ABSTRACT
Background: Porphyromonas gingivalis strain W83, one of the most widely investigated, is considered virulent in the context of periodontitis. The recently isolated P. gingivalis TDC60 has been reported to be highly pathogenic, although it has not yet been investigated in a mouse periodontitis model by oral gavage.
Aim: Our aim was to compare the virulence of both strains by evaluating their impact on alveolar bone loss and the composition of oral microbiota.
Methods: We inoculated by oral gavage C57BL/6 mice with either one of the two P. gingivalis strains and compared to a sham-treated group, without antibiotics pre-treatment. The mandibular alveolar bone of treated mice and controls were assessed, one month after the final inoculation, by microCT measurements. Moreover, at this time, we characterized their oral microbiota by 16S rRNA gene sequencing.
Results: While P. gingivalis W83 successfully initiated periodontitis, TDC60-treated mice only experienced moderate lesions. Furthermore, only W83-treated mice exhibited a specific distinct microbiota, with significantly lower richness and evenness than other samples, and decreased proportions of taxa usually found in healthy individuals.
Conclusion: This association between alveolar bone loss and a major persistent shift of the oral microbiota gives insights into virulence discrepancies among these bacterial strains.
Introduction
Periodontitis is a chronic inflammatory disease that affects tissues surrounding the teeth. Recent advances in the pathogenesis of periodontitis have suggested that polymicrobial synergy and microbiota dysbiosis together with a dysregulated immune response can induce inflammation-mediated damage in periodontal tissues [Citation1]. Porphyromonas gingivalis is still considered as a major driver for the dysbiosis, especially due to its ability to initiate periodontitis in animal models.
Initially applied in a non-human primate model [Citation2], P. gingivalis is widely used in murine models of periodontitis induced by oral gavage. Since its first description in 1994, this model allowed to explore the complex interactions between the bacteria and the host immunity [Citation3]. Variations in alveolar bone loss demonstrated the different virulence of several P. gingivalis strains in the mouse oral gavage model [Citation4], and one could hypothesize that specific virulent clones of periodontal pathogens may cause more severe periodontitis. The vast majority of animal studies used P. gingivalis W50 and W83. These two strains were isolated from clinical samples in the 1950s and have been found to be highly similar in genomic studies [Citation5]. Recently, the P. gingivalis TDC60 strain was isolated from a Japanese patient with severe periodontitis in the 2000s [Citation6]. According to the authors, TDC60 displayed a higher pathogenicity than other P. gingivalis strains, including W83.
Although TDC60 and W83 both express a majority of the virulence factors specific to P. gingivalis (), they are relatively distant in phylogeny [Citation5]. Moreover, the genome of P. gingivalis displays a great variability with functional and metabolic incidence, thus affecting the virulence of the strains [Citation7]. Currently, TDC60 is considered as the most aggressive P. gingivalis strain [Citation8]. However, to our knowledge, only the abscess model has been used to assess the pathogenicity of TDC60.
Table 1. Virulence factors of Porphyromonas gingivalis and their known genetic variation between strains W83 and TDC60. Kgp is a subtype of the proteinase gingipains; FimA is the major protein subunit of the major fimbriae; FimCDE are the accessory proteins of the major fimbriae; Mfa1 is the major protein subunit of the minor fimbriae; RagAB are outer membrane proteins and lipoproteins; the capsule is the capsular polysaccharide antigens (K-antigen).
Hajishengallis et al. showed that an indigenous bacterial community was required to induce periodontitis, as P. gingivalis failed to initiate bone loss in germ-free mice [Citation9]. The authors pinpointed the fact that the disease involved a disruption in the host-microbiota homeostasis caused by P. gingivalis inoculation. The discordance in bone loss obtained with different P. gingivalis strains could therefore be related to their variable ability to cause such imbalance. However, an extensive characterization of the oral microbiota after inoculation of P. gingivalis is lacking, despite the fact that molecular-based approaches to bacterial identification may give an easy access to its taxonomic composition, and could serve as a biomarker of the host homeostasis in the oral cavity.
In this study, our aim was to compare the virulence of W83 and TDC60 P. gingivalis strains by evaluating their impact on alveolar bone loss and the composition of the oral microbiota, after chronic inoculation by oral gavage, without antibiotics pre-treatment.
