ABSTRACT
Antimicrobial resistance is a growing public health problem and a threat to effective treatment and prevention of an array of infections caused by bacteria. Africa is already faced with many socio-economic and health crises. Many countries in Africa can seldom boast of a standardized health care facility comparable to those in developed countries. Yet, the non-therapeutic use of COL has been banned in developed countries. However, in Africa, except for South Africa, COL is an over-the-counter (OTC) medication sold and dispensed by non-professionals/without a veterinarian’s supervision. The ban of non-therapeutic COL in developed countries has proven to reduce the development of mobile colistin resistance (MCR) in humans and animals. The unregulated use of COL has been proven to select pathogenic and commensal bacteria resistance. A transmissible plasmid-mediated colistin determinant, mobile COL resistance (mcr) gene, which is rapidly transferred/acquired horizontally or laterally intra/inter-species/genera, has been reported. A highly promiscuous mobile genetic element like plasmids containing transposons, insertion sequences, and integrons aid the carriage/rapid transfer and acquisition of these mcr genes. Hence, we highlight the danger posed by escalating colistin (COL) resistance in the continent and the impetus to halt the indiscriminate and non-therapeutic use of COL to protect public health.
Introduction
For many decades, antibiotic has revolutionized healthcare globally, bringing respite to clinicians and veterinarians, especially regarding the treatment of stubborn infections. But the increasing problem of antimicrobial resistance threatens to reverse all the benefits of antibiotics in modern medicine. The damaging impact of antimicrobial resistance (AMR) is already manifesting around the globe. For instance, over 50,000 lives are lost each year across Europe and the US alone, and hundreds of thousands more are dying in other parts of the world[Citation1]. The global estimate of deaths due to AMR stands at 700,000, and it is projected to reach 10 million by the year 2050[Citation1]. In Africa, deaths attributable to AMR every year by 2050 are estimated to reach 4,150,000[Citation1]. The projected figure could even be higher in low- and middle-income countries and specifically in Africa, where there is a lack or absence of health infrastructure, poor hospital seeking habit, poor record-keeping, and generally weak or poor governance and regulatory framework. Hence it is time to sound the alarm on the trend and spread of colistin resistance in Africa and the need for every country in the continent to enhance its antibiotics stewardship programs.
Mechanisms of colistin resistance
Colistin (COL) is a polymyxin antibiotic used as a last-line agent for treating deadly infections in humans and animals. COL was discovered in 1947 and isolated in 1950 from the soil bacterium Paenibacillus (Bacillus) polymyxa subspecies colistinus [Citation2]. But its use in human medicine was largely abandoned in the 1970s due to its toxic effects on the kidney and nervous system. However, it continued to be used in the livestock sector in most parts of the world, including inAfrica [Citation3]. However, in the early 2000s, the use of COL in humans was reintroduced due to the rapid rise in deadly infections caused by multi- and extensively-drug resistant organisms [Citation4].
There was a low interest in the COL resistance concept because it was thought that the only mechanism for COL resistance is by mutation in chromosomal genes such as pmrAB, phoPQ, mgrB, and crrAB which are only vertically transferred to bacterial progenies (clones), and thus, by its very nature rare and self-limiting. This perception changed in late 2015 following the discovering of a transmissible plasmid-mediated colistin determinant, mobile COL resistance (mcr-1) gene (in Escherichia coli isolates from humans/meats in China), which is rapidly transferred/acquired horizontally/laterally intra/inter species/genera [Citation5]. After the discovery in China, subsequent detection of this plasmid-mediated mechanism was quickly reported worldwide [Citation6].However, retrospective studies have shown that mcr gene has been in existence since the 1980s. This coinciding with when COL use in livestock began in China but remained undetected [Citation7]. The rapid transfer and acquisition of mcr gene is because they are carried by highly promiscuous mobile genetic elements like plasmids containing transposons, insertion sequences, and integrons. Thus, with mobile colistin resistance (MCR) fast-spreading globally, this poses a considerable threat to antimicrobial therapy, especially in Africa where access to other good quality effective last-line antibiotics is limited.
