ABSTRACT
Recent studies on extracellular RNA raised awareness that extracellular vesicles (EVs) isolated from cultured cells may co-purify RNAs derived from media supplements such as fetal bovine serum (FBS) confounding EV-associated RNA. Defined culture media supplemented with a range of nutrient components provide an alternative to FBS addition and allow EV-collection under full medium conditions avoiding starvation and cell stress during the collection period. However, the potential contribution of serum-free media supplements to EV-RNA contamination has remained elusive and has never been assessed. Here, we report that RNA isolated from EVs harvested from cells under serum-replacement conditions includes miRNA contaminants carried into the sample by defined media components. Subjecting unconditioned, EV-free medium to differential centrifugation followed by reverse transcription quantitative PCR (RT-qPCR) on RNA isolated from the pellet resulted in detection of miRNAs that had been classified as EV-enriched by RNA-seq or RT-qPCR of an isolated EV-fraction. Ribonuclease (RNase-A) and detergent treatment removed most but not all of the contaminating miRNAs. Further analysis of the defined media constituents identified Catalase as a main source of miRNAs co-isolating together with EVs. Hence, miRNA contaminants can be carried into EV-samples even under serum-free harvesting conditions using culture media that are expected to be chemically defined. Formulation of miRNA-free media supplements may provide a solution to collect EVs clean from confounding miRNAs, which however still remains a challenging task. Differential analysis of EVs collected under full medium and supplement-deprived conditions appears to provide a strategy to discriminate confounding and EV-associated RNA. In conclusion, we recommend careful re-evaluation and validation of EV small RNA-seq and RT-qPCR datasets by determining potential medium background.
Introduction
Extracellular vesicle (EV) research has been strongly promoted by the discovery of different classes of RNAs, mainly mRNAs and miRNAs, which in association with EVs are delivered to target cells and are considered to modulate various target cell functions [Citation1–Citation3]. Since these pioneering findings, numerous studies reported RNA-seq and miRNA-profiling on EVs that were isolated from cell culture supernatants. Most cell types and in particular tumour cells are cultured in the presence of foetal bovine serum (FBS). However, FBS contains EVs as well as other macromolecular complexes such as ribonucleoprotein and lipoprotein particles, which carry RNAs and co-isolate in common EV-isolation protocols, introducing potential artefacts leading to misinterpretation of RNA topology during the analysis of extracellular RNA [Citation4–Citation6]. Furthermore, miRNAs share high sequence homology between species resulting in erroneous species annotation. In fact, a considerable number of FBS-associated miRNAs are indistinguishable from their human and rodent orthologs [Citation7]. Despite the use of EV-depleted FBS during the EV-collection period, adverse effects on cell growth were observed and a substantial proportion of EVs and RNA still persist after depletion [Citation7–Citation10]. Therefore, it was suggested to carefully consider potential mis-annotation of FBS-derived RNAs, avoiding them being wrongly perceived as genuine EV-RNAs.
A promising strategy to circumvent FBS-RNA contaminations in EV-isolates could be the substitution of FBS by chemically defined media supplements, allowing controlled and optimized growth conditions also during EV-collection periods. Several primary cultured cell types or stem cells are commonly cultured in the presence of commercially available media supplements. For example, the supplements B27 or NS21 are often used for cells of neural origin [Citation11,Citation12]. Here, we illustrate that small RNA-seq data obtained for EVs collected under serum-free, defined media conditions are still confounded by contaminating miRNAs that are carried in by media supplements. By careful analysis of individual media components, we identified the potential source of RNA contamination.
Material and methods
Cell culture
Primary oligodendrocytes were prepared from embryonic day (E) 14 or postnatal day (P) 7–9 mice of the C57Bl/6-N strain as described before [Citation13,Citation14]. Primary oligodendrocytes derived from E 14 mice were cultured in Sato 1% horse serum (Thermo Fisher Scientific), washed in PBS and switched to serum-free Sato supplemented with B27 [Citation11] (purchased from Gibco) before the harvesting period of EVs. Primary oligodendrocytes derived from P 7–9 mice were cultured in basal medium (MACS Neuro Medium, Miltenyi Biotec) supplemented with NS21 [Citation12] (NB21 supplement, Miltenyi Biotec), 1 mM L-glutamine and 10 ml/l 100x Penicillin-Streptomycin (Thermo Fisher Scientific).
Sm-NS16, sm-NS19 and sm-NS21 supplements
In order to compare self-made supplements to commercially available NS21 (Miltenyi Biotec), neural supplements were prepared by mixing individual components as described [Citation12]. Self-made (sm)-NS21 reflects the full supplement, while sm-NS19 is lacking SOD and Catalase, and sm-NS16 is lacking all protein components (components listed in Supplementary Table 1).
