptotic genes, including E-cadherin, Noxa, and Bax (Grooteclaes et al. Citation2003; Chen Citation2021). In the present study, CtBP1 ISGylation augmented the repression of these genes by CtBP1. In addition, USP18 relieved the enhanced transcriptional repression of the target genes by CtBP1 ISGylation. Enhanced binding affinity between the histone-modifying enzymes and CtBP1 by ISGylation increased the transcriptional corepressor activity of CtBP1. As CtBP1 is a potential target for cancer treatment, inhibitors of the assembly of CtBP1-associated complexes to reverse target gene repression are being developed (Dcona et al. Citation2017; Chen Citation2021). From this perspective, inhibiting CtBP1 ISGylation might provide a new therapeutic strategy for CtBP1-mediated cancer treatment.
In humans, there are two CtBP isoforms, CtBP1 and CtBP2, which share 76% sequence homology. While these two proteins have many overlapping functions, they also perform distinct and unique roles in different biological processes (Chinnadurai Citation2007; Chen Citation2021). Although they are highly conserved proteins, there are some distinct differences in their PTM regulation. CtBP1 is SUMOylated at Lys428 in the consensus SUMOylation motif (ΨKxE), whereas CtBP2 lacks a SUMO consensus site. CtBP1 SUMOylation increases the nuclear localization and corepressor activity of CtBP1 (Chen Citation2021). The N-terminus of CtBP1 and CtBP2 are somewhat different. CtBP1 is not acetylated, whereas CtBP2 is acetylated at three Lys residues (Lys6, Lys8, and Lys10) at the N-terminus by nuclear acetylase p300. Acetylation increases the nuclear retention of CtBP2 (Zhao et al. Citation2006). It remains to be identified if CtBP2 is ISGylated and the effect of ISGylation on its function.
The functions and properties of CtBP1, including protein stability, subcellular localization, and transcriptional activity, are regulated by various PTMs. In this study, ISGylation was added to the PTM of CtBP1. ISGylation regulates the affinity between the binding partners of CtBP1 and its transcriptional activity. The interplay between PTMs for CtBP1 is unknown, and elucidating the crosstalk between diverse PTMs will provide a better insight into the regulatory mechanisms of CtBP1.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Barnes CJ, Vadlamudi RK, Mishra SK, Jacobson RH, Li F, Kumar R. 2003. Functional inactivation of a transcriptional corepressor by a signaling kinase. Nat Struct Biol. 10(8):622–628.
- Blevins MA, Huang M, Zhao R. 2017. The role of CtBP1 in oncogenic processes and its potential as a therapeutic target. Mol Cancer Ther. 16(6):981–990.
- Boyd JM, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G. 1993. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 12(2):469–478. doi:10.1002/j.1460-2075.1993.tb05679.x.
- Byun JS, Gardner K. 2013. C-Terminal binding protein: a molecular link between metabolic imbalance and epigenetic regulation in breast cancer. Int J Cell Biol. 2013:647975.
- Chen Z. 2021. The transrepression and transactivation roles of CtBPs in the pathogenesis of different diseases. J Mol Med (Berl). 99(10):1335–1347.
- Chinnadurai G. 2007. Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol. 39(9):1593–1607. doi:10.1016/j.biocel.2007.01.025.
- Chinnadurai G. 2009. The transcriptional corepressor CtBP: a foe of multiple tumor suppressors. Cancer Res. 69(3):731–734. doi:10.1158/0008-5472.CAN-08-3349.
- Dao CT, Zhang DE. 2005. ISG15: a ubiquitin-like enigma. Front Biosci. 10:2701–2722.
- Dcona MM, Morris BL, Ellis KC, Grossman SR. 2017. CtBP- an emerging oncogene and novel small molecule drug target: advances in the understanding of its oncogenic action and identification of therapeutic inhibitors. Cancer Biol Ther. 18(6):379–391.
- Grooteclaes M, Deveraux Q, Hildebrand J, Zhang Q, Goodman RH, Frisch SM. 2003. C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc Natl Acad Sci A. 100(8):4568–4573. doi:10.1073/pnas.0830998100.
- Jeon YJ, Choi JS, Lee JY, Yu KR, Kim SM, Ka SH, Oh KH, Kim KI, Zhang DE, Bang OS, et al. 2009. ISG15 modification of filamin B negatively regulates the type I interferon-induced JNK signalling pathway. EMBO Rep. 10(4):374–380.
- Kim D, Seo SU, Zeng MY, Kim WU, Kamada N, Inohara N, Nunez G. 2017. Mesenchymal cell-specific MyD88 signaling promotes systemic dissemination of salmonella typhimurium via inflammatory monocytes. J Immunol. 199(4):1362–1371.
- Kim JH, Choi SY, Kang BH, Lee SM, Park HS, Kang GY, Bang JY, Cho EJ, Youn HD. 2013. AMP-activated protein kinase phosphorylates CtBP1 and down-regulates its activity. Biochem Biophys Res Commun. 431(1):8–13.
- Kwon J, Kim J, Kim KI. 2023. Crosstalk between endoplasmic reticulum stress response and autophagy in human diseases. Anim Cells Syst (Seoul). 27(1):29–37.
