ABSTRACT
Introduction: Leptospirosis is a bacterial zoonotic disease with wide geographical spread. Its presence in Kenya and some of the neighbouring countries has been documented before and it is thought to contribute significantly to the number of febrile cases in human populations and abortions in livestock. This study investigated Leptospira spp. presence in rodents collected in both a pastoral and irrigated region of Kenya.
Materials and methods: Blood and kidney samples were screened for leptospiral DNA by PCR, and ELISA was used to detect antibodies in tissue fluid.
Results and discussion: Almost 42% (28/67) of the rodents were found to be PCR positive and 25% (14/56) by the ELISA test. Focus group discussions revealed that the local population perceived an increase in the rodent population and febrile illnesses not responsive to malarial treatment, a possible attestation of importance of non-malarial acute febrile illnesses such as leptospirosis in the communities.
Conclusion: While the study was small, it indicated that rodents could play an important role as reservoir hosts for the bacteria in these areas.
Introduction
Leptospirosis is a neglected zoonotic disease caused by pathogenic bacteria of the genus Leptospira [Citation1]. Rodents are known to be important reservoirs of the bacteria, with rats being shown to be important reservoirs of the bacteria in urban environments [Citation2] and field mice important for occupationally-acquired leptospirosis in other parts of the world [Citation3]. The burden in Africa is far from being fully understood to date, due to the lack of proper surveillance and diagnosis [Citation4]. It is most likely that leptospirosis is a common, but usually non-diagnosed, cause of febrile illness in large areas of the African continent, which is now more noticable when an increasing number of these cases are being regarded as non-malarial [Citation5].
In Kenya, seroprevalence studies have been performed in various parts of the country to investigate the disease in human hosts in outbreak investigations [Citation6,Citation7] and in at-risk populations [Citation8,Citation9]. One study has so far included both human and livestock hosts in its study population, an important consideration in integrating one-health approaches in research on the disease [Citation10]. Few studies in the country have investigated rodents for the causative bacteria [Citation11].
Kenya has seen increased efforts in placing its large portion of arid and semi-arid land (ASAL) under agriculture over the recent past with an aim of making it more food secure. Changes in human activity are common drivers of disease, and the increased irrigation may contribute to increased disease incidence [Citation12,Citation13]. The Tana River area of Kenya has seen an increase in irrigated land, with a concomitant drop in vegetation cover. This has led to a decline in population densities of larger wildlife and an increase in small ruminants. The creation of novel mosquito breeding sites due to irrigation has resulted in an increase in the exposure to mosquito-borne pathogens like Rift Valley fever virus, Plasmodium spp. as well as West Nile and dengue fever viruses, changing the risks for vector-borne zoonotic diseases [Citation9,Citation14–Citation16]. Less is known about the impact of land-use changes as a driver of disease on rodent-borne diseases. Increased clearing of bushes and use of land for cultivation may contribute to the increase in synanthropic rodent populations in these areas [Citation9,Citation17] and therefore create a possible risk of rodent borne pathogens to human and animal populations. This study investigated the presence of Leptospira spp. in rodents collected from ASAL areas of Kenya with the aim of demonstrating the public health importance of these carriers and to inform future research efforts.
Materials and methods
Study area
The study was performed in Bura and Hola irrigation schemes in Tana River County, and Ijara and Sangailu towns in Garissa County (). Many inhabitants in Tana River county live under the absolute poverty line, which is estimated at a little over a one thousand shillings earnings a month. The county had a population density of 383.5 people per square kilometer from the last population census in 2010 and receives a low amount (long-term annual average between 220 and 500 mm) of rainfall with bimodal distribution in the months of April to June and November to December. The Bura and Hola irrigation schemes were set up to support economic activities in the region; mainly crop farming and to a lesser extent, extensive livestock production. The biggest irrigation schemes are the Bura Irrigation Scheme, the Tana Delta Irrigation Project, and the Hola Irrigation Scheme, and are used for production of important cereals like rice and maize [Citation18–Citation20].
Figure 1. Map of Kenya and the two study areas, Tana River and Garissa Counties, with towns involved in the study highlighted.