Isolation of EVs
EVs were collected from primary cultured oligodendrocytes in serum-free media either supplemented with B27, NS21, or without supplement over a period of 24–48 h and isolated by differential centrifugation as described with minor modifications [Citation15,Citation16]. Briefly, supernatants (derived from 5–15 × 106 cells in a volume of 12–30 ml) were subjected to centrifugation at 130 x g for 10 min (4°C), followed by 10,000 x g for 30 min (4°C) in a fixed angle rotor (220.78, Hermle, Wehingen, Germany) and ultracentrifugation at 29,000 rpm for 2 h at 4°C using a Beckman SW40 Ti rotor (RCF (avg) 106,154, RCF (max) 149,576, k-factor 260) and 14 ml polyallomer tubes (Beckman Coulter). Unconditioned media (25 ml input) was handled equally. Pellets were resuspended in 700 µl QIAzol (Quiagen) and stored at −80°C before RNA-isolation.
For depletion of NS21, 0.2 ml of NS21 supplement was subjected to ultracentrifugation for 2 h at 47,000 rpm at 4°C (Beckman TLA-55 rotor, RCF (avg) 98,963, RCF (max) 130,000, k-factor 90.4). Pellet and supernatant were subjected to RNA-isolation.
RNA isolation
EVs (obtained from culture-conditioned supernatants) as well as ultracentrifugation pellets derived from unconditioned media were resuspended in 700 µl QIAzol (Qiagen). Total cells, from which EVs were harvested, were washed in ice-cold PBS before scraping in the recommended volume of QIAzol. RNA was isolated using the miRNeasy micro Kit (Qiagen) according to the manufacturer’s instructions, including on-column DNase digest and <200 nt cut-off. In case of unconditioned medium (25 ml supplemented with 0.5 ml of NS21, sm-NS19, or sm-NS21), total RNA was isolated and DNAse digestion was omitted (as RT-qPCR is designed to only detect mature miRNAs).
For direct RNA extraction from media supplements, sm-NS16, NS21 or B27 (0.5 ml supplement, equivalent to 25 ml supplemented culture medium) were mixed with QIAzol Lysis Reagent (Qiagen) according to the manufacturer’s instructions. The following media components were individually analysed and subjected to direct RNA isolation (input normalized to their quantitative representation in 25 ml supplemented culture medium): BSA, superoxide dismutase (SOD), insulin, holo-Transferrin, and catalases of different origin (see details in Supplementary Table 2).
Small RNA-seq profiling
Oligodendrocyte-derived EVs, which were characterized previously [Citation13,Citation15,Citation16], were subjected to quality control by NTA and Western-blotting. Total RNA of three independent biological replicates (n = 3) was isolated from 100.000 x g EV-pellets as well as from corresponding cells as described above (without 200 nt cut-off). RNA was size-selected between 15 and 200 nt on 10% TBE-urea gel. Gel-purified RNA was eluted overnight in 0,3 M NaCl and then precipitated with 100% isopropanol and Glycoblue for 1 h at −20°C. The pellet was washed once with 75% ethanol and dissolved in nuclease-free water. The purified fraction was confirmed by Bioanalyzer Small RNA chip (Agilent). Library preparation was based on the NEBNext Multiplex Small RNA Library Prep Set for Illumina (New England BioLabs) with slight modification. In brief, RNA was first ligated to the 3ʹ adapter and then to the 5ʹ adapter, both of which contained four random bases and were chemically synthesized by Bio Scientific. Adapter-ligated RNA was reverse-transcribed and PCR-amplified for 14 cycles using index primers. The PCR-amplified cDNA construct was purified using AMPure XP beads (Beckman Coulter). The purified PCR reaction was checked on the Bioanalyzer using High Sensitivity DNA chip (Agilent). Size selection of the RNA library was performed on LabChip XT instrument (Perkin Elmer) using DNA 300 assay kit in the range 140–300 bp, the libraries were pooled in equal molar ratio and sequenced on Illumina HiSeq 2500 for 51 cycles plus 7 cycles index read. Sample demultiplexing and FastQ file generation was performed using bcl2fastq v.1.8.4 (Illumina). For miRNA-tailored analysis, the raw sequence reads were cleaned from adapter sequences and size-selected in the range 19–25 nucleotides (plus 8 random bases from the adapters) using Cutadapt v.1.7.1 (http://cutadapt.readthedocs.org). Reads containing low-quality bases (Phred score below 20) were filtered with the FastX toolkit (http://hannonlab.cshl.edu/fastx_toolkit) and deduplicated based on full sequence identity (library insert plus 8 random bases from the library adapters) using shell commands. Quality control of the raw and processed data was performed with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Mapping to the mouse reference genome NCBIM37 (mm9) with concomitant trimming of the random adapter bases was performed using Bowtie v.0.12.8 (http://bowtie-bio.sourceforge.net) with parameters “-v 3-M 1 -y --sam --best --strata --nomaqround --trim5 4 --trim3 4”. Mature miRNA annotation from miRbase (http://www.mirbase.org/) was used to estimate the number of reads per miRNA using Subread featureCounts v.1.4.6-p4 (http://subread.sourceforge.net/). Differential expression analysis of EVs vs. Cells was performed using LIMMA v. 3.24.0 (https://bioconductor.org/packages/release/bioc/html/limma.html) as recommended for RNA-seq data and Volcano plots were prepared using ggplot2 (https://ggplot2.tidyverse.org). Similar results were obtained using miRNA gene annotation from UCSC (http://genome.ucsc.edu/cgi-bin/hgTables) and performing differential expression analysis with edgeR (https://bioconductor.org/packages/release/bioc/html/edgeR.html). The NGS data have been deposited in the NCBI’s Gene Expression Omnibus (GEO) under accession number GSE134096 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134096).