Distribution of colistin resistance in Africa and its implications
Available evidence shows that these genes have been detected in isolates from all regions of Africa (). There are ten mcr gene types (mcr-1 to mcr-10), and of these, include mcr-1 the predominant one, other genes such as mcr-2, mcr-3, mcr-4, mcr-5, mcr-8, and mcr-9. All these have been detected in Africa (). These mcr genes evolved and are disseminated by various plasmids such as IncHI2, IncP, IncI, IncX4, IncN, IncR, transposons/insertion sequences (especially ISApl1), and class 1 integrons (). The genes are trafficked by a diversity of bacteria, including predominantly Escherichia coli, Salmonella, Klebsiella, Citrobacter, Enterobacter, Pseudomonas, Acinetobacter, and Alcaligenes faecalis isolated from humans, animals, food of animal origin, and environment [Citation8–11]. This is not surprising because while COL use in humans is uncommon in Africa, its use in livestock is largely unregulated [Citation12].
Table 1. Ecological niches in which mobile colistin resistance gene has been detected in Africa
Figure 1. Mobile colistin resistance mechanisms in diverse ecosystems in Africa. This map was created using an online service (https://mapchart.net/)
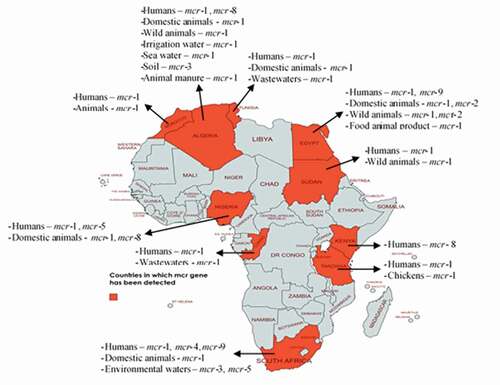
Moreover, it has been established that antibiotics induce selective pressure in bacteria prompting them to acquire resistance genes. Thus, there is a high likelihood that in Africa, MCR originated in the veterinary sector from where they disseminated to other ecosystems. Like other drugs in most African countries except for South Africa, COL is an over-the-counter (OTC) medication sold and dispensed by non-professionals/without a veterinarian’s supervision. Majority of Africans, including livestock farmers, have poor knowledge about antibiotics and antibiotic resistance. Hence they patronize quacks (non-professionals) and engage in self-medication thereby increasing the problem of resistance. Besides, it is also possible that some of the COL that is sold on the market in these nations are counterfeit or of low quality. Moreover, many antimicrobials used in human and animal medicine in Africa are mostly unregulated [Citation13]. Yet, these non-polymyxins also facilitate COL resistance [Citation14]. Thus, MCR tends to occur in conjunction with resistance against many other antimicrobial agents, making organisms multi to pandrug resistant. This further leads to limited options for therapy. This situation is exacerbated when an incriminated organism contains mcr and gene coding resistance against other last-resort antimicrobials such as fluoroquinolones, extended-spectrum cephalosporins, fosfomycin, and tigecycline. Unfortunately, such organisms, also referred to as ‘superbugs,’ have already been isolated from humans [Citation15], animals [Citation16], and food of animal origin [Citation17] in Africa.
Of noteworthy is that in Africa (), mcr genes (mcr-1 and mcr-2) have been detected in wildlife (terrestrial and aquatic) where antibiotics are not used [Citation18,Citation19], thus implying that MCR has disseminated to all ecological niches in Africa. Wildlife in Africa has also been associated with disseminating mcr genes into the environment (surface waters) and the human population [Citation19]. The latter may happen through water used for cooking, laundry, bathing, drinking, and food processing. The genes in the organism from the surface waters could also come from other parts of the world. For example, mcr-1 has been detected in isolates from the sea in Africa [Citation20].