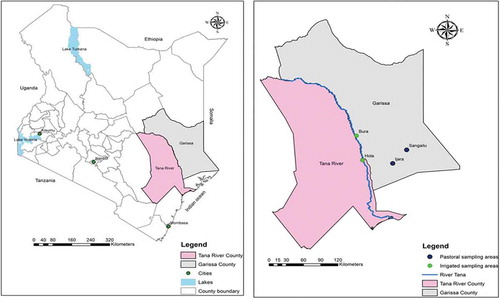
Ijara and Sangailu towns are located in Garissa County of Kenya which has a population density of 723.7 people per square kilometer. The majority of inhabitants practice transhumant pastoralism. The River Tana is an important source of water for the county, with mean rainfalls of between 250 and 300 mm. Rainfall also typically presents in bimodal distribution from March to May and from September to October [Citation18]. The county is considered among the poorest in the country and has been shown to have severe gaps in access to water [Citation21,Citation22].
Study design and ethical approval
The work was designed as a pilot study to investigate the occurrence of rodents at the different sites and to detect Leptospira spp. in rodents. The design was cross-sectional in nature. The study was implemented in a small area within the region used for the Dynamic Drivers of Disease in Africa: Ecosystems, livestock/wildlife, health and wellbeing: RVF case study in Kenya project (REF. NE/J001422/1). Ethical approval for this project was provided by the Ethics and Scientific Review Committee (ESRC) of the Africa Medical and Research Foundation (AMREF) (REF: AMREF-ESRC P65/2013).
Focus group discussions
Focus group discussions (FGDs) were conducted in order to collect qualitative data on people’s perceptions, historical and recent observation of rodent populations, unusual diseases and the sources of such diseases. Multi-level stratified random sampling of the participants was done. All the participants had to be residents of the irrigation schemes in Tana River County or the dryland communities in the study sites in Garissa County. Within the irrigation schemes, communities lived in villages which was considered a cluster. Individuals within each village were randomly picked to be part of the focus group. Stratification was also made based on gender because of cultural considerations. Initial discussions with key informants, who were either community leaders or government agricultural extension officers helped refine the focus group discussions guides based on their knowledge of local norms. The focus groups in the study ended up having 8 participants on average. The discussions were recorded and later transcribed. The data were analyzed using conventional content analysis method [Citation23]. To ensure active participation of all participants and accurate answers, the moderators tested the level of knowledge of wildlife species and disease symptoms.
Setting up of rodent traps
The procedure for trapping rodents followed standard protocol as outlined by Eymann et al. [Citation24] and was implemented by mammalogy experts at the National Museums of Kenya (NMK). Forty Sherman rodent live traps (H. B. Sherman Traps Inc., Tallahassee, FL) were used for trapping rodents, on paceline transect established in the field traversing farms and fallow bush. Within settlements, traps were set in houses, stores and along fences of homesteads, from around 5.30 pm to 7.30 pm and checked for trapped rodents at 7 am the next morning. Traps were closed during the day, and rebaited the following evening in the same location, for two consecutive trap nights per site. Each trap was baited using oats and peanut butter and set one per trapping station spaced at 10 meters apart in the bush and farms but irregulalrly in settlements. Rodent collection was performed in Bura and Hola in October 2013 and March 2014. Rodents from Ijara were collected in December 2013 and March 2014. Rodents in Sangailu were collected in December 2013 and during January and February 2014. In total there were 80 trap nights per site.
Sample collection
The rodent species were identified morphologically by mammalogists from NMK. Collected rodents were euthanized using an intraperitoneal injection of 0.1–0.5 ml Ketaminol (50 mg/ml). Blood samples were collected via cardiac puncture and urine samples were obtained when possible. Rodent morphometric data was recorded (weight, total length, and lengths of the tail, hind foot, forearm and ear). Internal organs (heart, liver, kidneys, lungs, spleen, caecum and bladder) were collected from each rodent. The sex and estimated age (whether adult, juvenile or sub-adult) of the rodents were noted, as well as autopsy comments made.
All organ samples were collected in sterile cryovials with unique barcode identifiers and immediately preserved in liquid nitrogen from where they were transported to the International Livestock Research Institute (ILRI) headquarters in Nairobi, Kenya.
PCR testing and phylogenetic analysis
Blood and/or kidney samples from 67 rodents were analyzed. Extraction of DNA was performed using the high-performance MagNA Pure Total Nucleic Acid isolation kit (Roche Diagnostics; Basel, Switzerland) according to the manufacturer’s recommendations. Nucleic acid concentration was determined using the NanoDrop 2000 spectrophotometer (Thermo Scientific; Wilmington, Delaware).
