ABSTRACT
Consumption of contaminated meat, milk, and water are among the major routes of human campylobacteriosis. This study aimed to determined the genetic diversity of C. coli and C. jejuni isolated from meat, milk, and water samples collected from different locations. From the 376 samples (meat = 248, cow milk = 72, and water = 56) collected, a total of 1238 presumptive Campylobacter isolates were recovered and the presence of the genus Campylobacter were detected in 402 isolates, and from which, 85 and 67 isolates were identified asC. jejuni and C. coli respectively. Of which, 71 isolates identified as C. coli (n = 35) and C. jejuni (n = 36) were randomly selected from meat, milk, and water samples and were genotyped using enterobacterial repetitive intergenic consensus PCR (ERIC-PCR). The digital images of the ERIC-PCR genotype were analyzed by GelJ v.2.0 software. The diversity and similarity of the isolates were assessed via an unweighted-pair group method using average linkages clustering algorithm. The results showed that the 36 C. jejuni strains separated into 29 ERIC-genotypes and 4 clusters while the 35 C. coli were delineated into 29 ERIC-genotypes and 6 clusters. The study revealed the genetic diversity among C. coli and C. jejuni strains recovered from different matrices characterized by Gelj.
Introduction
Campylobacters are commensal members of the gut microbiota of livestock, poultry [Citation1], wild birds [Citation2], and are also found in aquatic milieu [Citation3]. Several Campylobacter species are known to cause infections and campylobacteriosis are largely caused by C. coli and C. jejuni [Citation4,Citation5]. Campylobacteriosis cases have been reported in several parts of the world including South America, Africa, North America, Asia, and Europe [Citation6]. Campylobacteriosis is usually associated with consumption of contaminated vegetables, fruits, raw milk, untreated water, foodstuffs, and undercooked meats [Citation4]. About 50%–80% of human campylobacteriosis cases are caused by chicken consumption [Citation7]. Nevertheless, cattle-related campylobacteriosis cases through consumption of unpasteurized milk or undercooked beef have also been reported [Citation8]. Campylobacter species including C. coli and C. jejuni are often isolated and detected in sewage, milk, water, and meats samples [Citation9].
In developing countries, C. jejuni and C. coli are the two major Campylobacter species known as potential etiological agents of acute diarrhea especially in children [Citation10]. Campylobacter infections are ecologically diverse [Citation11] and as a result of the burdens and public health impact of Campylobacter species, studies on epidemiological analyses, genetic diversity, and similarity of these bacteria species are very significant [Citation4]. Hence, molecular typing techniques are used for the determination of the similarities of isolates and these have shown in better detection of Campylobacter strains [Citation12]. Molecular typing techniques with high sensitivity and specificity are recognized as gold standard test for epidemiological investigations of some pathogens including Campylobacter species [Citation13,Citation14]. Molecular typing of bacteria isolates is useful to characterize the genetic similarity of the isolates [Citation15] and also helpful in conducting epidemiological studies designed at source tracking of sporadic campylobacteriosis cases by providing information on the genetic Campylobacter subtypes in circulation [Citation12]. Molecular genotyping procedures have been used for the detection of intra-species variability of a specific microorganism [Citation16]. Molecular typing methods are assessed base on their performance and ease in usage [Citation17]. Some molecular typing techniques used for genotyping of bacteria isolates include multilocus sequence typing (MLST), gene sequencing-based methods, phylogenetic analysis [Citation18], PCR-ribotyping, PCR sequencing, and enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) [Citation19,Citation20]. ERIC-PCR technique is a simple tool used to differentiate bacteria strains isolated from diverse sources. This technique is a strong tool for the exploration of prokaryotic genomes and has been reported to have improved reproducibility and high discriminatory power [Citation21]. ERIC-PCR is a repetitive element-based PCR technique [Citation22]. The pros of ERIC-PCR over other molecular typing techniques include the capacity to distinguish between closely related bacteria strains as well as being quick, simple, cheap, dependable, and high-throughput genotyping method [Citation23]. Molecular typing of Campylobacter species by ERIC-PCR has been reported to show high discriminatory power in strains diversity [Citation24], and the explanation of banding patterns of ERIC-PCR DNA fingerprinting by visual judgment is an onerous task particularly when relating to multiple band patterns, distant, and diverse bands. However, GelJ is a tool that simplify this task and is used in analyzing DNA fingerprint gel images to overcome these limitations [Citation25]. In South Africa, there is paucity of information on genotyping of C. coli and C. jejuni isolated from milk, water and meat samples and there is no report on molecular genotyping of Campylobacter species in the study area. Since it is not possible to determine the genetic relatedness of isolates using ordinary PCR approach; therefore, the study evaluated the genetic diversity of C. jejuni and C. coli isolated from water, milk, and meat samples using ERIC-PCR techniques.