Handling, coming in contact with, and consuming contaminated food of animal origin and fomites, is also a putative source for acquiring mcr genes. Unfortunately, unhygienic animal slaughter techniques are employed in the majority of the slaughterhouses in Africa, and worse still, in Africans, especially those in rural areas, humans live in very close contact with their livestock [Citation21].
Possible modes of colistin gene transfer in Africa
Improper disposal of livestock/slaughterhouse wastes into the environment and their use as organic fertilizer in farmlands and aquaculture can spread mcr genes into the environment. For example, mcr-1 has been detected in irrigation and sea waters in Africa [Citation20,Citation22]. Through the environment, mcr genes are reincorporated into the food chain through contaminated irrigation water, soil, crops, and wildlife [Citation23]. Furthermore, poor personal hygiene, poor waste management, and poor environmental sanitation are prevalent in most African countries and households. This facilitates the spread of mcr genes () within the human population and other ecosystems due to practices like open air defecation that are still common in Africa.
How prepared is Africa?
MCR has an immense One Health ramification in Africa and therefore warrants urgent intervention. With the world gradually turning into ‘one village’ due to ease of travel between countries and continents, the inability to urgently control the spread of MCR in Africa can potentially result in an outbreak of difficult-to-treat diseases in humans and animals worldwide, which could have catastrophic impacts. The seriousness of this is appreciated when one considers that some of the mcr-harbouring organisms from Africa, especially the E. coli, belong to the high-risk zoonotic pandemic extraintestinal pathogenic E. coli (ExPEC) clones [Citation22,Citation24,Citation25]. Furthermore, globally, diseases associated with multidrug-/extensively drug-resistant organisms were recently reported to cause an estimated 700,000 human deaths annually [Citation1]. The question that arises is whether Africa is ready to face a threat posed by MCR in the absence of effective antibiotics that can destroy superbugs.
Antimicrobial stewardship (AST) is a proven way to curb antimicrobial resistance. Education of human and animal health workers and the public on prudent/judicious use of antibiotics, conduct of antimicrobial sensitivity test before prescription of antibiotics, and infection prevention and control (IPC) practices like hand hygiene, forms the core of AST program. Unfortunately, Africa is far behind regarding establishing and practicing AST programs [Citation26]. For example, the ban on the non-therapeutic COL has proven to reduce MCR development in humans and animals [Citation27]. But this policy is not yet in view in Africa because weak national drug regulatory authorities characterize most countries in Africa except a few like South Africa, inefficient antibiotic policies, and erratic access to antibiotics [Citation26]. Getting involved in globally-coordinated antimicrobial surveillance programs is needed to control the importation and exportation of mcr genes into/from Africa. Assessing for mcr genes among travelers returning to Africa and quarantine and testing animals imported into the continent is vital to control MCR’s spread. The justification for evaluating and testing both travelers and imported animals is not far-fetched. Some mcr-harboring isolates from sick individuals in South Africa were reported to be unrelated to any African strain [Citation28]. The later suggests, that medical tourism could be promoting acquisition of MCR.
Furthermore, through the food animal trade route, mcr-1 was imported from France into Tunisia [Citation29]. On the other hand, wild animals captured in Sudan were vectors for exporting mcr-1 to China [Citation30]. Human movement also plays an important role in the spread of mcr-1. For example, Moroccans and Algerians who traveled to Mecca for Hajj pilgrimage imported mcr-1 on returning to their home countries [Citation31]. A citizen of Algeria transported mcr-1 to France [Citation32]. An American and English person who visited Kenya and Egypt, respectively, carried mcr-1 back home [Citation32,Citation33], and mcr-1 was detected in isolates from hotel workers in Tanzania that often make contact with foreign tourists [Citation34].