cDNA synthesis and RT-qPCR
As input for reverse transcription of cDNA, 5 ng of cellular RNA and corresponding EV-RNA was used (determined by Nanodrop ND3300 using the RiboGreen Assay for RNA quantitation (Thermo Fisher Scientific)). In case of EVs harvested in presence or absence of NS21, equal input of 1.7 ng EV-RNA was used (determined by Bioanalyzer, RNA 6000 Pico Chip (Agilent)). As RNA was below the detection limit in samples derived from media and media supplements, the maximal input volume of 3.7 µl of RNA was included in the cDNA synthesis reaction of these samples. A spike-in of cel-miR-39-3p (UCACCGGGUGUAAAUCAGCUUG, 0.994 amol) was added to each RNA-sample as a control later used for normalization and inter-sample calibration of qPCR results. cDNA synthesis was carried out using the TaqMan Advanced miRNA cDNA Synthesis Kit (Thermo Fisher Scientific). For RT-qPCR, TaqMan Fast Advanced Master Mix and TaqMan Advanced miRNA Assays detecting miR-122-5p, miR-451a, miR-30d-5p, let-7g or cel-miR-39-3p (Thermo Fisher Scientific) were used. RT-qPCR was performed with the StepOne Real-Time PCR System (Applied Biosystems).
Inter-sample calibration of RT-qPCR using spike-in
Spike-in (cel-miR-39-3p) Cq-values were determined automatically and individually for all samples via StepOne Software 2.2.2 (Applied Biosystems). Since spike-in quantity is constant in all samples, an average spike-in Cq-value was calculated (23.628 ± 0.168, average ± SEM Cq over 46 reactions). To correct for technical variations between samples, the determined spike-in Cq mean of 23,628 was used to set the spike-in threshold, which was then applied to adjust the threshold for the target miRNAs of the sample accordingly (example given in Supplementary Fig. 1). Cq-cut-off was set to 35.
RNase-A and TritonX-100-treatment
One hundred-microlitre NS21 was treated with 20 μg/ml RNase A (Invitrogen) or with 1% (V/V) TritonX-100 (Sigma-Aldrich) followed by 20 μg/ml RNase A for 30 min at 37°C under gentle agitation. RNase digestion was stopped by addition of 1.25 μl/ml RNasin Plus RNase Inhibitor (Promega) and RNA-isolation was performed as described above.
Western blotting and antibodies
Unconditioned media supplemented with NS21, sm-NS21, or sm-NS19 (25 ml) were subjected to the EV isolation protocol including final ultracentrifugation as described above. Pellets were resuspended in SDS-PAGE sample buffer and stored at −20°C. Before loading on a 12% SDS-PAGE (Bio-Rad Mini PROTEAN electrophoresis system), the samples were boiled 10 min at 70°C. Proteins were transferred onto a PVDF membrane for 3 h and 300 mA (Bio-Rad Mini Trans-Blot Electrophoretic Transfer Cell system). The membrane was blocked with 4% milk powder/0.1% Tween (Roth) in PBS. Proteins were detected by sequential incubation of the membrane with primary antibodies – rabbit-anti-Catalase (sc-271803, Santa Cruz), rabbit-anti-SOD (Abcam ab51254) – and horseradish peroxidise-coupled secondary antibodies anti-rabbit (Dianova). Signal detection was performed using self-made enhanced chemiluminescence reagents and exposure of ECL Hyperfilms (GE healthcare), which were developed with the OptiMax XRay film processor (MS Laborgeräte Wiesloch).
MTT-assay
As readout of cell viability, metabolic activity of primary oligodendrocytes cultured in NS21 or sm-NS19 was measured by MTT-assay. Briefly, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Thiazolyl Blue Tetrazolium Bromide, Sigma) was added to the culture medium in a final concentration of 0.5 mg/ml and incubated for 1,5 h (37°C, 5% CO2). Subsequently, medium was removed and formed formazan crystals were dissolved in isopropanol under gentle shaking for 30 min. Absorbance was measured at 562 nm using a plate reader (Tecan Infinite 2000).
EV-TRACK
We have submitted all relevant data of our experiments to the EV-TRACK knowledgebase (EV-TRACK ID: EV190027) [Citation17].