Conventional PCR was used to detect leptospiral DNA using primers described by Gravekamp et al., (1993) that target the Leptospira spp. secY gene of pathogenic leptospires with the exception of L. kirschneri. A 25 µl reaction assembly was comprised of 1× DreamTaq Green PCR Master Mix (ThermoFisher Scientific; Waltham, Massachusetts), 400 nM of both G1 and G2 primers, 2 µl of extracted DNA and nuclease-free water. Thermocycling was performed on the GeneAmp® PCR system 9700 Thermocyler (ThermoFisher Scientific; Waltham, Massachusetts) using the following conditions; initial denaturation at 95°C for 3 minutes, with 40 cycles of 95°C for 30 seconds, 55°C for 1 min, and 72°C for 1 minute. Final extension was done at 72°C for 10 min. Both positive and negative controls were included in the PCR runs. PCR bands of interest (at 285bp) were detected using agarose gel electrophoresis. DNA purification was performed using the Wizard®SV Gel and PCR Clean-Up System (Promega; Madison, Wisconsin). Positive rodents were determined as those with positive amplification from kidney tissue, blood or both samples (if present). A negative rodent was determined when there were no amplification in any of the samples used for analysis.
PCR products from 5 positive samples were purified and sent to Macrogen Inc. (Amsterdam, The Netherlands) for Sanger sequencing. Sequence analysis was performed using the CLC Main Workbench software package version 6 (Qiagen; Aarhus, Denmark) and sequences searched using the BLASTn tool on the NCBI website. Sequences were deposited to GenBank and allocated accession numbers MG963181, MG963182, MG963183, MG963184 and MG963185. Phylogenetic analysis was performed by the MEGA6 software [Citation25]. Nineteen partial sequences of the secY gene representative of several leptospiral species were chosen as reference sequences, including non-pathogenic leptospires as outliers. The phylogenetic tree was generated using the Maximum Likelihood method using the Tamura-Nei model [Citation26] and the tree with the highest log likelihood was generated and drawn to scale. Initial tree(s) for the heuristic search were obtained automatically by applying the Maximum Parsimony method. The tree was drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA6 [Citation27]. Twenty four (24) nucleotide sequences were included in total ().
Figure 2. The phylogenetic analysis was performed on isolates from both study counties (highlighted with black dots) and compared with sequences of the secY gene of known leptospiral species. The evolutionary history was infered using the Maximum Likelihood method based on the Tamura-Nei model [Citation26]. The tree with the highest log likelihood (−1108.1041) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches.
![Figure 2. The phylogenetic analysis was performed on isolates from both study counties (highlighted with black dots) and compared with sequences of the secY gene of known leptospiral species. The evolutionary history was infered using the Maximum Likelihood method based on the Tamura-Nei model [Citation26]. The tree with the highest log likelihood (−1108.1041) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches.](/cms/asset/b429176a-8795-4af5-a837-e12753c650c0/ziee_a_1547093_f0002_b.gif)
Serological testing
The volume of blood obtained from the rodents was too small to be used directly in a serological assay. Blood or body fluids from rodent organs (lungs or liver) were absorbed on Nobuto filter paper strips (Toyo Roshi Kaisha Ltd, Tokyo, Japan), dried and sent to the Zoonosis Science Center at Uppsala University for analysis. The paper strips were re-suspended in 400 µl phosphate buffer saline (PBS). However, since different blood volumes were initially available, the final concentration varied between the samples. When the entire strip had absorbed fluids, this was estimated to be equal to 4 µl, giving a final dilution of 1:100. If the filter paper had only absorbed blood or body fluids to 25% of its surface area, the dilution was estimated to be as high as 1:400, increasing the risk of false negative samples. Heat inactivation of the PBS dilutions were done at 56ºC for 30 minutes.
The diluted samples were then screened using a commercial enzyme-linked immunosorbent assay (ELISA) kit (Qualitative Rat Leptospira IgG (LS-IgG), MyBioSource, San Diego, California) designed for usage on blood, serum, tissue homogenates and body fluids. Since the test samples were diluted at least 1:100 during resuspension from filter paper, the sensitivity was expected to be decreased. According to the instructions for the kit, samples are not supposed to be diluted unless OD values higher than the highest standard are obtained. Apart from this deviance, the instructions of the manufacturer were followed. Samples with optical densities higher than the average of the negative controls +0.15 were regarded as positive for leptospiral-reactive IgG, according to the instructions of the ELISA manufacturers.
Statistical analysis
Laboratory data was initially entered into MS Excel (Microsoft Corporation, USA) and later imported to STATA 14.0 (StataCorp Ltd, US) for analysis. Univariable analyses were performed using Chi2 test, or Fisher’s exact test (when the assumptions for Chi2 test were not met).