Since as mcr gene has been isolated from sea waters in Africa [Citation20], it can be carried by water current to other parts of the world. Therefore, efforts need to be stepped up in Africa to control the spread of MCR to preserve the efficacy of COL.
Conclusion
In conclusion, mobile COL resistance in Africa is a substantial public health issue with the potential to spread to other continents. However, the continent is not yet ready for this threat. This is a health catastrophe in the making if aggressive efforts to control the development and spread of MCR in Africa are not implemented immediately.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Notes on contributors
Madubuike Umunna Anyanwu
Madubuike Umunna Anyanwu, is a veterinarian and lecturer in the Department of Veterinary Pathology and Microbiology, University of Nigeria, Nsukka, Nigeria. He obtained his DVM, MSc and PhD from the University of Nigeria, Nsukka.
Ishmael Festus Jaja
Ishmael Festus Jaja, is a veterinarian in the Department of Livestock and Pasture Science, University of Fort Hare, Alice, South Africa. He obtained his DVM from the University of Nigeria, Nsukka, MSc and PhD from the University of Fort Hare, South Africa. He specializes in Veterinary public health, food safety, especially foodborne pathogen, and antimicrobial resistance.
James Wabwire Oguttu
James Wabwire Oguttu, is a veterinarian and Professor in the College of Agriculture & Environmental Sciences, School of Agriculture and Life Sciences, Department: Agriculture and Animal Health. He obtained his BVM from the Makerere University, Kampala, Uganda, and BVSc (Honors), MSc and PhD from the University of Pretoria, South Africa. He also holds an MSc in Epidemiology (Population based Field Epidemiology) from the University of Witwatersrand, Johannesburg. His main area of research is on Veterinary Public Health and Food Safety. He has expertise in applied epidemiology, Disease surveillance, Monitoring and evaluation, and Risk analysis.
Chinwe Juliana Jaja
Chinwe Juliana Jaja, is a Pharmacist and a public health consultant and post-doctoral research fellow in the Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa. She obtained her B.Pharm degree from the University of Jos, Nigeria, MSc from the University of Fort Hare, South Africa, and her PhD from the Stellenbosch University, South Africa. Her main research niche and special interests are infectious diseases such as food borne infections, STIs, TB, HIV/AIDS; antimicrobial resistance/stewardship; epidemiology and vaccines.
Kennedy Foinkfu Chah
Kennedy Foinkfu Chah, is a veterinarian and Professor in the Department of Veterinary Pathology and Microbiology, University of Nigeria, Nsukka, Nigeria. He obtained his DVM, MSc and PhD from the University of Nigeria, Nsukka.
Vincent Shodeinde Shoyinka
Vincent Shodeinde Shoyinka is a veterinarian and Professor in the Department of Veterinary Pathology and Microbiology, University of Nigeria, Nsukka, Nigeria. He obtained his DVM from the University of Ibadan, MSc and PhD from the University of Nigeria, Nsukka.
References
- O’Neill J. Antimicrobial Resistance: tackling a crisis for the health and wealth of nations. L, UK: 2014.
- Vaara M. Polymyxins and their potential next generation as therapeutic antibiotics. Front Microbiol. 2019;10:1.
- Webb HE, Angulo FJ, Granier SA, et al. Illustrative examples of probable transfer of resistance determinants from food animals to humans: streptothricins, glycopeptides, and colistin. F1000Res. 2017;6:1805.
- Wang S, Shen J. Active surveillance of the spread of mcr-1-positive E coli. Lancet Microbe. 2020;1(1):e4–7.
- Liu -Y-Y, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168.
- Skov RL, Monnet DL. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill. 2016;21(9):87–96.
- Shen Z, Wang Y, Shen Y, et al. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect Dis. 2016;16(3):293.
- Ngbede EO, Poudel A, Kalalah A, et al. Identification of mobile colistin resistance genes (mcr-1.1, mcr-5 and mcr-8.1) in Enterobacteriaceae and Alcaligenes faecalis of human and animal origin, Nigeria. Int J Antimicrob Agents. 2020;56(3):106108.