Material and methods
Sources of bacteria strains
A total of 376 samples (Meat = 248, cow milk = 72, and water = 56) were collected in Chris Hani and Amathole District Municipalities, South Africa between March 2019–August 2019. The meat samples [beef, turkey, pork, mutton, and chicken) were obtained from supermarkets, retail shops, and butcheries, the milk samples were obtained from farms, cars/roads side, retail shops, and butcheries while the water samples were obtained from rivers and pond used for irrigation. The meat samples were aseptically packed into different sterile plastic bags while the water and milk samples were collected in sterile 1 L and 250 mL polypropylene bottles, transported to the laboratory for examination in a cooler box with ice packs within six hours of collection. For the microbiological analysis of the samples, the methods described by [Citation26 Citation27], was adopted for isolation of Campylobacter from water, milk, and meat samples, respectively, incubated under microaerophilic conditions in 10% CO2 in HF151UV incubator for 48 h at 42°C.
Extraction of bacterial DNA
DNA was extracted by boiling method following the procedure described by [Citation28], with slight modification as reported by [Citation29].
Molecular confirmation and identification of C. jejuni and C. coli
Molecular identification of the genus Campylobacter was carried out by PCR assay using the primer sets listed in supplementary Table 1 as reported by [Citation30], targeting part of the 16S rRNA gene at 439 bp. The identified genus Campylobacter were further delineated into C. coli and C. jejuni using the primer sets reported by [Citation31], targeting asK and cj0414 genes, respectively, as shown in .
Table 1. Primers sets used for the identification of Campylobacter species.
ERIC-PCR typing of C. jejuni and C. coli
ERIC-PCR is a molecular-based method that has been well applied for the discrimination of Campylobacter species [Citation32]. From the identified isolates detected as C. coli and C. jejuni, 71 strains [C. coli = 35 and C. jejuni = 36) were randomly selected from different sources and the identified isolates were subjected to PCR using the ERIC primer sets R1: ATGAAGCTCCTGGGGATTCAC and R2: AAGTAAGTGACTGGGGTGAGCG following the method described by [Citation33]. The PCR reactions were verified by resolving them in 3% agarose gel in a 5x TBE buffer, stained with ethidium bromide at 90 volts for 240 min and viewed as stated before.
Clustering analysis and determination of discriminatory power (D]
The DNA fingerprints obtained from the ERIC-PCR technique were analyzed with computer-assisted pattern analysis using the GelJ v.2.0. software [Citation25]. The relatedness of the isolates was compared and dendrograms were constructed by UPGMA and cluster analysis were used to determine the relationships between each isolate. The value of discriminatory power [D) was determined using online calculator for discriminatory power as reported by [Citation34].
Determination of reproducibility of ERIC-PCR
The capacity of a technique to give the same result when repeated tests are carried out on the same isolate is recognized as reproducibility [Citation22]. The ERIC-PCR analysis of the DNA fingerprints result can be directly correlated within a single PCR experiment. However, to determine the reproducibility of these techniques, the clustered result of the 35 C. coli and 36 C. jejuni isolates were repeated twice in further ERIC-PCR assay.