Results and discussion
Small RNA-seq suggests contamination with foreign miRNAs
With the goal to identify miRNAs and other small RNAs involved in EV-mediated oligodendrocyte-neuron communication, we performed deep sequencing (RNA-seq) of small RNAs derived from EVs isolated from culture supernatants of primary cultured oligodendrocytes by differential centrifugation. The efficiency of the isolation protocol and the resulting EVs were validated and characterized before [Citation13,Citation15,Citation16,Citation18]. To avoid serum-derived RNA contamination, EVs were harvested under defined medium conditions using B27 supplement. Bioinformatic analysis of RNA-seq data from EVs and their corresponding cells of origin revealed several miRNAs enriched in EVs versus cells or vice versa. The two most significantly enriched miRNAs in oligodendrocyte-derived EVs were miR-122-5p and miR-451a (), which are highly expressed in liver and erythrocytes, respectively. Other miRNAs such as miR-30d-5p and let-7g appeared equally represented in EVs and cells and were intended to serve as normalization reference during validation. RT-qPCR detection of these miRNAs on newly generated RNA samples (now derived from oligodendrocytes cultured in presence of the advanced NS21 supplement) verified the anticipated distribution of these miRNAs between EVs and cells (). In the absence of a validated miRNA that could serve for normalization between cells and EVs, RT-PCRs were normalized to RNA-input (small RNA <200 nt). miR-122-5p and miR-451a were detected with lower RT-qPCR cycle of quantification (Cq) in EVs versus cells (miR-122-5p: Cq EVs 21.2/cells 29.9; miR-451a: Cq EVs 24.3/cells 26.9) and thus appeared more abundant in EV-RNA. On the other hand, Cq-values of miR-30d-5p and let-7g suggested that these two miRNAs are more equal or slightly stronger represented in cellular as compared to EV-RNA (miR-30d-5p: Cq EVs 25.0/cells 23.4; let-7g Cq EVs 24.7/cells 21.9). The strong enrichment of miR-122-5p and miR-451a in oligodendroglial EVs was startling, as miR-122-5p appears largely liver-specific and absent from brain and miR-451a has highest abundance in blood [Citation19,Citation20]. However, both miRNAs were suspected before to reflect serum contaminants in other EV samples and it has been recommended to carefully control their association with EVs in individual data sets [Citation6,Citation7].
Figure 1. RNA-seq results and RT-qPCR verification of EV-associated miRNAs.
(a) Volcano plot of relative representation of miRNAs in oligodendrocytes and oligodendrocyte-derived EVs. MiR-122-5p and miR-451a are strongly enriched in EVs (two top left dots marked), while miR-30d-5p and let-7g are equally represented in cells and EVs (n = 3). Red dotted line: significance level of 1% FDR (adjusted pValue = 0.01). logFC: log2 Fold Change, AveExpr: Average Expression. (b) RT-qPCR verification of miR-122-5p, miR-451a, miR-30d-5p and let-7g with independent biological replicates of cells and EVs (n = 3). Dashed line, cut-off Cq 35 defining background qPCR signal; error bars, SEM. (C) RNA-seq results indicate enrichment of the bovine- and primate-specific miR-1246 in EVs samples versus cells in all three replicates. RPM: Reads Per Million.
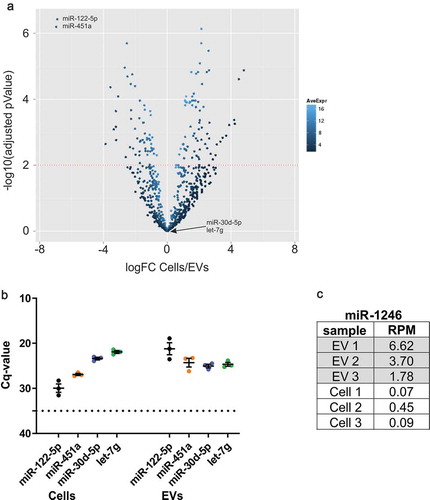
We further checked the RNA-seq data set for the presence of miR-1246, which is generated through non-canonical biogenesis from a small nuclear RNA of the U2 spliceosome complex [Citation21]. However, miR-1246 appears highly abundant in FBS and is not produced in rodent cells according to the present literature [Citation22]. Furthermore, miR-1246 was detected in diverse mouse cell lines that were cultured in FBS, suggesting uptake of miR-1246 by these cells during culture [Citation7,Citation22]. Surprisingly, despite the absence of FBS in our samples, miR-1246 was robustly detected in all RNA-seq replicates of EVs and less abundantly in cells (). As miR-1246 cannot be derived from mouse oligodendrocytes, its detection is most likely due to contamination of the EV-samples with miRNAs originating from bovine- or primate-sourced components of the media supplement.