Results
A total of 119 rodents representing six identified species () were collected at the four sites, over the two trapping periods. Trap success rates were 28.1% (45/160) in Bura, 24.4% (39/160) in Hola, 17.5% (14/80) in Ijara and 13.1% (21/160) in Sangailu. The most abundant species was the common house mouse, Mus musculus (74) followed by the multimammate rat, Mastomys natalensis (33). In spite using the same number of traps the same number of nights on all sites, the total capture was higher in the irrigated areas, Bura and Hola, than in the two non-irrigated sites of Ijara and Sangailu. Mus musculus was most frequently trapped adjacent or inside houses, while Mastomys natalensis was more often trapped in bushes. Majority of the rodents collected were adults (n = 108, 90.8%), with sub-adults (n = 7, 5.9%) and juveniles (n = 4, 3.4%) being fewer.
Table 1. Distribution of rodents collected with the towns and diagnostic test results in the study sites.
Qualitative analysis
There were a total of 20 FGDs held across the study site. The community members provided a historical timeline with major environmental changes and accompanying changes in human health and a summary of the results from the various study sites is presented in below. There was a general perception by the local population that there was a correlation between increased irrigation in the area over the years and cases of non-malarial fever.
Table 2. Summary findings of focus group discussion of the perceptions of human diseases and wildlife in the community.
Molecular analyses
In total, blood and kidney tissues from 67 rodents were available for testing in this study (from both wet and dry seasons in Ijara and Sangailu, and the dry season in Bura and Hola). DNA was extracted and analyzed by PCR for the presence of leptospira DNA, with positive results obtained on samples from 28 rodents (41.8%, 95% CI = 29.8% – 54.5%). The percent positivity was highest in Ijara with 82% (95% CI 48.22 −97.72), followed by Sangailu with 58% (95% CI 27.67–84.83). In the irrigated areas, the percent positive was 16% (95% CI 4.54 −36.08) in Bura and 42% (95% CI 20.25–66.50) in Hola ().
There were significantly (p = 0.001) higher proportions of rodents that were PCR positive among samples from pastoral areas collected in the wet season (69.6%), than those collected in the dry season in the irrigation areas (27.3%), with the odds ratio being 6.25.
Comparing adults to the juvenile and sub-adults, more young were found PCR positive (60.0%) than the adults (31.3%), but the difference was not significant (p = 0.2). Comparing mice and multimammate rats, there was no significant difference in PCR results. Phylogenetic analysis showed that all 5 sequences from this study clustered with the partial sequences from the species Leptospira interrogans. Other leptospiral species also clustered closely with each other, corroborating the ability of the secY gene of the bacteria in differentiating the various species within the leptospiral genotypic classification system [Citation11,Citation28–Citation30].
Serological analyses
Serological analysis revealed only 14 out of 56 tested rodents (25.0%, 95% CI 14.4–38.4%) as being seropositive, with 9 (36%, 95% CI 18.0–57.5%) from Bura, 2 (11.8%, 95% 1.5–36.4%) from Hola, 1 (12.5%, 95% CI 0.3–52.7%) from Ijara and 2 (33.3%, 95% CI 4.3–7.8%) from Sangailu. There were no significant differences (p = 0.25) between the sites. Multimammate rats were more likely (p < 0.001) to be found seropositive (58.8%) than mice (11.4%). More adults were seropositive (26.7%) compared to young (11.1%), though the difference was not significant.
There was no significant difference in seropositivity when comparing irrigated (Bura and Hola) with the non-irrigated, pastoral sites (Ijara and Sangailu). A comparison of the samples tested both with PCR and ELISA is shown in .
Table 3. Rodent samples from Garissa and Tana river county tested for Leptospira spp. using both ELISA and PCR.