Results
Incidence of Campylobacter and Campylobacter species in the matrices
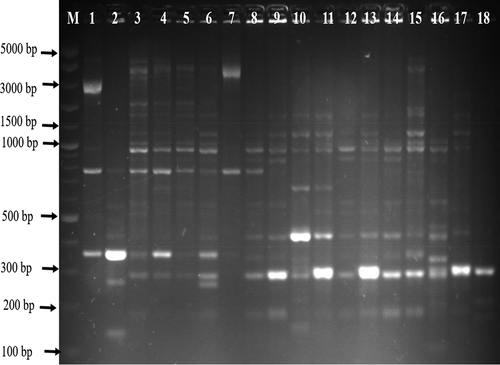
From culture, a total of 1238 presumptive Campylobacter isolates were recovered and the presence of the genus Campylobacter was detected in 402 isolates, and from which 85 and 67 isolates were detected to be C. jejuni and C. coli respectively. Of which 71 isolates identified as C. coli (n = 35) and C. jejuni (n = 36) were randomly selected from meat, milk, and water samples. Of the 71 identified selected C. jejuni and C. coli fingerprint, 23 (65.71%) C. coli isolates were from meat samples (chicken = 4, pork = 6, beef offals = 13), 11 (31.43%) C. coli from milk samples, 1 (2.88%) C. coli from water samples, 23 (63.89%) C. jejuni were from water samples, 13 (36.11%) C. jejuni from meat samples (chicken = 9, beef = 2, and beef offals = 2). shows a gel image of some of the PCR identified C. coli and C. jejuni isolates.
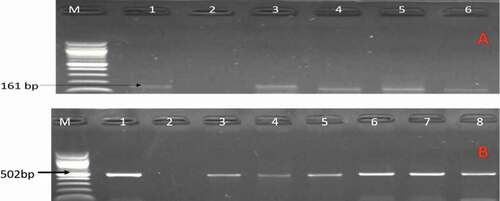
Figure 1. A gel image of PCR confirmed C. jejuni. Lane M: 100 bp DNA ladder, lane 1: positive control (C. jejuni ATCC 33560), lane 2: negative control, lane 3–6: positive C. jejuni isolates (161 bp) while image B are some confirmed C. coli. Lane 1: positive control (C. coli ATCC 33559), lane 2: negative control, lane 3–8: positive C. coli isolates (502 bp).
Genetic diversity of C. jejuni and C. coli isolates
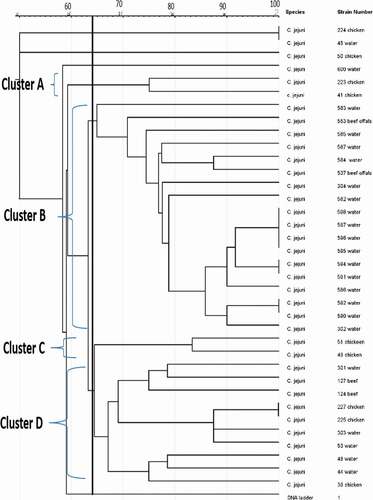
The genetic fingerprints of C. jejuni and C. coli from meat, water, and milk were characterized by ERIC-PCR and the dendrogram images obtained from the analysis of ERIC-PCR results were constructed using the GelJ v.2.0. software to define the genetic similarity of the isolates. The ERIC-PCR condition that was previously developed and reported for fingerprints analysis of Campylobacter species was used to analyze 71 Campylobacter isolates identified as C. jejuni (36) and C. coli (35). The gel image band patterns of C. jejuni and C. coli strains varied in relation of the distribution of polymorphic bands ranging from 100 to 4500 bp ( and ). The variances among the two Campylobacter species analyze were evaluated on the basis of the migration arrangements of the amplified bands. From the 36 C. jejuni isolates from water and meat samples genotyped using ERIC-PCR assay, 35 isolates out of the 36 isolates produced 1–14 bands and the UPGMA dendrogram clustering image separated all the 36 isolates into 29 ERIC-genotypes and the 29 ERIC-genotypic profiles generated were grouped into four clusters at a similarity cutoff of 65% ().