Defined media supplements carry exogenous miRNAs into EV-samples
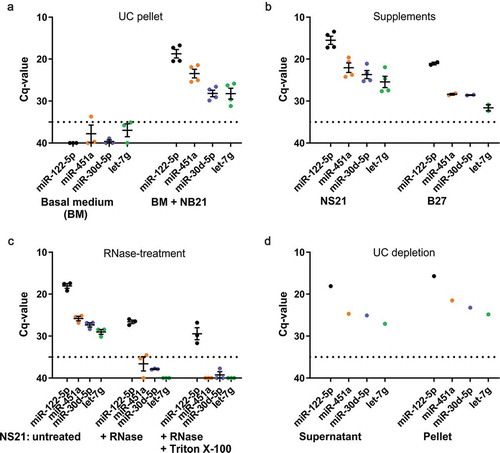
We further investigated the possibility of contaminating miRNAs present in the basal medium or the supplements that may be misinterpreted as EV-associated miRNAs. To this end, we performed differential ultracentrifugation (UC) with unconditioned basal medium lacking or containing NS21 supplement and checked the resulting pellets for the presence of miRNAs by RT-qPCR, indicating their potential co-purification with EVs. The input medium volume (25 ml) was in the same range of that used for EV-isolation and miRNA detection (maximum of 30 ml). UC-pellets of basal medium lacking supplement did not show miRNA signals above background. In contrast, UC-pellets of medium containing NS21 supplement revealed clear signals for all miRNAs examined (), which appear related to NS21-addition. To provide a direct proof, we subjected undiluted NS21 as well as B27 supplements to RNA-isolation and RT-qPCR (). Supplement input corresponded to the amount represented in full medium samples that were subjected to UC pelleting. In both NS21 as well as B27 supplements, all tested miRNAs were robustly detected. Together, the results demonstrate that media supplements contain miRNAs that can co-purify with EVs and carry exogenous miRNAs into EV-samples. Thus, the term “defined” for such supplements appears incorrect (they are rather “ill-defined”) and should be avoided.
Figure 2. RT-qPCR analysis indicates miRNA contamination of defined media supplements.
(a) RT-qPCR of miRNAs derived from a pellet after differential ultracentrifugation (UC) of unconditioned basal medium (n = 3) or NS21 supplemented medium (n = 4, two different lots included, 2n each). Supplemented medium contains miR-122-5p, miR-451a, miR-30d-5p and let-7g, while basal medium is free of these miRNAs. Input volume for all samples: 25 ml. (b) RT-qPCR of miRNAs directly isolated from media supplements NS21 (n = 4, two different lots included, 2n each) and B27 (n = 2). Supplement input volume of 500 µl reflects amount represented in 25 ml used for UC-pelleted samples in (A). (c) RNase A treatment or combined Triton X-100/RNase A treatment of NS21 indicates partial protection of miR-122-5p to both treatment conditions. (d) RT-qPCR of miRNAs derived from supernatant and pellet fraction after depletion of NS21 supplement by ultracentrifugation. All miRNAs are detected in the pellet and supernatant indicating that depletion does not efficiently remove miRNAs from the supplement (n = 1). Dashed line, cut-off Cq 35 defining background qPCR signal; error bars, SEM.
It is important to note that Cq-values achieved for the miRNAs largely deliver qualitative information. Although the sample input amount was similar and a spike-in miRNA (cel-miR-39) was used to correct for all steps of the RT-qPCR, a stringent normalization parameter such as an internal miRNA reference is lacking. Notably, cell-conditioned containing EVs and non-conditioned media samples were quite distinct regarding RNA-content (below detection limit in unconditioned samples) and variations in RNA-isolation efficiency remained uncorrected. We thus refrain from a quantitative comparison of the different sample types. The cel-miR-39 spike-in served for threshold setting and normalization of similar samples and replicates (Supplementary Fig. 1). We would like to emphasize that the qualitative proof of miRNA contaminations in the medium should be viewed largely as an alert, which requires follow up of specific miRNAs in question. However, due to the reasons outlined above (absence of internal sample normalization), quantitative strategies of background subtraction are considered difficult to achieve.
Supplement-associated miR-122-5p is partly resistant to RNase treatment
We further explored potential strategies to get rid of the contaminating miRNAs in supplements that can be employed for EV-harvesting under full-media conditions. Resistance of miRNAs to RNase digestion and, furthermore, sensitivity to successive Triton-X100 and RNase treatment are often used as an argument for EV-association of miRNAs, as these are enclosed by a lipid bilayer and only accessible to digestion after detergent treatment [Citation23]. To test how supplement-associated miRNAs perform under these conditions, we treated NS21 supplement with RNase A followed by RNA isolation and RT-qPCR (). Although all miRNAs were affected by RNase-digestion and miR-451a, miR-30d-5p and let-7g were below the detection threshold, miR-122-5p was still detected to a significant extent (Cq-value 26,6 ± 0.34). Consecutive Triton-X100 and RNase A treatment of NS21 had only slight effects on miR-122-5p resistance (Cq-value 29.4 ± 1.16) indicating that its protection is not based on the presence of a lipid layer. Hence, miR-122-5p is partly resistant to RNase digestion, but RNase protection is most likely not due to association with EVs that could be present in the supplement. These results suggest that some miRNAs are present in macromolecular complexes together with proteins, that are not dissolved by amphipathic detergents, and therefore are inaccessible to RNase digestion [Citation24]. Most likely, proteinase treatment would render miR-122-5p sensitive to RNAse digestion. Nonetheless, we abstained from performing this experiment as it would destroy the supplement components and not provide a sensible approach to deplete it from contaminating miRNAs.