Discussion
Results from the FGDs were consistent with the little tree cover noted in the pastoral Garissa county and the high human settlements and irrigation activities noted in the irrigation area of Tana River county. These anthropogenic activities in the area have also been linked with low biodiversity [Citation9]. Frequent rodent sightings reported by the participants could also be an indicator of leptospirosis exposure in humans as has been shown in a slum area in the country [Citation11]. Participants also reported infection with non-severe malaria or sleeping sickness that did not respond to conventional drugs administered in the local health facilities being common in the area. This is despite the diseases being recently demonstrated in the study area [Citation9]. Poor knowledge of leptospirosis and other non-malarial acute febrile illnesses has been shown to be more prevalent in rural populations as opposed to urban populations in other parts of the world [Citation31,Citation32]. However, there has been an increased number of non-malarial acute undifferentiated fevers in the African continent due to arboviral diseases and others such as brucellosis and leptospirosis [Citation4,Citation33]. This therefore could corroborate the perceptions of the study participants of the importance of ‘malaria’ and such similar febrile illnesses being important considerations in the community’s health. This also underpins the usefulness of participatory epidemiological methods in future studies in a bid to produce research outputs more relevant to the local communities as well as assist in disease surveillance in rural communities [Citation34,Citation35]
This study shows the presence of leptospires in rodents in two counties in North-Eastern Kenya, which is an ASAL and rural area of Kenya. Phylogenetic analysis showed that the bacteria detected are pathogenic species. Rodents are regarded as important maintenance hosts of leptospires and through them, other animals and human hosts acquire the bacteria. Different rodents also are linked to specific serovars, with rats generally linked to serovars of the serogroup Icterohaemorrhagiae and mice to those of the serogroup Ballum [Citation36]. Demonstration of pathogenic leptospires through PCR therefore indicates risk of disease to human and animal populations, especially in poor communities [Citation37]. The majority of the rodents identified in this study were from areas under irrigation. Even though this study cannot determine the difference in rodents trapped between the irrigated and pastoral areas with certainty, it is well known that increased vegetation and food availability from increased rainfall in other parts of the world can lead to an increase in rodent population [Citation38], a situation that is plausible with irrigation activities. PCR percent positivity in this study was found to be higher than in other studies done in the continent which ranged from 1.5% to 18.9% in rodent populations and the serological results being within the limits noted in other African countries (0% to 62.5%) based on various serological tests [Citation5]. This study however utilized a small sample size and therefore may not have be enough to state the prevalence of the bacteria in the counties.
This study also shows that land-use may have an effect in the percent positivity of Leptospira in rodent carriers. Rodents collected from the pastoral regions (Ijara and Sangailu) had six times the odds of carrying the bacteria than those from the irrigated regions (Bura and Hola). It is likely that the presence of synanthropic rodent populations habouring the bacteria could lead to more maintenance of the bacteria and likely transmission of human and animal leptospirosis as has been hypothesized in other parts of the world [Citation39]. However, due to insecurities in the area, this study was onlyable to analyze samples from the dry season from the irrigated areas, and samples from the wet and dry seasons in the pastoral areas. The confounding effects of season between irrigated and pastoral areas are therefore not possible to separate. This is especially so because both rodent abundance and leptospirosis carriage in rodents have both been shown to be affected by seasonality of weather, with warm and wet seasons seeing higher rodent populations and survival of the bacteria in the moist and warm environment [Citation39]. It has however been proposed that drier environments may encourage for more circulation of leptospires among hosts due to rodents living closer to human housing areas due to lack of food in their habitats or even sharing the few watering points in the locality that may be contaminated by infected urine [Citation9,Citation38].
The serological testing was also expected to have a lower sensitivity, and there may be therefore many false negatives affecting this result. Serologically positive animals were often negative by PCR, which is not unusual since the antibodies indicate a past exposure, and not necessarily an ongoing infection. The PCR and ELISA tests did also not agree well when a Cohen’s kappa was done (data not shown), which can be explained by the fact that the two tests detect DNA and antibodies respectively. Future studies that factor in seasonality in the sampling by including both wet and dry seasons over a longer period of time would also reveal seasonality of the disease, if any. This would greatly inform control measures in these ASAL areas. There is a need to show the presence of the bacteria in various animal hosts, including both domestic and wild animals. Knowledge on locally circulating serovars is also very needful as this will inform diagnosis and control strategies in the country, especially since immunity in humans is serovar-specific and so are vaccines available for human use currently [Citation40]. MAT testing, though very important in leptospirosis research, was not performed in this study to confirm the results obtained due to lack of resources.
This study did not show any association between the rodent species or age and the percent positivity of the bacteria using PCR analyses, although that has been shown by others [Citation11,Citation41–Citation44]. This study was however very small, and the power may not have been sufficient to detect this.
Multimammate rats were found to have a higher percent positivity than the other species by the ELISA test. Demonstration of leptospiral antibodies in the rodents is further proof of exposure of the rodents to the bacteria. The use of diluted samples in the ELISA test may have underestimated the seroprevalence. It did however show that the filter paper method can be used for analyses by this ELISA kit, although probably with a lower sensitivity. This can be important in circumstances that would not allow the direct use of blood or tissue fluid.