- Shabban M, Fahim NAE, Montasser K, et al. Resistance to colistin mediated by mcr-1 among multidrug resistant gram negative pathogens at a tertiary care hospital, Egypt. J Pure Appl Microbiol. 2020;14(2):106108–106132. https://doi.org/https://doi.org/10.22207/JPAM.14.2.07
- Ali A, Nouraldein M, Hamad M. Molecular detection of colistin resistance gene mcr-1 in gram- negative rods isolated from hospitalized patients in khartoum state molecular detection of colistin resistance gene mcr-1 in gram- from hospitalized patients in khartoum state. Saudi J Pathol Microbiol. 2020;380–384. DOI:https://doi.org/10.36348/sjpm.2020.v05i08.005
- Gabriel A, Tarcisse BN, Rachel M, et al. Study of Colistin Resistance Encoded by the mcr-1 Gene in Community and Clinical Pseudomonas in Brazzaville, Republic of Congo. J Microb Biochem Technol. 2019;11:422.
- Maamar E, Alonso CA, Hamzaoui Z, et al. Emergence of plasmid-mediated colistin-resistance in CMY-2-producing Escherichia coli of lineage ST2197 in a Tunisian poultry farm. Int J Food Microbiol. 2018;269:60–63.
- Van TTH, Yidana Z, Smooker PM, et al. Antibiotic use in food animals worldwide, with a focus on Africa: pluses and minuses. J Glob Antimicrob Resist. 2020;20:1125–1132.
- Lentz SA, De Lima-morales D, Cuppertino VM, et al. Letter to the editor: escherichia coli harbouring mcr-1 gene isolated from poultry not exposed to polymyxins in Brazil. Eurosurveillance. 2016;21(26):1.
- Soliman AM, Maruyama F, Zarad HO, et al. Emergence of a Multidrug-Resistant Enterobacter hormaechei Clinical Isolate from Egypt Co-Harboring mcr-9 and blaVIM-4. Microorganisms. 2020;8(4):595.
- Hassen B, Saloua B, Abbassi MS, et al. mcr-1 encoding colistin resistance in CTX-M-1/CTX-M-15- producing Escherichia coli isolates of bovine and caprine origins in Tunisia. First report of CTX-M-15-ST394/D E. coli from goats. Comp Immunol Microbiol Infect Dis. 2019;67:101366.
- Sadek M, Poirel L, Nordmann P, et al. Draft genome sequence of an mcr-1/IncI2-carrying multidrug-resistant Escherichia coli B1:ST101 isolated from meat and meat products in Egypt. J Glob Antimicrob Resist. 2020;20:41–42.
- Bachiri T, Lalaoui R, Bakour S, et al. First Report of the Plasmid-Mediated Colistin Resistance Gene mcr-1 in Escherichia coli ST405 Isolated from Wildlife in Bejaia, Algeria. Microb Drug Resist. 2018;24(7):890–895.
- Ahmed ZS, Elshafiee EA, Khalefa HS, et al. Evidence of colistin resistance genes (mcr-1 and mcr-2) in wild birds and its public health implication in Egypt. Antimicrob Resist Infect Control. 2019;8(1):197.
- Drali R, Berrazeg M, Zidouni LL, et al. Emergence of mcr-1 plasmid-mediated colistin-resistant Escherichia coli isolates from seawater. Sci Total Environ. 2018;642:90–94.
- Alonso CA, Zarazaga M, Ben Sallem R, et al. Antibiotic resistance in Escherichia coli in husbandry animals: the African perspective. Lett Appl Microbiol. 2017;64(5):318–334.
- Touati M, Hadjadj L, Berrazeg M, et al. Emergence of Escherichia coli harbouring mcr-1 and mcr-3 genes in North West Algerian farmlands. J Glob Antimicrob Resist. 2020;21:132–137.