- Lee JS, Lee SK, Youn HD, Yoo SJ. 2012. C-terminal binding protein-mediated transcriptional repression is regulated by X-linked inhibitor of apoptosis protein. Biochem Biophys Res Commun. 417(1):175–181.
- Lee JS, Yoo SJ. 2013. C-terminus of Hsc70-interacting protein regulates C-terminal binding protein 2 and the expression of its target genes. Biochem Biophys Res Commun. 432(3):418–424.
- Lin X, Sun B, Liang M, Liang YY, Gast A, Hildebrand J, Brunicardi FC, Melchior F, Feng XH. 2003. Opposed regulation of corepressor CtBP by SUMOylation and PDZ binding. Mol Cell. 11(5):1389–1396.
- Mroz EA, Baird AH, Michaud WA, Rocco JW. 2008. COOH-terminal binding protein regulates expression of the p16INK4A tumor suppressor and senescence in primary human cells. Cancer Res. 68(15):6049–6053. doi:10.1158/0008-5472.CAN-08-1279.
- Okumura F, Zou W, Zhang DE. 2007. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev. 21(3):255–260. doi:10.1101/gad.1521607.
- Paliwal S, Kovi RC, Nath B, Chen YW, Lewis BC, Grossman SR. 2007. The alternative Reading frame tumor suppressor antagonizes hypoxia-induced cancer cell migration via interaction with the COOH-terminal binding protein corepressor. Cancer Res Res. 67(19):9322–9329. doi:10.1158/0008-5472.CAN-07-1743.
- Park JH, Yang SW, Park JM, Ka SH, Kim JH, Kong YY, Jeon YJ, Seol JH, Chung CH. 2016. Positive feedback regulation of p53 transactivity by DNA damage-induced ISG15 modification. Nat Commun. 7:12513.
- Shao Y, Ren W, Dai H, Yang F, Li X, Zhang S, Liu J, Yao X, Zhao Q, Sun X, et al. 2023. Skp2 contributes to AKT activation by ubiquitination degradation of PHLPP1, impedes autophagy, and facilitates the survival of thyroid carcinoma. Mol Cells. 46(6):360–373.
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y, Shi Y. 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 422(6933):735–738.
- Stankiewicz TR, Gray JJ, Winter AN, Linseman DA. 2014. C-terminal binding proteins: central players in development and disease. Biomol Concepts. 5(6):489–511.
- Takeuchi T, Kobayashi T, Tamura S, Yokosawa H. 2006. Negative regulation of protein phosphatase 2Cbeta by ISG15 conjugation. FEBS Lett. 580(18):4521–4526.
- Tecalco-Cruz AC. 2021. Molecular pathways of interferon-stimulated gene 15: implications in cancer. Curr Protein Pept Sci. 22(1):19–28.
- Turner J, Crossley M. 2001. The CtBP family: enigmatic and enzymatic transcriptional co-repressors. Bioessays. 23(8):683–690.
- Um H, Jeong H, Lee B, Kim Y, Lee J, Roh JS, Lee SG, Park HR, Robinson WH, Sohn DH. 2023. FAT10 induces cancer cell migration by stabilizing phosphorylated ABI3/NESH. Anim Cells Syst (Seoul). 27(1):53–60.
- Wang SY, Iordanov M, Zhang Q. 2006. c-Jun NH2-terminal kinase promotes apoptosis by down-regulating the transcriptional co-repressor CtBP. J Biol Chem. 281(46):34810–34815.
- Xue X, Tian X, Zhang C, Miao Y, Wang Y, Peng Y, Qiu S, Wang H, Cui J, Cao L, et al. 2022. YAP ISGylation increases its stability and promotes its positive regulation on PPP by stimulating 6PGL transcription. Cell Death Discov. 8(1):59.
- Yoo L, Yoon AR, CO Y, Chung KC. 2018. Covalent ISG15 conjugation to CHIP promotes its ubiquitin E3 ligase activity and inhibits lung cancer cell growth in response to type I interferon. Cell Death Dis. 9(2):97. doi:10.1038/s41419-017-0138-9.
- Zhang D, Sun F, Yao H, Wang D, Bao X, Wang J, Wu J. 2022. Generation of urothelial cells from mouse-induced pluripotent stem cells. Int J Stem Cells. 15(4):347–358. doi:10.15283/ijsc21250.
- Zhang D, Zhang DE. 2011. Interferon-stimulated gene 15 and the protein ISGylation system. J Interferon Cytokine Res. 31(1):119–130. doi:10.1089/jir.2010.0110.
- Zhang Q, Yoshimatsu Y, Hildebrand J, Frisch SM, Goodman RH. 2003. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell. 115(2):177–186.
- Zhao LJ, Subramanian T, Zhou Y, Chinnadurai G. 2006. Acetylation by p300 regulates nuclear localization and function of the transcriptional corepressor CtBP2. J Biol Chem. 281(7):4183–4189.
- Zhou J, Yan X, Liu Y, Yang J. 2023. Succinylation of CTBP1 mediated by KAT2A suppresses its inhibitory activity on the transcription of CDH1 to promote the progression of prostate cancer. Biochem Biophys Res Commun. 650:9–16.
- Zou W, Zhang DE. 2006. The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J Biol Chem. 281(7):3989–3994.
