ABSTRACT
For people living in developed countries life span is growing at a faster pace than ever. One of the main reasons for such success is attributable to the introduction and extensive use in the clinical practice of antibiotics over the course of the last seven decades. In hospital settings, Klebsiella pneumoniae represents a well-known and commonly described opportunistic pathogen, typically characterized by resistance to several antibiotic classes. On the other hand, the broad wedge of population living in Low and/or Middle Income Countries is increasing rapidly, allowing the spread of several commensal bacteria which are transmitted via human contact. Community transmission has been the original milieu of K. pneumoniae isolates characterized by an outstanding virulence (hypervirulent). These two characteristics, also defined as “pathotypes”, originally emerged as different pathways in the evolutionary history of K. pneumoniae. For a long time, the Sequence Type (ST), which is defined by the combination of alleles of the 7 housekeeping genes of the Multi-Locus Sequence Typing, has been a reliable marker of the pathotype: multidrug-resistant clones (e.g. ST258, ST147, ST101) in the Western world and hypervirulent clones (e.g. ST23, ST65, ST86) in the Eastern. Currently, the boundaries separating the two pathotypes are fading away due to several factors, and we are witnessing a worrisome convergence in certain high-risk clones. Here we review the evidence available on confluence of multidrug-resistance and hypervirulence in specific K. pneumoniae clones.
Introduction
Klebsiella pneumoniae stands up among other members of the Enterobacterales order, both for the speed and for the tenacity of its spread. These fermentative, non-motile, gram-negative bacteria can be considered ubiquitous microorganisms: members of the genus Klebsiella are widely represented in soil, water, and vegetation [Citation1]. They are also natural inhabitants of the gut microbiota of healthy humans and animals [Citation1–3]. Within the genus, K. pneumoniae represents an emerging worldwide public health issue, being one of the most frequent bacterial species associated with nosocomial infections [Citation4,Citation5].
While it can be a silent bowel colonizer for long periods of time [Citation6,Citation7], the pooled mortality rate for K. pneumoniae infections is high even nowadays, ranging from 21% to 42% based on its susceptibility to antimicrobial agents [Citation8].
As a matter of fact, K. pneumoniae ‘sensu stricto’ (not to be mistaken with other members of the related species complex, which seem to be more often linked with the environment) [Citation5] emerged as an epidemically successful bacterium via two different evolutionary pathways, often defined as pathotypes: Multidrug-Resistant K. pneumoniae (MDR-Kp) and hypervirulent K. pneumoniae (hvKp) [Citation9].
In both cases, infections often start from colonization of the gut microbiota [Citation10], yet the pathophysiology of the host–microorganism interaction is fundamentally different [Citation4].
In Western countries MDR-Kp is a leading cause of hospital acquired infections [Citation11,Citation12] and it is the subject of frequent risk assessments by health authorities [Citation13–15]. On the other hand, in Asia, and more specifically in the Asian Pacific Rim, several cases of severe and invasive infections caused by hvKp in otherwise healthy individuals were reported since the mid-late 1980s [Citation16].
Notwithstanding a 30-year history, hvKp in the scientific literature is way less represented than MDR-Kp, and its characteristics are less defined; yet, its spread is highly worrisome due to its ability to cause disease in healthy subjects [Citation17].
In this review, we focus on specific Clonal Groups (CGs) [Citation18], and on their associated Sequence Types (STs) [Citation19], which are considered the leading causes of the spread of MDR- and of hvKp, respectively. In particular, GC258, CG147, and CG101 were considered as representatives of the MDR-Kp type, and CG23, CG65, and CG86 as those of the hvKp. These lineages spread globally from different geographical areas over various time periods, and they are for this reason referred to as high-risk clones. Isolates/strains belonging to the same high-risk clone have similar phenotypic and genotypic traits and phylogenetic relatedness. By comparing common and discriminating genetic features, this review aims to summarize these characteristics in a selection of relevant CGs.