The dendrogram image obtained from GelJ cluster analysis of C. jejuni, the highest ERIC-genotype cluster of C. jejuni profiles generated was found in cluster B (composed of 18 isolates) followed by cluster D (composed of 10 isolates) and cluster A and C (composed of 2 isolates each). Some isolates had no polymorphic band and were not group into any cluster. Also, some of the C. jejuni strains showed a high degree of relatedness with similarity indices of 100%. Furthermore, the dendrogram image constructed showed that some of the C. jejuni isolates recovered from chicken, beef and water were clustered into the same group and this indicates high genetic relatedness among the isolates. Likewise, the ERIC-PCR genotyping result of 35 C. coli isolates produced 1–10 bands (), and the dendrogram clustering image constructed separated the 35 C. coli isolates into 29 ERIC-genotypic profiles at a similarity cutoff of 70%. Furthermore, the dendrogram image shown an important intra-species diversity of C. coli strains irrespective of the isolates origins. The 29 ERIC-genotypes generated were grouped into six clusters () and among the clusters, the most prevalent genotypes were in cluster F represented by 10 isolates, followed by cluster C composed of 11 isolates, cluster A composed of 4 isolates, cluster B composed of 3 isolates and Cluster D and E composed of 2 isolates each. The six clusters of C. coli generated were visibly separated and in cluster C, C. coli isolate recovered from pork and milk showed a high level of genetic relatedness (100%). Similarly, the DNA fingerprints of some isolates obtained from water samples were clustered into the same group with those isolated from meat and milk samples obtained from different locations while some isolates had no band and were not clustered into any group with others.
Figure 2. Gel ERIC-PCR amplification results of C. jejuni isolates. M, DNA ladder, 1–18: some amplified C. jejuni isolates.
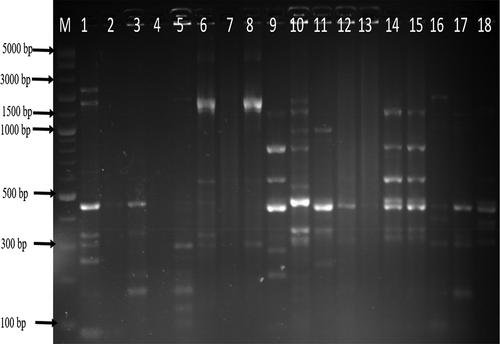
Figure 3. UPGMA dendrogram image obtained from cluster analysis showing the relationship and diversity of 36 C. jejuni isolates from different sources using ERIC-PCR technique.
Reproducibility
Reproducibility of the experiments was confirmed by the UPGMA clustering results of the 35 (C. coli) and 36 (C. jejuni) isolates that had clustered together on preceding ERIC-PCR assay, which were observed to continuing to cluster on the following ERIC-PCR assay.
Discussion
Molecular typing techniques including ERIC-PCR have been revealed to shown high genomic diversity of Campylobacter species, reflecting its quick adaptive changes during infection [Citation21]. Some Campylobacter species are well known to cause infections and it is vital that Campylobacter species are correctly differentiated into intra-species diversity. The present study revealed the genetic diversity of C. coli and C. jejuni obtained from different sources using ERIC-PCR DNA fingerprinting assay. DNA fingerprinting assay is used for comparing DNA patterns, which allows the analysis of the genomic similarity among different samples and categorizes them into clusters [Citation25]. From the PCR results, a total of 71 isolates identified as C. jejuni and C. coli from milk, water, and meats were randomly selected and genotyped by ERIC-PCR. ERIC-PCR study has been used to amplify different DNA regions to generate genetic patterns that are precise for specific isolates [Citation35]. The result from the ERIC-PCR fingerprinting of both C. coli and C. jejuni clustered the isolates into six and four clusters, respectively, and this result suggests that the isolates from meats, milk, and water sources have high genetic diversity.