Though we utilized a small sample size, this study is one of the few that has demonstrated the presence of leptospira in rodent carriers in these remote, areas of Kenya and, by extension, the risk that it poses in these poor and marginalized communities. This demonstrates the understated public health importance of leptospirosis in these areas. The use of a sample size that is more statistically representative of the number and species of rodents in the area will also reveal more on these rodent carriers of the disease.
Conclusion
While the study was small to illustrate the epidemiological situation in Kenya, it has shown molecular and serological evidence of leptospiral infections in rodents from north-eastern Kenya. This finding will therefore underline the risk posed by rodent carriers in the area and the need for control strategies.
Acknowledgments
This study was part of a project entitled Dynamic Drivers of Disease in Africa:Ecosystems, livestock/wildlife, health and wellbeing NE-J001570-1 that was jointly funded by the Ecosystem Services for Poverty Alleviation Programme (ESPA) and the CGIAR Research Program Agriculture for Nutrition and Health. The ESPA programme is funded by the Department for International Development (DFID), the Economic and Social Research Council (ESRC) and the Natural Environment Research Council (NERC). The contribution of JL was supported by the Swedish Research Council (348-2013-167). This paper is published with permission from the Director of the Kenya Medical Research Institute and was developed in a write shop supported by the Washington State University, Kenya program.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
Notes on contributors
Martin Wainaina
Martin Wainaina is currently a PhD student (Biomedical Sciences) at the Free University of Berlin, Germany where he is focused on metagenomic studies of brucellosis in human and animal hosts in partnership with the German Institute of Risk Assessment (BfR) and International Livestock Research Institute (ILRI). He has an MSc in Global Health and Infectious Diseases from the University of Edinburgh, UK and a BSc in Biomedical Sciences from the University of Nairobi, Kenya. He has been a research assistant at ILRI for more than 4 years where he was focused on laboratory diagnostics for a variety of zoonotic diseases in East Africa in the animal and human health department.
Bernard Bett
Bernard Bett is a veterinarian with a postgraduate training on epidemiology, and co-leads research on emerging infectious diseases at the International Livestock Research Institute (ILRI). His competencies include infectious disease modeling, participatory epidemiology, design and implementation of analytical studies and disease surveillance. His current research focusses on the impacts of climate, land use and biodiversity changes on the occurrence and transmission of infectious diseases, especially those that are transmissible between animals and people. He works with various actors in public and private sectors in the development of decision support tools for managing disease risks based on information generated from research.
Enoch Ontiri
Enoch Ontiri is a research technician in social science at the People, Livestock and Environment team at ILRI” since this bio is not grammatically sound.
Kim Picozzi
Kim Picozzi is a senior lecturer in Global Health at University of Edinburgh with extensive experience in vector-borne and zoonotic diseases.
Bernard Agwanda
Bernard Agwanda is a mammologist working at National Museums of Kenya and involved in several international projects involving screening for diseases in wild animals
Tanja Strand
Tanja Strand is Coordinator of One Health Sweden and Scientific administrator at National Veterinary Institute. She defended her PhD in population biology at Uppsala University. Since then, Tanja Strand has performed two postdocs in infection biology. Besides veterinary and public health, her interests include communication regarding zoonotic infections and One Health.
Delia Grace
Delia Grace is an epidemiologist and veterinarian with 20 years experience in developing countries. She is a graduate from several leading universites including the National University of Ireland, Edinburgh University, the Free University Berlin and Cornell University. She leads research on zoonoses and foodborne disease at the International Livestock Research Institute in Kenya and the CGIAR Research Program on Agriculture for Human Nutrition and Health. Her main research focus is food safety in the domestic markets of developing countries. Research interests include emerging diseases, participatory epidemiology, gender and animal welfare. Her career has spanned the private sector, field-level community development and aid management, as well as research. She has lived and worked in Asia, west and east Africa and authored or co-authored more than 200 peer-reviewed publications as well as training courses, briefs, films, articles and blog posts.
Åke Lundkvist
Åke Lundkvist is a professor in Virology at Uppsala University and one of the founders of the Zoonosis Science Center (ZSC) at the Department of Medical Biochemistry and Microbiology, Uppsala University, Sweden. His research group is focused on viral zoonoses, with a specific interest in hanta-, flavi- and avian influenza viruses. His research has included basic virology, immunology, genetics, molecular epidemiology, animal models, virus evolution, antivirals and diagnostics.