To investigate, whether NS21 can be depleted from miRNA contaminants, we further subjected NS21 to UC (20 h, 100.000 x g) and examined the pellet and supernatant fractions for the distribution of NS21-associated miRNAs. All miRNAs were present in the pellet as well as the supernatant, demonstrating that depletion of miRNAs by UC was not efficient (). Recent studies indicated that UC-depletion is not sufficient to generate RNA-free FBS [Citation7,Citation8] and that the quantity and type of depleted RNAs are affected by variations of the depletion protocol [Citation8]. Therefore, it is not surprising that UC-depletion of supplements is not appropriate to remove RNA-contaminants and we did not explore this strategy further. However, RNase treatment of defined media supplements might be considered as an approach to minimize associated RNA-contaminants, but similar to UC-depletion for FBS, should not be expected to eliminate this problem completely.
Catalase is a source of miRNA contamination in NS21 supplement
Next, we tried to identify the supplement components responsible for the miRNA-contamination, to explore the possibility of formulating a culture medium free of contaminants. Since the most prominently detected NS21-contaminants, miR-122-5p and miR-451a, are highly abundant in liver and blood, we checked the original NS21 formulation [Citation12] for animal-derived components that are purified from these tissues. In addition to bovine serum albumin (BSA), NS21 contains Superoxide dismutase (SOD) and Catalase derived from bovine erythrocytes and liver, respectively. Of note, both SOD and Catalase (as well as BSA, not depicted) are clearly detected in the 100.000 x g pellet, when medium supplemented with commercially available NS21 is subjected to UC (). Hence, these components will co-isolate with EVs in UC-pellets and could be responsible for carrying exogenous miRNAs into EV-samples. With the notion of creating a miRNA-free supplement, we generated a self-made defined supplement (sm-NS19) lacking SOD and Catalase but not BSA, which is an essential component of the medium (see Supplementary Table 1). Similar to commercial NS21, Catalase and SOD were detected in the UC-pellet when added to the self-made supplement (sm-NS21). RT-qPCR analysis of these self-made supplements demonstrated that sm-NS19 was clean from the miRNAs tested, while sm-NS21 included miR-122-5p (), indicating that either SOD or Catalase are responsible for this contamination. To examine the influence of contaminant-free sm-NS19 lacking SOD and Catalase on the growth of primary oligodendrocytes, we measured the metabolic activity of cells cultured in sm-NS19 compared to sm-NS21. The metabolic activity of cells cultured in sm-NS19 was significantly reduced by 13 ± 2%, indicating that growth conditions are suboptimal (). SOD and Catalase are enzymes neutralizing reactive oxygen species most likely protecting cultured cells from oxidative damage.
Figure 3. Analysis of NS21 supplement components reveals Catalase as the main source of miRNA contamination.
(a) Western blot analysis of pellets after application of differential ultracentrifugation (EV-isolation protocol) to non-conditioned media supplemented with commercial NS21, sm-NS19 lacking SOD and Catalase, or sm-NS21 including SOD and Catalase. Both SOD and Catalase (CAT) are detected in the pellet. (b) RT-qPCR of miRNAs derived from a pellet after differential ultracentrifugation of non-conditioned sm-NS19 lacking SOD and Catalase and sm-NS21 including SOD and Catalase. Sm-NS21 is positive for miR-122-5p indicating that SOD or Catalase addition is linked to miRNA contamination (n = 1). (c) MTT-assay to measure metabolic activity of primary oligodendrocytes cultured in sm-NS19 and sm-NS21. Lack of SOD and Catalase is associated with significantly reduced metabolic activity of the cells (n = 3, SEM, *p = 0.017, two-way ANOVA). (d) RT-qPCR analysis of miRNAs associated with Catalase. Mammalian Catalase derived from different suppliers and tissue origin is contaminated to different degrees with miRNAs, while Catalase from Aspergillus niger is miRNA-free (n = 3). (e) Differential RT-qPCR analysis of miRNAs associated with EVs collected in the presence (EV+NS21) or absence (EV-NS21) of NS21 supplement. miRNA-122-5p and −451a do only appear in EVs+NS21, while miR-30d-5p and let-7g are also present in EVs-NS21.