Though clustering of C. jejuni and C. coli isolates from different sampling sites and source propose evolutionary relatedness of the isolates. This result revealed that investigation on genotyping of different C. jejuni and C. coli strains using ERIC-PCR showed better detection of the diverse strains and these results is in accordance with the reported of [Citation36–Citation38]. Furthermore, C. coli and C. jejuni isolates obtained from different matrices were grouped either in the same or different clusters. The ERIC-PCR analysis separated distinguishingly the 36 C. jejuni isolates from water and meat samples into 34 ERIC-genotypes and this indicates genetic diversity among C. jejuni isolates recovered from the two sources. This result is in akin with the reports of [Citation39–Citation41], who have also reported high diversity of Campylobacter species obtained from porcine, avian, turkey, and broiler, respectively. The UPGMA dendrogram clustering image of the 36 C. jejuni isolates genotyped grouped the isolates into four clusters and this finding is in line with the report of [Citation42,Citation43]. Result obtained from C. jejuni fingerprinting is contrary to the report of [Citation44], who in their report revealed that 15 C. jejuni genotyped were grouped into 10 genotypes with 3 clusters. C. jejuni have a natural capacity for gene rearrangements and gene transfer in the genome which might likely explain the increase in its genomic heterogeneity [Citation45]. The UPGMA dendrogram image generated from the genotyping analysis of C. jejuni isolates revealed high level of genetic diversity among C. jejuni isolates and our result corresponds with the report of [Citation46]. The dendrogram image of C. jejuni isolates showed that some isolates were highly diverse while some were 100% genetically related and this shows the genetic multiplicity of C. jejuni strains analyzed. This result corresponds with the report of [Citation47], who also revealed the genetic multiplicity of C. jejuni isolates recovered from aquatic sources. In the report of [Citation48], multiple strains of C. jejuni belonging to four distinct clades were revealed to be implicated in human campylobacteriosis outbreak as a result of the consumption of undercooked chicken liver pâte. Multiple strains or co-infection of human campylobacteriosis cases involving more than one C. jejuni subtype might occur in up to 10% of most Campylobacter infection cases [Citation49].
In the study conducted by [Citation50], multiple C. jejuni subtypes were detected in chickens. In addition, some of the C. jejuni subtypes causing human infection may be minor strains in the chicken microflora [Citation48]. Furthermore, study has also shown that C. jejuni has caused the highest zoonosis menace to public health compared to other Campylobacter species [Citation51]. Another Campylobacter species reported to cause high rate of campylobacteriosis is C. coli and the ERIC-PCR profile of C. coli genotyped showed that some of the C. coli strains were genetically diverse, demonstrating the occurrence of various genotypes of C. coli in the study area. It was also observed that some C. coli strains had no polymorphic band while some showed a high level of genetic similarity of 100%. Furthermore, the 35 C. coli isolates analyzed were grouped into 29 genotypes and this result is similar with the report of [Citation52], who in their study obtained 22 genotypes from 65 C. coli isolates analyzed. Also, the dendrogram revealed the genetic diversity among the C. coli isolates recovered from the same or different sources. Simultaneously, high genetic relatedness was also observed among some C. coli isolates recovered from milk and pork as seen in cluster C in and this result corroborates with the report of [Citation8]. The finding also revealed that C. coli isolate recovered from aquatic milieu was closely related with those from other sources and this result corresponds with the report of [Citation53]. Detection of genotypic relatedness among some C. coli and C. jejuni isolates recovered from aquatic milieu and meat sources confirmed that livestock and poultry are sources of environmental spread of Campylobacter species and these results is in accordance with the report of [Citation54]. Genetic diversity is one of the different mechanisms that helps pathogens to thrive in unfriendly circumstances within the environment or in the host given them the ability to colonize multiple hosts [Citation55]. Genomic rearrangement and horizontal gene transfer are among the diverse phenomena that can cause genetic diversity in C. jejuni and C. coli [Citation45]. The observed various band patterns suggest the presence of genetically diverse strains of C. coli and C. jejuni analyzed.
This method used in Campylobacter genotyping is very sensitive in identifying or detecting variances between isolates of the same species and also provide discerning way for discriminating C. coli and C. jejuni strains and this result is in line with the report of [Citation56]. In this study, the ERIC-PCR result gave a discriminatory power [D) value of 0.78 for C. coli and 0.60 for C. jejuni and this is a good value and the result is akin with the report of [Citation46]. This is the first report from the study area on the application of ERIC-PCR for genotyping of Campylobacter species from different sources. The DNA profiles were visibly noticeable through precise fingerprint arrangements. The ERIC-PCR is therefore suggested to be used as simple and cheap tool for the determination of diverse strains of bacterial species. The primer sets used in this study aided in differentiating closely related strains within the same Campylobacter species and the results from this study revealed a wide genomic multiplicity of C. jejuni and C. coli recovered from water, milk and meat samples. However, the isolates analyzed in this present study are not representative of the Campylobacter populace and we are guarded that no generalization is deduced from the study. More investigation would provide more data on the epidemiology of Campylobacter in the study area.
Author contribution
AI design, carried out the experiment, analyze the result, and drafted the manuscript. AIO supervises the work, corrected, and proofread the manuscript.