Methods
Animals
The study was approved by the ethical committee of Rennes for animal experimentation. All experimented mice were specific pathogen-free male C57BL/6JRj bred and raised in Janvier Labs (Saint-Berthevin, France). The animals were purchased at 6 weeks of age and kept in the animal colony at UMS Biosit in the University of Rennes 1 (Arche), and given free access to tap water and food (Teklad 19% Protein Rodent Diet). The animals were group-housed (three per cage) and maintained under standard conditions of temperature, atmosphere and light. All experimental procedures were performed in agreement with European law and regulations. This study conforms to the ARRIVE guidelines [Citation10].
Bacteria
P. gingivalis strain W83 was directly obtained from the American Type Culture Collection (ATCC) and strain TDC60 was obtained from the Japan Collection of Microorganisms (Riken BioResource Research Center JCM). These strains are maintained at the University of Rennes 1, frozen at – 80°C in cryobeads. For the experiments, the bacteria were transferred and maintained on Columbia 3 agar plates supplemented with 5% (v/v) defibrinated horse blood (bioMérieux, France), 25 mg.L−1 of hemin, and 10 mg.L−1 of menadione. The cultures were incubated at 37°C in an anaerobic chamber Macs-VA500 (Don Whitley) flooded with 80% N2, 10% H2 and 10% CO2.
Periodontitis mouse model by oral gavage
Following one-week acclimatization, the experiments started with 7 weeks old animals and were developed according to Baker et al. with slight modifications [Citation3]. Animals were randomly divided into three groups according to their treatment: Control, P. gingivalis W83 and P. gingivalis TDC60 (n = 6 mice/group). Animals were cohoused according to their group to avoid horizontal transmission (3 mice/cage). Prior to each infection, the bacteria were grown in enriched brain-heart infusion (BHI) broth containing, per liter, 37 g of BHI powder (AES Chemunex, France), 5 g of yeast extract (Conda, Dutscher), 25 mg of hemin (Sigma), and 10 mg of menadione (Sigma), placed in an AnaeroPack™ rectangular jar (Mitsubishi Gas Chemical Co. Inc., Tokyo, Japan) with an AnaeroGen™ 3.5 L (Mitsubishi Gas Chemical Co. Inc.) for 24 h at 37°C to reach the mid-exponential phase. The W83- and TDC60-infected groups received 109 CFU of live P. gingivalis of the corresponding strain, resuspended in 100 µL of PBS. The infection was made by direct inoculation of the oral cavity, three times a week, during 5 weeks. Controls were sham-infected mice which received the PBS, but no P. gingivalis. The oral administrations were made at the same time for all groups.
Sample collection
Oral swab samples were taken during the third week of the infection phase and stored at – 80°C, before DNA extraction. One month after the final oral gavage, mice were anaesthetised and sacrificed at 16 weeks of age, and mandibles were dissected. One hemi-mandible was quickly frozen in liquid nitrogen and stored at – 80°C, before performing the microbiota analyses. The other hemi-mandible was fixed in 70% ethanol 10% formaldehyde for 24 h at 4°C, and then stored in absolute acetone at 4°C until micro-computed tomography (micro-CT).
Micro-computed tomography
Micro-CT was performed on the hemi-mandible in the GEROM research unit (Angers, France) with a Skyscan 1272 (Bruker). The samples were placed in microtubes, filled with water to prevent desiccation. The tubes were fixed on brass stubs with plasticine and scanned with the following parameters: 9 µm resolution, X-ray energy of 70 kV and 142 µA for 1.9 s exposure, 0.2° rotation step, 5.000097 μm image pixel size. The data were reconstructed with NRecon (v. 1.7.0.4, Bruker-MicroCT)
Quantification of alveolar bone loss
The DataViewer software (v. 1.5.6, Bruker-MicroCT) was used to visualise bone and produce the sagittal slices. All images were reoriented such that the cemento-enamel junction (CEJ) and the root apex (RA) appeared [Citation11]. Among the sagittal images, the image which showed the most recession of alveolar bone was selected for measurement. The distance from the alveolar bone crest (ABC) to the CEJ was measured in µm with the CTAn software (v. 1.18.8, Bruker-MicroCT) at two interdental sites: between the first and the second mandibular molars (M1M2) or between the second and the third mandibular molar (M2M3). As described elsewhere, the CEJ-ABC distance was the shortest distance from ABC to the line connecting the adjacent CEJs [Citation11]. Measurements of root lengths from the CEJ to the RA were also taken to assess the percentage of remaining alveolar bone using an equation previously described: Percent remaining bone (%) = ([root length – CEJ-ABC]/root length) × 100 [Citation12].