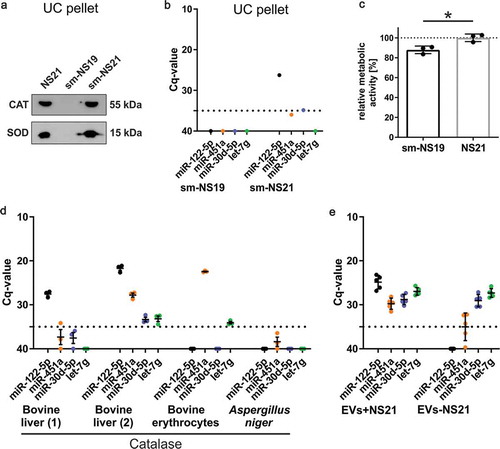
We further analysed individual NS21-components for the presence of miRNAs and focussed on protein components purified from animal tissues such as BSA, holo-Transferrin, SOD, and Catalase as well as Insulin, which is a recombinant product (Supplementary Table 2). All other non-protein components of the supplement were mixed and analysed as pool (sm-NS16-pool). Intriguingly, the sm-NS16-pool, BSA, holo-Transferrin, SOD, and Insulin were free of all miRNAs tested (not shown). BSA of different sources was consistently free of miRNA signals (not shown), which is remarkable as albumin is heavily abundant and a known carrier protein in blood [Citation25]. The only component that reproducibly revealed miRNA signals of varying degrees was Catalase (). Consistent with the tissue of origin, liver-derived Catalase contained significant levels of liver miR-122-5p, while erythrocyte-derived Catalase contained blood miR-451a. In fact, Catalase can be heavily contaminated by several miRNAs including significant levels of miR-451a and miR-30d, depending on the lot or the supplier (). Why Catalase appears to be prone to miRNA contamination, whether it has a biological meaning or is associated with its biophysical characteristics, remains elusive. Notably, Catalase derived from the fungus Aspergillus niger appeared to be free of the tested miRNAs, although it is possible that other miRNAs not included in our RT-qPCR panel could be detected.
Differential analysis of EVs collected in presence or absence of NS21 supplement
Another possibility to avoid supplement-associated impurities is to perform EV collection under supplement-free conditions. Comparison of EV-RNA collected under conditions with or without supplement may be informative regarding the discrimination of supplement-derived and EV-associated RNAs. Primary cultured oligodendrocytes that are differentiated to a mature state do well manage supplement-deprivation without entering apoptosis (not shown), although it is unknown to what level it affects the cellular metabolism. The same batch of cells was used to harvest EVs either in the presence (EVs+NS21) or absence of NS21 supplement (EVs-NS21) and RT-qPCR was performed with equal amounts of EV-RNA isolated from UC-pellets (). EVs+NS21 exhibited the expected miRNA pattern that is also detected in unconditioned NS21 supplemented media. In EVs-NS21, miR-122-5p was clearly undetectable and miR-451a was down to background level, again confirming these two miRNAs as contaminants of EV-RNA. Intriguingly, miR-30d-5p and let-7g were detected at similar levels in EVs+NS21 and EVs-NS21. This demonstrates that, despite their presence in the NS21 supplement, 30d-5p and let-7g may reflect genuine EV-miRNAs. However, it should be also noted that supplement-deprivation may influence miRNA expression or sorting in the cells affecting the miRNA pattern of the EVs.
Overall, the differential analysis of EVs collected in presence or absence of supplement delivers two types of outcomes: First, it allows the identification of miRNAs that are only medium-derived and can be excluded from the analysis. Second, it recognizes supplement-associated miRNAs that are also present in EVs and could be lost by simple exclusion of all miRNAs detected in the medium control.
Conclusion
In summary, our results raise unexpected and serious concerns about miRNA contaminations associated with so-called chemically defined supplements, which co-purify with EVs and confound EV miRNA analysis. Replacing FBS by addition of formulated supplements to culture media does not avoid the problem of potential exogenous miRNA contaminants in EV RNA data sets. We would like to emphasize that the study largely is of qualitative nature and, due to the absence of an internal reference miRNA, has its limitations in providing precise quantitative information. In general, the repeated detection of miR-122-5p and miR-451a in media supplements, as well as FBS, suggests that these miRNAs could serve as indicators, which may alert EV-researchers to perform a rigorous verification analysis at least of specific miRNAs that are studied further as biomarkers or in a functional context. Although the panel of miRNAs analysed in this study was restricted, the source of contamination was attributed to only a single factor within the supplement, the antioxidant enzyme Catalase. In fact, Catalase (and SOD) co-isolate with EVs indicating that media-associated proteins can be misinterpreted as EV-components too. Formulation of media lacking Catalase or making use of Catalases derived from recombinant sources or lower eukaryotes, such as that of the fungus Aspergillus niger, may help avoiding impurities through exogenous miRNAs in media and EV-samples. However, it will be difficult, if not impossible to consistently exclude contamination from supplements, as the quality of the components varies between supplier and production lots. Depletion of RNAs by UC or RNase-digestion does not reliably remove contaminating RNAs. Supplements free of animal-derived components could provide an alternative solution.