Acknowledgments
The authors are grateful to South Africa Medical Research Council (SAMRC] for financial support.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Sheppard SK , Colles FM , Mccarthy ND , et al. Niche segregation and genetic structure of Campylobacter jejuni populations from wild and agricultural host species. Mol Ecol. 2011;20(16):3484–9.
- Atterby C , Mourkas E , Méric G , et al. The potential of isolation source to predict colonization in avian hosts: a case study in Campylobacter jejuni strains from three bird species. Front Microbiol. 2018;9:591.
- Mourkas E , Florez‐Cuadrado D , Pascoe B , et al. Gene pool transmission of multidrug resistance among Campylobacter from livestock, sewage and human disease. Environ Microbiol. 2019;21(12):4597–4613.
- Gomes CN , Souza RA , Passaglia J , et al. Genotyping of Campylobacter coli strains isolated in Brazil suggests possible contamination amongst environmental, human, animal and food sources. J Medical Microbiol. 2016;65(1):80–90.
- Ma L , Wang Y , Shen J , et al. Tracking Campylobacter contamination along a broiler chicken production chain from the farm level to retail in China. Int J Food Microbiol. 2014;181:77–84.
- Paruch L , Paruch AM , Sørheim R. DNA-based faecal source tracking of contaminated drinking water causing a large Campylobacter outbreak in Norway 2019. Int J Hygiene Environ Health. 2020;224:113420.
- González-Hein G , Huaracán B , García P , et al. Prevalence of virulence genes in strains of Campylobacter jejuni isolated from human, bovine and broiler. Brazilian J Microbiol. 2013;44(4):1223–1229.
- Sheppard SK , Dallas JF , MacRae M , et al. Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6. Int J Food Microbiol. 2009;134(1–2):96–103.
- Pallavi , Kumar A , Kumar. MS , et al. Comparative analysis of cultural isolation and PCR based assay for detection of Campylobacter jejuni and Campylobacter coli in foods of animal origin. J Animal Res. 2015;5:21–25.
- Silva J , Leite D , Fernandes M , et al. Campylobacter spp. as a foodborne pathogen: a review. Front Microbiol. 2011;2:200.
- Lu X , Huang Q , Miller WG , et al. Comprehensive detection and discrimination of Campylobacter species by use of confocal micro-Raman spectroscopy and multilocus sequence typing. J Clin Microbiol. 2012;50(9):2932–2946.
- Taboada EN , Clark CG , Sproston EL , et al. Current methods for molecular typing of Campylobacter species. J Microbiol Methods. 2013;95(1):24–31.
- Amar CF , East CL , Gray J , et al. Detection by PCR of eight groups of enteric pathogens in 4627 faecal samples: re- examination of the English case-control infectious intestinal disease study (1993–1996). Eur J Clinical Microbiol Infect Dis. 2007;26(5):311–323.
- Taylor EV , Herman KM , Ailes EC , et al. Common source outbreaks of Campylobacter infection in the USA, 1997– 2008. Epidemiol Infect. 2013;141(5):987–996.
- Behringer M , Miller WG , Oyarzabal OA . Typing of Campylobacter jejuni and Campylobacter coli isolated from live broilers and retail broiler meat by flaA-RFLP, MLST, PFGE and REP-PCR. J Microbiol Method. 2011;84:194–201.
- Wassenaar TM , Newell DG . Genotyping of Campylobacter species. Appl Environ Microbiol. 2000;66(1):1–9.
- Adzitey F , Huda N , Ali GRR . Molecular techniques for detecting and typing of bacteria, advantages and application to foodborne pathogens isolated from ducks. 3 Biotech. 2013;3(2):97–107.
- Merga JY , Leatherbarrow AJH , Winstanley C , et al. Comparison of Arcobacter isolation methods and diversity of Arcobacter spp. in Cheshire, UK. Appl Environ Microbiol. 2011;77:1646–1650.
- Anjay AK , Agarwal RK , Ramees TP , et al. Molecular typing of Salmonella Typhimurium and S. Enteritidis serovars from diverse origin by ERIC-PCR. J Pure Appl Microbiol. 2015;9(3):2627–2634.