Extraction and amplification of bacterial DNA
Total DNA was extracted from frozen oral swabs and hemi-mandibles using the QIAamp DNA Mini Kit (Qiagen, France) according to the manufacturer’s recommendations. The DNA was then kept frozen at – 80°C prior to amplification. DNA from oral swabs was pooled together according to their group, and then amplified to assess the colonization of the administered P. gingivalis in the oral cavity by using specific primers for the 16S rRNA gene of the bacterium (forward: 5ʹ-TGG-GTT-TAA-AGG-GTG-CGT-AG-3ʹ; reverse: 5ʹ-CAA-TCG-GAG-TTC-CTC-GTG-AT-3ʹ) [Citation13] with 35 cycles of PCR at an annealing temperature of 57°C. The PCR products were resolved by electrophoresis in 3% agarose gel in Tris-acetate-EDTA buffer. The V3-V4 regions of the 16S rRNA gene were amplified with the primers 338F (5′-ACT-CCT-ACG-GGA-GGC-AGC-AG-3′) and 802R (5′-TAC-NVG-GGT-ATC-TAA-TCC-3′) using 25 amplification cycles with an annealing temperature of 45°C. The PCR products were sequenced with the Illumina MiSeq at the GeT-PlaGe facility (Toulouse, France).
Oral microbiota analysis
The FASTQ files from the GeT-PlaGe facility are archived at NCBI Sequence Read Archive under the BioProject Accession Number PRJNA667316. The FASTQ files were processed with the QIIME2 software (v. 2018.4, https://qiime2.org/) following a pipeline adapted from the ‘Moving Pictures’ tutorial (https://docs.qiime2.org/2018.4/tutorials/moving-pictures/) [Citation14]. The files were imported as ‘PairedEndFastqManifestPhred33’ format. The pipeline DADA2 was used to control the sequence quality and construct the feature table. The forward and reverse sequences were truncated at 250 bases, with all other parameters set to default. Sequences count per sample ranged between 4,041 and 17,201. Prior to the taxonomic assignation at the genus level, reference reads were extracted from the ‘Ribosomal Database Project’ based on matches to the primers pair (338 F/802 R) and trained as a Naive Bayes classifier (RDP release 11, training set No. 16, http://rdp.cme.msu.edu/misc/rel10info.jsp). The reads that were classified as Archaea or Unassigned were removed from the feature table. The core diversity analysis was performed with a specific sampling depth (4,041 reads). The core diversity included alpha (Sobs, Shannon–Weaver) and beta (weighted UniFrac) diversity metrics.
Statistics
Data were analysed using the R (v. 3.5.0) and RStudio softwares (v. 1.1.383, https://www.rstudio.com/). Non-parametric tests were used and considered significant for p < 0.05. The results were expressed as mean ± standard error of the mean. The Kruskal–Wallis with Dunn’s multiple comparisons tests were used to compare means for quantitative data related to weight, alveolar bone parameters, alpha diversity indices and taxa relative abundances. The PERMANOVA and ANOSIM tests were performed on beta diversity metric with the QIIME2 diversity plugin [Citation15]. A linear discriminant analysis of the taxa relative abundance was computed with the LEfSe algorithm (parameters set to default) in Galaxy (https://huttenhower.sph.harvard.edu/galaxy/). All plots were made with ‘ggplot2ʹ for RStudio [Citation16], except for the LEfSe results and the 3D PCoA plot (EMPeror) [Citation17].
Results
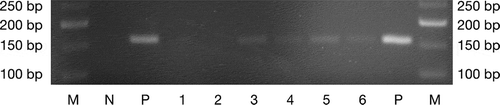
The body weight was not significantly different between the three groups (n = 6 mice/group) and no adverse event has been observed at any time of the experiments (). The PCR amplification made on oral swabs taken during the third week of infection confirmed the colonization of the oral cavity of mice by both P. gingivalis strains compared to controls ().
Table 2. Evolution of the animals’ weight during the experiments. Data are presented as mean ± standard error (n = 6 mice/group). The p-value of the Kruskal–Wallis test is indicated.
Figure 1. P. gingivalis specific 16S rRNA gene amplicons from DNA of oral swabs after electrophoresis on a 3% agarose gel. 1–2: sham-infected mice (controls); 3–4: W83-treated mice; 5–6: TDC60-treated mice; N: negative control; P: positive control (expected amplicon size: 161 bp). M: molecular weight marker (Quick-Load Low Molecular Weight DNA Ladder, NEB®).