Johanna Lindahl
Johanna Lindahl is a veterinary epidemiologist working at International Livestock Research Institute (ILRI) in Nairobi, Kenya. Johanna graduated from Swedish University of Agricultural Sciences after doing her PhD working on Japanese encephalitis virus in Vietnam. Since her PhD she has been focusing her research on food safety, and vector-borne, zoonotic and emerging infectious diseases in developing countries, mainly East Africa and South and Southeast Asia. In addition to this, she is coordinating a number of aflatoxin projects within ILRI.
References
- Ellis WWA. Animal leptospirosis. In: Adler B, editor. Leptospira and Leptospirosis. 1st ed. Berlin: Springer; 2015. p. 99–10.
- Costa F, Ribeiro GS, Felzemburgh RD, et al. Influence of household rat infestation on Leptospira transmission in the urban slum environment. PLoS Negl Trop Dis. 2014;8(12):e3338.
- Zhang C, Wang H, Yan J. Leptospirosis prevalence in Chinese populations in the last two decades. Microbes Infect. 2012 Apr;14(4):317–323.
- Costa F, Hagan JE, Calcagno J, et al. Global morbidity and mortality of leptospirosis: a systematic review. PLoS Negl Trop Dis. 2015. p. 1–19.
- de Vries SG, Visser BJ, Nagel IM, et al. Leptospirosis in Sub-Saharan Africa: a systematic review. Int J Infect Dis. 2014 Nov;28:47–64.
- Woods CW, Karpati AM, Grein T, et al. An outbreak of Rift Valley fever in northeastern Kenya, 1997–98. Emerg Infect Dis. 2002;8(2):138–144.
- Ari MD, Guracha A, Fadeel MA, et al. Challenges of establishing the correct diagnosis of outbreaks of acute febrile illnesses in Africa: the case of a likely Brucella outbreak among nomadic pastoralists, Northeast Kenya, March-July 2005. Am J Trop Med Hyg. 2011;85(5):909–912.
- Cook EAJ, De Glanville WA, Thomas LF, et al. Risk factors for leptospirosis seropositivity in slaughterhouse workers in western Kenya. Occup Environ Med. 2017;74(5):357-365.
- Bett BK, Said MY, Sang R, et al. Effects of flood irrigation on the risk of selected zoonotic pathogens in an arid and semi-arid area in the eastern Kenya. PLoS One. 2017;12(5):1–15.
- Nakeel M, Arimi S, Kitala P, et al. A sero-epidemiological survey of brucellosis, Q-fever and leptospirosis in livestock and humans and associated risk factors in kajiado county-Kenya. J Trop Dis. 2016;4(3):1-8.
- Halliday JEB, Knobel DL, Allan KJ, et al. Urban leptospirosis in Africa: A cross-sectional survey of Leptospira infection in rodents in the Kibera urban settlement, Nairobi, Kenya. Am J Trop Med Hyg. 2013;89(6):1095–1102.
- Lindahl JF, Grace D. The consequences of human actions on risks for infectious diseases: a review. Infect Ecol Epidemiol. 2015;5.
- Lindahl J, Bett B, Robinson T, et al. “Rift valley fever and the changing environment: a case study in east africa.” In: Bouzid M, editor. Examining the role of environmental change on emerging infectious diseases and pandemics. Hershey, PA: IGI Global; 2016. p. 178–204.
- Nyamwaya D, Wang’ondu V, Amimo J, et al. Detection of West Nile virus in wild birds in Tana River and Garissa Counties, Kenya. BMC Infect Dis. 2016 Dec;16(1):696.
- Sang R, Lutomiah J, Said M, et al. Effects of irrigation and rainfall on the population dynamics of rift valley fever and other arbovirus mosquito vectors in the epidemic-prone tana river county, Kenya. J Med Entomol. 2017; 54(2):460-470. doi: https://doi.org/10.1093/jme/tjw206
- Mbotha D, Bett B, Kairu‐Wanyoike S, et al. Inter-epidemic Rift Valley fever virus seroconversions in an irrigation scheme in Bura, south-east Kenya. Transbound Emerg Dis. 2017;65(1):e55-e62.
- Young HS, McCauley DJ, Dirzo R, et al. Interacting effects of land use and climate on rodent-borne pathogens in central Kenya. Philos Trans R Soc B Biol Sci. 2017;372(1722):20160116.
- Kenya National Bureau of Statistics. Volume 1A-population distribution by administrative units. In: The 2009 Kenya population and housing census. 2010.
- Koech OK, Kinuthia RN, Karuku GN, et al. Field curing methods and storage duration affect the quality of hay from six rangeland grass species in Kenya. Ecol Process. 2016;5(1):3.
- KNBS and SID. Exploring Kenya’s inequality. Pulling apart or pooling together: Tana River county. Nairobi; 2013.