- Ramees TP , Rathore RS , Bagalkot PS , et al. Genotyping and genetic diversity of Arcobacter butzleri and Arcobacter cryaerophilus isolated from different sources by using ERICPCR from India. Veterinary Quarterly. 2014;34:211–217.
- Wieczorek K . Relationship between the molecular typing of Campylobacter strains and the prevalence of their virulence genes. Bull Veterinary Inst Pulawy. 2009;53:193–198.
- Bilung LM , Pui CF , Su’ut L , et al. Evaluation of BOX-PCR and ERIC- PCR as molecular typing tools for pathogenic. In: Leptospira. Disease markers. 2018; 1–9.
- Dombek PE , Johnson LK , Zimmerley ST , et al. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl Environ Microbiol. 2000;66(6):2572–2577.
- Wilson MK , Lane AB , Law BF , et al. Analysis of the pan genome of Campylobacter jejuni isolates recovered from poultry by pulsed-field gel electrophoresis, multilocus sequence typing (MLST), and repetitive sequence polymerase chain reaction (rep-PCR) reveals different discriminatory capabilities. Microbial Ecol. 2009;58(4):843–855.
- Heras J , Domínguez C , Mata E , et al. GelJ–a tool for analyzing DNA fingerprint gel images. BMC Bioinformatics. 2015;16(1):270.
- Van Dyke MI , Morton VK , McLellan NL , et al. The occurrence of Campylobacter in river water and waterfowl within a watershed in southern Ontario. Canada J Appl Microbiol. 2010;109(3):1053–1066.
- Habib I , Berkvens D , De Zutter L , et al. Campylobacter contamination in broiler carcasses and correlation with slaughterhouses operational hygiene inspection. Food Microbiol. 2012;29(1):105–112.
- Sierra-Arguello YM , Furian TQ , Perdoncini G , et al. Fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli from poultry and human samples assessed by PCR-restriction fragment length polymorphism assay. PloS One. 2018;13(7):0199974.
- Igwaran A , Okoh I . Campylobacteriosis agents in meat carcasses collected from two district municipalities in the eastern Cape Province, South Africa. Foods. 2020;9(2):1–18.
- Moreno Y , Botella S , Luis Alonso J , et al. Specific detection of Arcobacter and Campylobacter strains in water and sewage by PCR and fluorescent in situ hybridization. Appl Environ Microbiol. 2003;69(2):1181–1186.
- Yamazaki-Matsune W , Taguchi M , Seto K , et al. Development of a multiplex PCR assay for identification of Campylobacter coli, Campylobacter fetus, Campylobacter hyointestinalis subsp. hyointestinalis, Campylobacter jejuni, Campylobacter lari and Campylobacter upsaliensis . J Medical Microbiol. 2007;56(11):1467–1473.
- Zorman T , Heyndrickx M , Uzunović-Kamberović S , et al. Genotyping of Campylobacter coli and C. jejuni from retail chicken meat and humans with campylobacteriosis in Slovenia and Bosnia and Herzegovina. Inter J Food Microbiol. 2006;110(1):24–33.
- Versalovic J , Koeuth T , Lupski JR . Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19(24):6823–6831.
- Milton AAP , Rathore RS , Kumar A . Genotyping of Campylobacter jejuni and C. coli from different poultry sources and human by ERIC-PCR. J Vet Public Health. 2015;13(2):93–98.
- Ateba C , Mbewe M . Genotypic characterization of Escherichia coli O157: H7 isolates from different sources in the north-west province, South Africa, using enterobacterial repetitive intergenic consensus PCR analysis. Intern J Mol Sci. 2014;15(6):9735–9747.
- Tanil GB , Radu S , Nishibuchi M , et al. Characterization of Vibrio parahaemolyticus isolated from coastal seawater in peninsular Malaysia. Southeast Asian J Tropical Med Public Health. 2005;36(4):940.
- Kosek M , Yori PP , Gilman RH , et al. Facilitated molecular typing of Shigella isolates using ERIC-PCR. Am J Tropical Med Hygiene. 2012;86(6):1018–1025.
- Candan ED , Idil N , Bilkay S . Usefulness of REP and ERIC-PCR combination for tracking the spread of Staphylococcus aureus strains. Minerva Biotecnologica. 2013;25:245–250.