The micro-CT analysis and comparison between controls and infected mice showed that W83-treated mice had a significant alveolar bone loss (). This was illustrated by the increase of the distance from the cementoenamel junction to the alveolar bone crest (CEJ-ABC) in both M1M2 (), and M2M3 interproximal sites (). A significant decrease of the remaining bone support was observed when the CEJ-ABC distance was related to the root length in both M1M2 (), and M2M3 interproximal sites (). While the TDC60-treated mice exhibited a trend for alveolar bone loss, only one site reached the significance threshold. The bone loss induced by P. gingivalis TDC60 was lower than that induced by W83 in all measurements, although no difference was found between the two groups of infected mice.
Figure 2. Alveolar bone loss in mice. Measurements of bone levels were made by comparing the distance from the cementoenamel junction (CEJ) to the alveolar bone crest (ABC) (a, b) and the percentage of remaining bone (c, d) at two interdental sites: between the first and second mandibular molars (M1M2) and between the second and the third mandibular molars (M2M3). Root lengths (from the CEJ to the root apex) were also measured to assess the percentage of remaining alveolar bone: Percent remaining bone (%) = ([root length – CEJ-ABC]/root length) x 100. Representative images of micro-CT of control (e), TDC60-treated (f) and W83-treated (g) mice are presented. Control and P. gingivalis infected mice were 16 weeks old (n = 6 mice/group). Dunn’s Kruskal–Wallis multiple comparisons, statistically significant at: *p < 0.05; **p < 0.01.
![Figure 2. Alveolar bone loss in mice. Measurements of bone levels were made by comparing the distance from the cementoenamel junction (CEJ) to the alveolar bone crest (ABC) (a, b) and the percentage of remaining bone (c, d) at two interdental sites: between the first and second mandibular molars (M1M2) and between the second and the third mandibular molars (M2M3). Root lengths (from the CEJ to the root apex) were also measured to assess the percentage of remaining alveolar bone: Percent remaining bone (%) = ([root length – CEJ-ABC]/root length) x 100. Representative images of micro-CT of control (e), TDC60-treated (f) and W83-treated (g) mice are presented. Control and P. gingivalis infected mice were 16 weeks old (n = 6 mice/group). Dunn’s Kruskal–Wallis multiple comparisons, statistically significant at: *p < 0.05; **p < 0.01.](/cms/asset/11cc4586-34f3-4dea-aefa-e51ffdd9d115/zjom_a_1832837_f0002_oc.jpg)
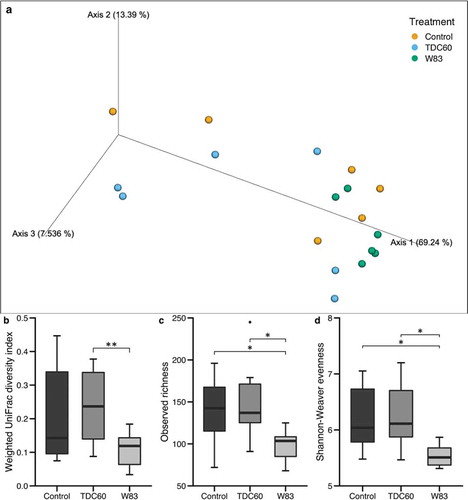
During the beta diversity analysis of oral microbiota, the Principal Component Analysis (PCoA) revealed an apparent homogeneity in the samples from W83-treated mice (). Moreover, they appeared to be clustered together, on the side-lines of the two other groups, which were more distributed in the 3D exploration. The PERMANOVA and ANOSIM tests, performed on weighted UniFrac distances plotted in PCoA, showed significant differences between the samples from W83- and TDC60-treated mice (). Moreover, the alpha diversity metrics confirmed that W83-treated mice had a distinct oral microbiota, with significantly lower richness and evenness than both controls and TDC60-treated mice (, d).
Figure 3. Alpha and beta diversity of the oral microbiota of mice. Bioinformatics processing of the microbiota samples from the oral cavity of control and P. gingivalis infected mice (n = 6 mice/group, 16 weeks old) allowed for Principal Coordinate Analysis (PCoA). The PCoA calculated with weighted UniFrac metric revealed clustering of W83-treated mice (a). Analysis of distances between samples showed significant differences between W83- and TDC60-treated mice, with both PERMANOVA and ANOSIM tests (**p < 0.01, b). Alpha diversity analysis showed significantly lower richness (Sobs, c) and lower evenness (Shannon-Weaver, d) in W83-treated mice; Dunn’s Kruskal–Wallis multiple comparisons, statistically significant at: *p < 0.05.