- Anyanwu MU, Jaja IF, Nwobi OC. Occurrence and Characteristics of Mobile Colistin Resistance (mcr) Gene-Containing Isolates from the Environment: a Review. Int J Environ Res Public Health. 2020;17(3):1028.
- Manges AR, Geum HM, Guo A, et al. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin Microbiol Rev. 2019;32. DOI:https://doi.org/10.1128/CMR.00135-18
- Hassen B, Abbassi MS, Ruiz-Ripa L, et al. High prevalence of mcr-1 encoding colistin resistance and first identification of blaCTX-M-55 in ESBL/CMY-2-producing Escherichia coli isolated from chicken faeces and retail meat in Tunisia. Int J Food Microbiol. 2020;318:108478.
- Iwu CJ, Jordan P, Jaja IF, et al. Treatment of COVID-19: implications for antimicrobial resistance in Africa. Pan Africa Med J. 2020;35:119.
- Wang Y, Xu C, Zhang R, et al. Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study. Lancet Infect Dis. 2020;20:30242–30245.
- Osei Sekyere J, Maningi NE, Modipane L, et al. Emergence of mcr-9.1 in Extended-Spectrum-β-Lactamase-Producing Clinical Enterobacteriaceae in Pretoria, South Africa: global Evolutionary Phylogenomics, Resistome, and Mobilome. MSystems. 2020;5. DOI:https://doi.org/10.1128/msystems.00148-20.
- Grami R, Mansour W, Mehri W, et al. Impact of food animal trade on the spread of mcr-1-mediated colistin resistance, tunisia, july 2015. Eurosurveillance. 2016;21:1–5.
- Feng C, Wen P, Xu H, et al. Emergence and Comparative Genomics Analysis of Extended-Spectrum-β-Lactamase-Producing Escherichia coli Carrying mcr-1 in Fennec Fox Imported from Sudan to China. MSphere. 2019;4. DOI:https://doi.org/10.1128/msphere.00732-19.
- Leangapichart T, Gautret P, Brouqui P, et al. Acquisition of mcr-1 plasmid-mediated colistin resistance in Escherichia coli and Klebsiella pneumoniae during Hajj 2013 and 2014. Antimicrob Agents Chemother. 2016;60:6998–6999.
- Henig O, Rojas LJ, Bachman MA, et al. Identification of four patients with colistin-resistant Escherichia coli containing the mobile colistin resistance mcr-1 gene from a single health system in Michigan. Infect Control Hosp Epidemiol. 2019;40:1059–1062.
- Doumith M, Godbole G, Ashton P, Larkin L, Dallman T, Day M, et al. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother. 2016;71:2300–2305.
- Büdel T, Kuenzli E, Clément M, et al. Polyclonal gut colonization with extended-spectrum cephalosporin- and/or colistin-resistant Enterobacteriaceae: a normal status for hotel employees on the island of Zanzibar, Tanzania. J Antimicrob Chemother. 2019;74:2880–2890.
- Saidani M, Messadi L, Chaouechi A, et al. High Genetic Diversity of Enterobacteriaceae Clones and Plasmids Disseminating Resistance to Extended-Spectrum Cephalosporins and Colistin in Healthy Chicken in Tunisia. Microb Drug Resist. 2019;mdr.2019.0138. DOI:https://doi.org/10.1089/mdr.2019.0138.
- Maamar E, Hammami S, Alonso CA, et al. High prevalence of extended-spectrum and plasmidic AmpC beta-lactamase-producing Escherichia coli from poultry in Tunisia. Int J Food Microbiol. 2016;231:69–75.
- Saidani M, Messadi L, Mefteh J, et al. Various Inc-type plasmids and lineages of Escherichia coli and Klebsiella pneumoniae spreading blaCTX-M-15, blaCTX-M-1 and mcr-1 genes in camels in Tunisia. J Glob Antimicrob Resist. 2019;19:280–283.