- Government of Kenya, The equalization fund appropriation act, 2017. Kenya: Kenya Gazette Supplement No. 51 (Acts No. 8), 2017, pp. 91–95.
- KNBS and SID, “Exploring Kenya’s inequality. Pulling apart or pooling together: Garissa county,” 2013.
- Hsieh H-F, Shannon SE. Three approaches to qualitative content analysis. Qual Health Res. 2005 Nov;15(9):1277–1288.
- Eymann J, Degreef J, Häuser C, et al. Manual on field recording techniques and protocols for all Taxa Biodiversity Inventories and Monitoring. Abc Taxa. vol. 8 i-iv; 330 pp; i-iv 322 p. 2010.
- Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013 Dec;30(12):2725–2729.
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993 May;10(3):512–526.
- Gravekamp C, Van de Kemp H, Franzen M, et al. Detection of seven species of pathogenic leptospires by PCR using two sets of primers. J Gen Microbiol. 1993;139(8):1691–1700.
- Mayer-Scholl A, Hammerl JA, Schmidt S, et al. Leptospira spp. in rodents and shrews in Germany. Int J Environ Res Public Health. 2014;11(8):7562–7574.
- WHO. Human leptospirosis: guidance for diagnosis, surveillance and control. World Health Organisation: Geneva; 2003.
- Ahmed A, Engelberts MFM, Boer KR, et al. Development and validation of a real-time PCR for detection of pathogenic leptospira species in clinical materials. Eurosurveillance. 2009;4(9):e7093.
- De Araújo WN, Finkmoore B, Ribeiro, GS, et al. Knowledge, attitudes, and practices related to leptospirosis among urban slum residents in Brazil. Am J Trop Med Hyg. 2013;88(2):359–363.
- Chipwaza B, Mugasa JP, Mayumana I, et al. Community knowledge and attitudes and health workers’ practices regarding non-malaria febrile illnesses in eastern tanzania. PLoS Negl Trop Dis. 2014;8(5).
- Naing C, Kassim AIBM. Scaling-up attention to nonmalaria acute undifferentiated fever. Trans R Soc Trop Med Hyg. 2012;106(6):331–332.
- Catley A, Irungu P, Simiyu K, et al. Participatory investigations of bovine trypanosomiasis in Tana River District, Kenya. Med Vet Entomol. 2002;16:55–66.
- Jost CC, Mariner JC, Roeder PL, et al. Participatory epidemiology in disease surveillance and research. Rev Sci Tech Int Des Epizoot. 2007;26:537–549.
- Levett PN. Leptospira. In: Jorgensen JH, Pfaller MA, Carroll KC, et al., editors. Manual of Clinical Microbiology, 11th Edition, 11th, 1028–1036. Washington D.C: ASM Press; 2015, no. 24.
- Meerburg BG, Singleton GR, Kijlstra A. Rodent-borne diseases and their risks for public health. Crit Rev Microbiol. 2009;35(3):221–270.
- Gubler DJ, Reiter P, Ebi KL, et al. Climate variability and change in the USA: potential impacts on vector- and rodent-borne diseases. Environ Health Perspect. 2001;109:223–233.
- Perez J, Brescia F, Becam J, et al. Rodent abundance dynamics and Leptospirosis carriage in an area of hyper-endemicity in new Caledonia. PLoS Negl Trop Dis. 2011;5(10):e1361.
- Haake DA, Levett PN. Leptospirosis in humans. In: Adler B, editor. Leptospirosis and Leptospira. 1st ed. Melbourne: Springer-Verlag Berlin Heidelberg; 2015. p. 65–98.
- Sarkar U, Nascimento SF, Barbosa R, et al. Population-based case-control investigation of risk factors for leptospirosis during an urban epidemic. Am J Trop Med Hyg. 2002;66(5):605–610.
- de Faria MT, Calderwood MS, Athanazio DA, et al. Carriage of Leptospira interrogans among domestic rats from an urban setting highly endemic for leptospirosis in Brazil. Acta Trop. 2008;108:1–5.
- Piedad AF, Londoño AF, Quiroz VH, et al. Prevalence of Leptospira spp. in urban rodents from a Groceries Trade Center of Medellín, Colombia. Am J Trop Med Hyg. 2009;81(5):906–910.
- Vanasco NB, Sequeira MD, Sequeira G, et al. Associations between leptospiral infection and seropositivity in rodents and environmental characteristics in Argentina. Prev Vet Med. 2003;60(3):227–235.