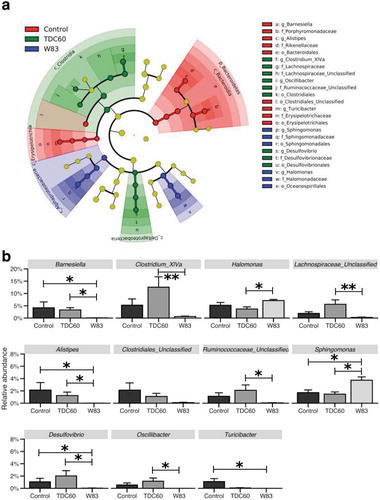
The taxonomic analysis of the oral microbiota showed differentially abundant taxa when assessed with LEfSe algorithm (). In high taxonomic levels, the samples from controls were found to be enriched in Bacteroidetes and Erysipelotrichia, while samples from TDC60-treated mice had increased abundance of Clostridia and Deltaproteobacteria. Samples from W83-treated mice had increased abundance of Alphaproteobacteria and Oceanospirillales. However, the lineages in the cladogram indicated that these variations were related with significant differences in taxa at the genus-level. These taxa were further filtered and assessed by Kruskal–Wallis and Dunn’s multiple comparisons tests (). As in the diversity analyses, significant variations were found between samples from W83-treated mice and the two other groups. Specifically, Alistipes, Barnesiella, Clostridium_XIVa, Desulfovibrio, Oscillibacter, Turicibacter, Lachnospiraceae_Unclassified and Ruminococcaceae_Unclassified were found significantly decreased in W83-treated mice; several of them were almost undetected. On the contrary, Halomonas and Sphingomonas were found significantly increased compared to controls and TDC60 groups.
Figure 4. Analysis of taxa relative abundances in the oral cavity according to the treatment of mice. The linear discriminant analysis with effect size (LEfSe) cladogram identified taxa that were differentially abundant across the groups (n = 6 mice/group; p < 0.05) (a). Taxa at the genus-level were then filtered (excluding those with a mean relative abundance < 1% in all groups), and plotted when the Kruskal–Wallis test returned significant result (p < 0.05) (b). Dunn’s Kruskal–Wallis multiple comparisons, statistically significant at: *p < 0.05; **p < 0.01.
Discussion
Despite the fact that some authors inoculated mice with other species from the Socransky’s red complex [Citation18,Citation19], most of the studies with the periodontitis mouse model used P. gingivalis, which is reported as a keystone pathogen in periodontitis [Citation20]. P. gingivalis TDC60 is reported to have a higher pathogenicity than P. gingivalis W83 [Citation6]. However, to our knowledge, the impact of oral gavage using TDC60 has not yet been investigated in a mouse periodontitis model. Unexpectedly, despite colonization of the oral cavity of mice by both strains, TDC60 exhibited a lower ability in causing alveolar bone loss, while the oral administration of W83 initiated periodontitis in both molar interproximal sites in our experiments.
Regarding the major virulence factors of the P. gingivalis, they are mostly cell surface proteins such as gingipains (RgpA, RgpB and Kgp), haemagglutinins, major and minor fimbriae (FimA and Mfa1), capsule (K-antigen) and the immunodominant surface antigens RagAB () [Citation21]. The cysteine peptidase gingipains account for 85% of the extracellular proteolytic activity of the pathogen [Citation22]. By degrading a variety of host proteins and taking part in the dysregulation of the host’s immune function, they are critical for bacterial colonization and the establishment of the disease [Citation20,Citation23]. According to a large in silico work, gingipains of W83 and TDC60 are similar for catalytic and C-terminal haemagglutinin domains [Citation24]. The fimbriae are involved in both bacterial adhesion and immune subversion [Citation25]. Moreover, the different genotypes imply various protein structures that demonstrated differential antigenicity [Citation26]. Hence, the genomic variations of P. gingivalis fimbriae are reported to be related to periodontitis initiation and progression. The major fimbriae (FimA), and the fimbrial accessory proteins (FimCDE), are different between TDC60 and W83 (). Interestingly, P. gingivalis W83 is poorly fimbriated which could be disadvantageous; however, Baker et al. showed that a mutant P. gingivalis W50, deficient for the fimbrial protein (strain DPG3), could also induce bone loss [Citation4]. In human and animal studies, type II (TDC60-like) and type IV fimA (W83-like) are more commonly found in periodontitis patients, and are more cytotoxic and invasive than strains with other types of fimbriae [Citation27,Citation28]. It is worth noting that type II fimA genotype is frequently reported to be more prevalent than type IV fimA in periodontitis-affected sites, while the latter was the most commonly found genotype among gingivitis patients [Citation28–30]; yet, another study found that the type II was the most widely distributed genotype, among both healthy and periodontitis sites [Citation31] suggesting that other virulence factors could be implicated and should be elucidated. In vitro experiments showed that the minor fimbriae (Mfa1) is implicated in coaggregation between P. gingivalis ATCC 33277 and Streptococcus gordonii [Citation32]. Due to an insertion element, W83 is disrupted for the mfaI gene, while TDC60 produces Mfa1 type I [Citation24]. This could be an advantage for colonizing the oral cavity of mice, as its indigenous microbiome is frequently dominated by Streptococcus [Citation33,Citation34]. RagA and