- Saidani M, Messadi L, Sahmin E, et al. ESBL- and mcr-1-producing Escherichia coli in veal calves in Tunisia. J Glob Antimicrob Resist. 2019;19:104–105.
- Hassen B, Abbassi MS, Ruiz-Ripa L, et al. Genetic characterization of extended-spectrum β-lactamase-producing Enterobacteriaceae from a biological industrial wastewater treatment plant in Tunisia with detection of the colistin-resistance mcr-1 gene. FEMS Microbiol Ecol. 2021;97. https://doi.org/10.1093/femsec/fiaa231.
- Olaitan AO, Chabou S, Okdah L, et al. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16:147.
- Hadjadj L, Riziki T, Zhu Y, et al. Study of mcr-1 Gene-Mediated Colistin Resistance in Enterobacteriaceae Isolated from Humans and Animals in Different Countries. Genes (Basel). 2017;8:394.
- Chabou S, Leangapichart T, Okdah L, et al. Real-time quantitative PCR assay with Taqman® probe for rapid detection of MCR-1 plasmid-mediated colistin resistance. New Microbes New Infect. 2016;13:71–74.
- Chabou S, Leulmi H, Rolain JM. Emergence of mcr-1-mediated colistin resistance in Escherichia coli isolates from poultry in Algeria. J Glob Antimicrob Resist. 2019;16:115–116.
- Berrazeg M, Hadjadj L, Ayad A, et al. First detected human case in Algeria of mcr-1 plasmid-mediated colistin resistance in a 2011 Escherichia coli isolate. Antimicrob Agents Chemother. 2016;60:6996–6997.
- Yanat B, Machuca J, Yahia RD, et al. First report of the plasmid-mediated colistin resistance gene mcr-1 in a clinical Escherichia coli isolate in Algeria. Int J Antimicrob Agents 2016;48:760–761.
- Nabti LZ, Sahli F, Hadjadj L, et al. Autochthonous case of mobile colistin resistance gene mcr-1 from a uropathogenic Escherichia coli isolate in Sétif Hospital, Algeria. J Glob Antimicrob Resist. 2019;19:356–357.
- Elnahriry SS, Khalifa HO, Soliman AM, et al. Emergence of plasmid-mediated colistin resistance gene mcr-1 in a clinical Escherichia coli isolate from Egypt. Antimicrob Agents Chemother. 2016;60:3249–3250.
- Abd El-Baky RM, Masoud SM, Mohamed DS, et al. Prevalence and Some Possible Mechanisms of Colistin Resistance Among Multidrug-Resistant and Extensively Drug-Resistant <em>Pseudomonas aeruginosa. Infect Drug Resist. 2020;13:323–332.
- Zafer MM, El-Mahallawy HA, Abdulhak A, et al. Emergence of colistin resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli strains isolated from cancer patients. Ann Clin Microbiol Antimicrob. 2019;18:40.
- Zakaria AS, Edward EA, Mohamed NM. Genomic Insights into a Colistin-Resistant Uropathogenic Escherichia coli Strain of O23: H4-ST641Lineage Harboring mcr-1.1 on a Conjugative IncHI2 Plasmid from Egypt. Microorganisms. 2021;9:799.
- Sadek M, JM ODLR, Abdelfattah MM, et al. Genomic Features of MCR-1 and Extended-Spectrum β-Lactamase-Producing Enterobacterales from Retail Raw Chicken in Egypt. Microorganisms. 2021;9:195.
- Lima Barbieri N, Nielsen DW, Wannemuehler Y, et al. mcr-1 identified in Avian Pathogenic Escherichia coli (APEC). PLoS One. 2017;12:e0172997.