In conclusion, we recommend rigorous validation of EV-RNA data by including accurate controls of unconditioned media samples and if applicable, by comparison of EVs collected under supplement-free conditions. The background generated by media supplements may be very different in individual EV-studies and thus needs to be addressed in quantitative terms in the specific context. However, quantitative subtraction of RNA-background miRNA levels based on algorithms will be difficult to achieve in the absence of a stringent normalization factor that allows correction between unconditioned (EV-free) and cell-conditioned (EV-containing) samples, or between EV-samples collected in presence or absence of supplement. Finally, our findings are relevant beyond the EV-RNA research field, as miRNAs in the medium may have biological impact and influence various cellular functions. Possibly, the development of completely animal-free media supplements generated on the basis of recombinant technologies may provide an avenue to optimized defined media supplements free of contaminating miRNAs in the future.
Author contributions
MA, DF, EMKA conceived and designed experiments. MA, DF performed experiments. MA, DF, OD, EK and EMKA collected and analysed data. MA, EMKA wrote the manuscript.
Supplemental Material
Download MS Word (8.4 MB)Acknowledgments
We thank the IMB Genomics Core Facility for NGS library preparation and sequencing.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–10.
- Ratajczak J, Miekus K, Kucia M, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856.
- Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659.
- Shelke GV, Lasser C, Gho YS, et al. Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J Extracell Vesicles. 2014;3.
- Mateescu B, Kowal EJ, van Balkom BW, et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J Extracell Vesicles. 2017;6:1286095.
- Tosar JP, Cayota A, Eitan E, et al. Ribonucleic artefacts: are some extracellular RNA discoveries driven by cell culture medium components? J Extracell Vesicles. 2017;6:1272832.
- Wei Z, Batagov AO, Carter DR, et al. Fetal bovine serum RNA interferes with the cell culture derived extracellular RNA. Sci Rep. 2016;6:31175.
- Driedonks TAP, Nijen Twilhaar MK, Nolte-’t Hoen ENM. Technical approaches to reduce interference of Fetal calf serum derived RNA in the analysis of extracellular vesicle RNA from cultured cells. J Extracell Vesicles. 2019;8:1552059.
- Lehrich BM, Liang Y, Khosravi P, et al. Fetal bovine serum-derived extracellular vesicles persist within vesicle-depleted culture media. Int J Mol Sci. 2018;19:3538.
- Eitan E, Zhang S, Witwer KW, et al. Extracellular vesicle-depleted fetal bovine and human sera have reduced capacity to support cell growth. J Extracell Vesicles. 2015;4:26373.
- Romijn HJ. Development and advantages of serum-free, chemically defined nutrient media for culturing of nerve tissue. Biol Cell. 1988;63:263–268.
- Chen Y, Stevens B, Chang J, et al. NS21: re-defined and modified supplement B27 for neuronal cultures. J Neurosci Methods. 2008;171:239–247.
- Fröhlich D, Kuo WP, Frühbeis C, et al. Multifaceted effects of oligodendroglial exosomes on neurons: impact on neuronal firing rate, signal transduction and gene regulation. Philos Trans R Soc London, Ser B. 2014;369:20130510.
- Krämer EM, Koch T, Niehaus A, et al. Oligodendrocytes direct glycosyl phosphatidylinositol-anchored proteins to the myelin sheath in glycosphingolipid-rich complexes. J Biol Chem. 1997;272:8937–8945.
- Krämer-Albers EM, Bretz N, Tenzer S, et al. Oligodendrocytes secrete exosomes containing major myelin and stress-protective proteins: trophic support for axons? Proteomics Clin Appl. 2007;1:1446–1461.
- Frühbeis C, Fröhlich D, Kuo WP, et al. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 2013a;11:e1001604.
- Van Deun J, Mestdagh P, Agostinis, P., et al. EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat Methods. 2017;14:228–232.
- Frühbeis C, Fröhlich D, Kuo WP, et al. Extracellular vesicles as mediators of neuron-glia communication. Front Cell Neurosci. 2013b;7:182.
- Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414.
- Ludwig N, Leidinger P, Becker K, et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44:3865–3877.
- Xu YF, Hannafon BN, Khatri U, et al. The origin of exosomal miR-1246 in human cancer cells. RNA Biol. 2019;16:770–784.
- Zhang Y, Liao JM, Zeng SX, et al. p53 downregulates down syndrome-associated DYRK1A through miR-1246. EMBO Rep. 2011;12:811–817.
- Hill AF, Pegtel DM, Lambertz U, et al. ISEV position paper: extracellular vesicle RNA analysis and bioinformatics. J Extracell Vesicles. 2013;2:22859.
- Wang K, Zhang S, Weber J, et al. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259.
- Majorek KA, Porebski PJ, Dayal A, et al. Structural and immunologic characterization of bovine, horse, and rabbit serum albumins. Mol Immunol. 2012;52:174–182.