RagB are outer membrane receptor linked to transporter TonB, and an associated lipoprotein, respectively. Serum analyses of periodontal patients revealed their immunogenicity, and RagB appeared to be an immunodominant surface antigen [Citation35]. Four distinct sequence types (rag-1 to rag-4) have been described and linked to the virulence of the bacterium. Most of the P. gingivalis rag-1 had higher scores for virulence in vivo than rag-4, and the rag-1 locus was associated with deep pockets in periodontitis patients [Citation36–38]. Thus, rag-1 could be beneficial for W83 [Citation24]. However, studies about the virulence of the different rag alleles were performed prior to the isolation of TDC60, which has therefore not yet been tested. Finally, most of the P. gingivalis strains are encapsulated with one of the six serotypes of the capsular polysaccharide antigens (K-antigens, K1 to K6), while some strains are not encapsulated at all [Citation39,Citation40]. The presence of the capsule and the differences in its chemical composition imply variations in the bacterial persistence, the host’s immune responses, the adhesion and coaggregation capacities [Citation40]. Encapsulated P. gingivalis strains, such as W83 (K1 [Citation41]), have demonstrated higher virulence than non-encapsulated strains, such as ATCC 33277 (K–) [Citation39,Citation40]. However, most of the P. gingivalis genomes contain a similar number of capsule-related genes, irrespective of the actual presence of a capsule, indicating that these genes may be subjected to gene expression controls [Citation5]. In summary, while Rag outer membrane proteins and capsule have not yet been characterized in TDC60, virulence mediated by FimA and Mfa1 should be in favor of TDC60. However, the impact of these proteins on the virulence is discussed in the literature and our results showed greater alveolar bone loss with W83 strain than with the TDC60 strain.
Regarding dysbiosis, a previous study showed that P. gingivalis triggers changes in the composition of the oral microbiota [Citation9]. Using aerobic/anaerobic cultures, the authors have identified alterations in six different genera spp. Our results suggest that these alterations could be a part of a major bacterial shift, detectable with high-throughput sequencing methods. This shift, which could be linked to a dysbiosis, appeared to be strain-dependent in our diversity analyses. Indeed, only W83-treated mice exhibited a poorly diversified oral microbiota, which tended to be clustered away from the two other groups. It is noteworthy that the samples were collected 1 month after the last oral inoculation; therefore, we cannot conclude about the ability of P. gingivalis TDC60 to alter the microbiota during the experiments. However, the strain W83 appeared to be able to stably disrupt the host-microbiota balance in a persistent way, and it was associated with a greater alveolar bone loss.
In an interesting way, the oral microbiota of W83-treated mice exhibited very low levels of several unclassified sequences in the Lachnospiraceae and Ruminococcaceae families, both belonging to the Clostridiales order. Noteworthy, samples from TDC60-treated mice showed increased, but not significant, proportions of these taxa. Although they may be less numerous than previously thought, these not-yet-cultured bacteria still represent a substantial part of the normal gut microbiota in mice, as well as Clostridium cluster XIVa, Barnesiella, Alistipes, Turicibacter and Oscillibacter that were also depleted in W83-treated mice [Citation42,Citation43]. A recent study pointed out the protective role of the Clostridia class, that is outcompeted by Desulfovibrio in an experimental knocked-out mouse model of metabolic disease [Citation44]. However, these taxa were significantly decreased in W83-treated mice. In these mice, the taxa with significantly higher proportions were Halomonas and Sphingomonas, two genera which are likely to be associated with pathological conditions. Halomonas was detected in lung tissue from mice with lung fibrosis [Citation45], while Sphingomonas spp. are known to carry ligands for NKT cells, an important member of the innate immune defence involved in the autoimmunity process [Citation46].
In conclusion, this study showed strain-dependent alveolar bone loss associated to a major alteration of the oral microbiota, in a mouse periodontitis model by oral gavage with P. gingivalis. Although described as highly pathogenic, P. gingivalis TDC60 had weaker consequences than W83. This could be due to their abilities to dysregulate the host’s immune responses. This study underlines the need for further clarifications about the various virulence and pathogenicity among P. gingivalis strains. Moreover, the lasting nature of the host-microbiota imbalance that we observed should encourage us to i) look for the occurrence of long-term deleterious inflammatory effects in various body sites of the experimental animals (gut microbiota, brain, joints, cardiovascular system, etc.), and ii) identify more precisely the P. gingivalis strains emerging in severe periodontitis in humans.