- Madden R , Moran L , Scates P . Diversity of Campylobacter coli genotypes in the lower porcine gastrointestinal tract at time of slaughter. Lett Appl Microbiol. 2007;45:575–580.
- Ridley A , Toszeghy M , Cawthraw S , et al. Genetic instability is associated with changes in the colonization potential of Campylobacter jejuni in the avian intestine. J Appl Microbiol. 2008;105:95–104.
- Staji H , Birgani SF , Raeisian B . Comparative clustering and genotyping of Campylobacter jejuni strains isolated from broiler and turkey faeces by using RAPD- PCR and ERIC-PCR analysis. Ann Microbiol. 2018;68(11):755–762.
- Ahmed HA , El Hofy FI , Ammar AM , et al. ERIC- PCR genotyping of some Campylobacter jejuni isolates of chicken and human origin in Egypt. Vector-Borne Zoonotic Dis. 2015;15(12):713–717.
- Zhong X , Wu Q , Zhang J , et al..Prevalence, genetic diversity and antimicrobial susceptibility of Campylobacter jejuni isolated from retail food in China. Food Cont. 2016;62:10–15.
- Zheng Y , Wu Q , Wu K , et al.. Virulence associated gene detection and ERIC-PCR typing of Campylobacter jejuni strains isolated from foods in four Southern Chinese provinces. Acta Microbiol Sinica. 2014;54:14–23.
- Siemer BL , Harrington CS , Nielsen EM , et al.. Genetic relatedness among Campylobacter jejuni serotyped isolates of diverse origin as determined by numerical analysis of amplified fragment length polymorphism (AFLP) profiles. J Appl Microbiol. 2004;96(4):795–802.
- El-Adawy H , Hotzel H , Tomaso H , et al.. . Detection of genetic diversity in Campylobacter jejuni isolated from a commercial turkey flock using flaA typing, MLST analysis and microarray assay. PloS One. 2013;8(2):51582.
- Le´vesque S , Frost E , Arbeit RD , et al.. Multilocus sequence typing of Campylobacter jejuni isolates from humans, chickens, raw milk, and environmental water in Quebec. Canada J Clinical Microbiol. 2008;46:3404–3411.
- Devane M , Gilpin B , Robson B , et al.. Identification of multiple subtypes of Campylobacter jejuni in chicken meat and the impact on source attribution. Agri. 2013;3(3):579–595.
- Gilpin B , Robson B , Lin S , et al.. Pulsed-field gel electrophoresis analysis of more than one clinical isolate of Campylobacter spp. from each of 49 patients in New Zealand. J Clin Microbiol. 2012;50:457–459.
- Skånseng B , Trosvik P , Zimonja M , et al.. Co-infection dynamics of a major food-borne zoonotic pathogen in chicken. PLoS Pathog. 2007;3(11):175.
- Gürtler M , Alter T , Kasimir S , et al.. The importance of Campylobacter coli in human campylobacteriosis: prevalence and genetic characterization. Epidemiol Infect. 2005;133:1081–1087.
- Aquino MHC , Filgueiras ALL , Matos R , et al.. Diversity of Campylobacter jejuni and Campylobacter coli genotypes from human and animal sources from Rio de Janeiro. Brazil Res Veterinary Sci. 2010;88(2):214–217.
- Denis M , Tanguy M , Chidaine B , et al.. Description and sources of contamination by Campylobacter spp. of river water destined for human consumption in Brittany, France. Pathol Biol. 2011;59(5):256–263.
- Meistere I , Ķibilds J , Eglīte L , et al.. Campylobacter species prevalence, characterisation of antimicrobial resistance and analysis of whole-genome sequence of isolates from livestock and humans, Latvia, 2008 to 2016. Eurosurveillance. 2019;24(31). DOI:10.2807/1560-7917.ES.2019.24.31.1800357
- Chuma IS , Nonga HE , Mdegela RH , et al.. Epidemiology and RAPD- PCR typing of thermophilic campylobacters from children under five years and chickens in Morogoro municipality, Tanzania. BMC Infectious Dis. 2016;16(1):692.
- Duan H , Chai T , Liu J , et al.. Source identification of airborne Escherichia coli of swine house surroundings using ERIC-PCR and REP-PCR. Environ Res. 2009;109:511–517.