- Moawad AA, Hotzel H, Neubauer H, et al. Antimicrobial resistance in Enterobacteriaceae from healthy broilers in Egypt: emergence of colistin-resistant and extended-spectrum β-lactamase-producing Escherichia coli 06 Biological Sciences 0604 Genetics 11 Medical and Health Sciences 1108 Medical Microbiology. Gut Pathog. 2018;10:39.
- Rahmatallah N, El RH, Laraqui A, et al. Detection of colistin encoding resistance genes MCR-1 in isolates recovered from broiler chickens in morocco. Saudi J Pathol Microbiol. 2018;DOI:https://doi.org/10.21276/sjpm.2018.3.12.10
- Hammad AM, Hoffmann M, Gonzalez-Escalona N, et al. Genomic features of colistin resistant Escherichia coli ST69 strain harboring mcr-1 on IncHI2 plasmid from raw milk cheese in Egypt. Infect Genet Evol. 2019;73:126–131.
- Sabala RF, Usui M, Tamura Y, et al. Prevalence of colistin-resistant Escherichia coli harbouring mcr-1 in raw beef and ready-to-eat beef products in Egypt. Food Control. 2021;119:107436.
- El Sayed Zaki M, Abou ElKheir N, Mofreh M. Molecular study of colistin resistant clinical isolates of Enterobacteriaceae species. J Clin Mol Med. 2018;1. https://doi.org/https://doi.org/10.15761/jcmm.1000103
- Rabie R, Lotfy A. Plasmid mediated colistin resistant genes mcr-1 and mcr-2 among Escherichia coli and Klebsiella Pneumoniae isolates at Zagazig University Hospitals, Egypt. Egypt J Med Microbiol. 2020;1:1–7.
- Bontron S, Poirel L, Real-time NP. PCR for detection of plasmid-mediated polymyxin resistance (mcr-1) from cultured bacteria and stools. J Antimicrob Chemother. 2016;71:2318–2320.
- Perreten V, Strauss C, Collaud A, et al. Colistin resistance gene mcr-1 in avian-pathogenic Escherichia coli in South Africa. Antimicrob Agents Chemother. 2016;60:4414–4415.
- Founou LL, Founou RC, Allam M, et al. Extended-spectrum beta-lactamase-producing escherichia coli harbouring mcr-1 gene isolated from pigs in south africa. S Afr Med J. 2018;108:796–797.
- Poirel L, Jayol A, Polymyxins: NP. Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30:557–596.
- Coetzee J, Corcoran C, Prentice E, et al. Emergence of plasmid-mediated colistin resistance (MCR-1) among Escherichia coli isolated from South African patients. S Afr Med J. 2016;106:449–450.
- Newton-Foot M, Snyman Y, Maloba MRB, et al. Plasmid-mediated mcr-1 colistin resistance in Escherichia coli and Klebsiella spp. clinical isolates from the Western Cape region of South Africa. Antimicrob Resist Infect Control. 2017;6:1–7.
- Igwaran A, Iweriebor BC, Okoh AI. Molecular characterization and antimicrobial resistance pattern of Escherichia coli recovered from wastewater treatment plants in Eastern Cape South Africa. Int J Environ Res Public Health. 2018;15. DOI:https://doi.org/10.3390/ijerph15061237
- Snyman Y, Reuter S, Whitelaw AC, et al. Characterisation of mcr-4.3 in a colistin-resistant Acinetobacter nosocomialis clinical isolate from Cape Town, South Africa. J Glob Antimicrob Resist. 2021;25:102–106.
- Adam N, Altayb HN. Proteomics analysis of proteins associated with urinary tract infections. J Proteomics Bioinform. 2017;10:98.
- Kyany’ C, Musila L. Colistin Resistance Gene mcr-8 in a High-Risk Sequence Type 15 Klebsiella pneumoniae Isolate from Kenya. Microbiol Resour Announc. 2020;9:e00783–20.
- Olaitan AO, Diene SM, Kempf M, et al. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int J Antimicrob Agents. 2014;44:500–507.