Acknowledgments
The authors would like to thank everyone at the Arche animal facility (UMS Biosit, Université de Rennes 1, France), and the GEROM research unit (Pr Chappard, Angers, France) for the micro-computed tomography. We also thank the GeT-PlaGe facility and the NED team (Toulouse, France) for sequencing the samples.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Van Dyke TE, Bartold PM, Reynolds EC. The nexus between periodontal inflammation and dysbiosis. Front Immunol. 2020;11:511.
- Holt SC, Ebersole J, Felton J, et al. Implantation of Bacteroides gingivalis in nonhuman primates initiates progression of periodontitis. Science. 1988;239(4835):55–9
- Baker PJ, Evans RT, Roopenian DC. Oral infection with Porphyromonas gingivalis and induced alveolar bone loss in immunocompetent and severe combined immunodeficient mice. Arch Oral Biol. 1994;39(12):1035–1040.
- Baker PJ, Dixon M, Evans RT, et al. Heterogeneity of Porphyromonas gingivalis strains in the induction of alveolar bone loss in mice. Oral Microbiol Immunol. 2000;15(1):27–32
- Chen T, Siddiqui H, Olsen I In silico comparison of 19 Porphyromonas gingivalis strains in genomics, phylogenetics, phylogenomics and functional genomics. Front Cell Infect Microbiol [Internet]. 2017 Feb 14 [cited 2018 Apr 23]; 7. Available from: http://journal.frontiersin.org/article/10.3389/fcimb.2017.00028/full
- Watanabe T, Maruyama F, Nozawa T, et al. Complete genome sequence of the bacterium Porphyromonas gingivalis TDC60, which causes periodontal disease. J Bacteriol. 2011;193(16):4259–4260
- Olsen I, Chen T, Tribble GD. Genetic exchange and reassignment in Porphyromonas gingivalis. J Oral Microbiol. 2018;10(1):1457373.
- Eltigani SA, Eltayeb MM, Bito T, et al. Argeloside I inhibits the pathogenicity of Porphyromonas gingivalis TDC60. J Biosci Bioeng. 2020 Aug 23. DOI: 10.1016/j.jbiosc.2020.07.016.
- Hajishengallis G, Liang S, Payne MA, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10(5):497–506
- Percie Du Sert N, Ahluwalia A, Alam S, et al. Reporting animal research: explanation and elaboration for the ARRIVE guidelines 2.0. PLoS Biol. 2020;18(7):e3000411.
- Cho Y-J, Song HY, Ben Amara H, et al. In vivo inhibition of Porphyromonas gingivalis growth and prevention of periodontitis with quorum-sensing inhibitors. J Periodontol. 2016;87(9):1075–1082
- Park CH, Abramson ZR, Taba M, et al. Three-dimensional micro-computed tomographic imaging of alveolar bone in experimental bone loss or repair. J Periodontol. 2007;78(2):273–281
- Ho M-H, Huang L, Goodwin JS, et al. Two small molecules block oral epithelial cell invasion by Porphyromons gingivalis. PloS One. 2016;11(2):e0149618
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26(1):32–46.
- Wickham H, Chang W, RStudio. ggplot2: create elegant data visualisations using the grammar of graphics [Internet]. 2016 [cited 2017 Jun 21]. Available from: https://cran.r-project.org/web/packages/ggplot2/index.html
- Vázquez-Baeza Y, Pirrung M, Gonzalez A, et al. EMPeror: a tool for visualizing high-throughput microbial community data. GigaScience. 2013;2:16.
- Lee SF, Andrian E, Rowland E, et al. Immune response and alveolar bone resorption in a mouse model of Treponema denticola infection. Infect Immun. 2009;77(2):694–698
- Sharma A, Inagaki S, Honma K, et al. Tannerella forsythia-induced alveolar bone loss in mice involves leucine-rich-repeat BspA protein. J Dent Res. 2005;84(5):462–467
- Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10(10):717–725.
- Darveau RP, Hajishengallis G, Curtis MA. Porphyromonas gingivalis as a potential community activist for disease. J Dent Res. 2012;91(9):816–820.
- Guevara T, Rodríguez-Banqueri A, Lasica AM, et al. Structural determinants of inhibition of Porphyromonas gingivalis gingipain K by KYT-36, a potent, selective, and bioavailable peptidase inhibitor. Sci Rep. 2019 20; 9(1): 4935
- Klein BA, Tenorio EL, Lazinski DW, et al. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics. 2012;13:578.
- Dashper SG, Mitchell HL, Seers CA, et al. Porphyromonas gingivalis uses specific domain rearrangements and allelic exchange to generate diversity in surface virulence factors. Front Microbiol. 2017;8:48.
- Enersen M, Nakano K, Amano A. Porphyromonas gingivalis fimbriae. J Oral Microbiol. 2013;5:1.
- Nagano K, Hasegawa Y, Abiko Y, et al. Porphyromonas gingivalis FimA fimbriae: fimbrial assembly by fimA alone in the fim gene cluster and differential antigenicity among fimA genotypes. PloS One. 2012;7(9):e43722
- Nakano K, Kuboniwa M, Nakagawa I, et al. Comparison of inflammatory changes caused by Porphyromonas gingivalis with distinct fimA genotypes in a mouse abscess model. Oral Microbiol Immunol. 2004;19(3):205–209
- Missailidis CG, Umeda JE, Ota-Tsuzuki C, et al. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral Microbiol Immunol. 2004;19(4):224–229
- Enersen M, Olsen I, Kvalheim Ø, et al. fimA genotypes and multilocus sequence types of Porphyromonas gingivalis from patients with periodontitis. J Clin Microbiol. 2008;46(1):31–42
- Nagano K, Hasegawa Y, Iijima Y, et al. Distribution of Porphyromonas gingivalis fimA and mfa1 fimbrial genotypes in subgingival plaques. PeerJ. 2018;6:e5581.
- Moon J-H, Herr Y, Lee H-W, et al. Genotype analysis of Porphyromonas gingivalis fimA in Korean adults using new primers. J Med Microbiol. 2013;62(9):1290–1294
- Park Y, Simionato MR, Sekiya K, et al. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect Immun. 2005;73(7):3983–3999
- Abusleme L, O’Gorman H, Dutzan N, et al. Establishment and stability of the murine oral microbiome. J Dent Res. 2020;99(6):721-729.
- Payne MA, Hashim A, Alsam A, et al. Horizontal and vertical transfer of oral microbial dysbiosis and periodontal disease. J Dent Res. 2019;98(13):1503-1510.
- Curtis MA, Hanley SA, Aduse-Opoku J. The rag locus of Porphyromonas gingivalis: a novel pathogenicity island. J Periodontal Res. 1999;34(7):400–405.
- Hall LMC, Fawell SC, Shi X, et al. Sequence diversity and antigenic variation at the rag locus of Porphyromonas gingivalis. Infect Immun. 2005;73(7):4253–4262
- Hanley SA, Aduse-Opoku J, Curtis MA. A 55-kilodalton immunodominant antigen of Porphyromonas gingivalis W50 has arisen via horizontal gene transfer. Infect Immun. 1999;67(3):1157–1171.
- Liu Y, Zhang Y, Wang L, et al. Prevalence of Porphyromonas gingivalis four rag locus genotypes in patients of orthodontic gingivitis and periodontitis. PloS One. 2013;8(4):e61028
- Laine ML, van Winkelhoff AJ. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol Immunol. 1998;13(5):322–325.
- Xu W, Zhou W, Wang H, et al. Roles of Porphyromonas gingivalis and its virulence factors in periodontitis. Adv Protein Chem Struct Biol. 2020;120:45–84.
- Tribble GD, Kerr JE, Wang B-Y. Genetic diversity in the oral pathogen Porphyromonas gingivalis: molecular mechanisms and biological consequences. Future Microbiol. 2013;8(5):607–620.
- Lagkouvardos I, Pukall R, Abt B, et al. The mouse intestinal bacterial collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat Microbiol. 2016;1(10):16131
- Clavel T, Lagkouvardos I, Blaut M, et al. The mouse gut microbiome revisited: from complex diversity to model ecosystems. Int J Med Microbiol IJMM. 2016;306(5):316–327
- Petersen C, Bell R, Klag KA, et al. T cell-mediated regulation of the microbiota protects against obesity. Science. 2019 26;365:6451.
- D’Alessandro-Gabazza CN, Méndez-García C, Hataji O, et al. Identification of halophilic microbes in lung fibrotic tissue by oligotyping. Front Microbiol. 2018;9:1892.
- Vas J, Mattner J, Richardson S, et al. Regulatory roles for NKT cell ligands in environmentally induced autoimmunity. J Immunol Baltim Md 1950. 2008;181(10):6779–6788.