Multidrug-Resistance
The infections caused by MDR-Kp, often described as the ‘Classical K. pneumoniae’, represent a huge threat to medical care of the growing number of critically ill patients [Citation20]. This pathotype is of great concern because of i) its fast and vast dissemination, especially in critical settings such as Intensive Care Units (ICUs), ii) its ability to acquire multiple antimicrobial resistance (AMR) genes, and iii) the scarcity of effective antibiotics for extensively drug resistant K. pneumoniae. In fact, MDR-Kp has been listed as a priority bacterial pathogen for which new antibiotics are urgently needed by the World Health Organization [Citation21].
Even though in K. pneumoniae a proportion of AMR (e.g. toward colistin [Citation22] or tigecycline [Citation23]) emerges occasionally through chromosomal mutations [Citation24], the vast majority of AMR in this pathogen results from Horizontal Gene Transfer (HGT) of large, conjugative plasmids [Citation4]. In this scene, the acquisition of β-lactamases set the pace for the expansion of antimicrobial resistance in K. pneumoniae.
The blaSHV-1 gene is a chromosomally encoded β-lactamase gene which is found across members of the species and which provides resistance to ampicillin; the relationship between this specific AMR gene and K. pneumoniae is so tight that this bacterium has been proposed as the original source of the SHV β-lactamase family [Citation25].
The detection of Extended Spectrum β-lactamases (ESBLs) genes in K. pneumoniae began shortly after the introduction of third-generation cephalosporins in the clinical practice during the first half of the 1980s [Citation26,Citation27] While originating from Kluyvera spp., an environmental bacterium [Citation28,Citation29], β-lactamases of the CTX-M family are the most prevalent ESBLs in the Enterobacterales order; in addition, these ESBLs were at the basis of the success of some K. pneumoniae STs [Citation30].
Nevertheless, the K. pneumoniae trump card has been the acquisition of carbapenemases [Citation31]. These specific β-lactamase enzymes, which can hydrolyze a last-resort class of drugs named carbapenems, are often associated with Mobile Genetic Elements (MGEs), which mediate their horizontal diffusion within and between bacterial species. Since 1996, the Klebsiella Pneumoniae Carbapenemase (KPC) became the most prevalent carbapenemase in K. pneumoniae [Citation32].
More than 100 different acquired AMR genes have been identified in K. pneumoniae [Citation33], and they are mostly carried by AMR plasmids [Citation34,Citation35]. The K. pneumoniae plasmids pKpQIL and pKPN were initially identified in the genomes of the earliest carbapenemase-resistant isolates belonging to the ST258 clone from Israel and the U.S.A [Citation31,Citation36]. Both plasmids have FII-related replicons, subcategorized as FIIk [Citation37], and highly different FIB-replicons, named as FIB-pKpQIL and FIB-pKPN, respectively. pKpQIL and pKPN plasmids are compatible with each other, being co-resident in most of the ST258 bacterial cells. They can recombine, and, in some cases, they can create plasmid fusions [Citation38]. Besides the IncFIIk, many different plasmid families encoding for AMR genes have been described in K. pneumoniae, with IncA, IncC, IncHI1, IncX3, and IncN being the most prevalent ones [Citation5,Citation31]. These have been associated with the acquisition and spread of other carbapenemase genes, such as blaOXA-48 and blaNDM.
Hypervirulence
The first description of hvKp dates back in the 1980s, when this pathotype became endemic in the Pacific Rim region [Citation16].
While the phenotype needed to define a MDR strain is clear and well described [Citation39], the same cannot be said for the hypervirulent one [Citation40]. The string test, in which a colony cultured in standard conditions can be stretched into a string of at least 5 mm, was initially believed to be pathomognomonic of hvKp [Citation41]. However, a positive result to this test only states a hypermucoviscous phenotype. Furthermore, given the semi-qualitative nature of this test, sedimentation tests are currently preferred over the string-test due to their higher robustness as hypermucoviscosity detection methods [Citation42,Citation43].
The presence of a hyper-expressed capsule mediated by upregulators of gene expression (regulator of the mucoid phenotype rmpA/rmpA2-rmpD [Citation44,Citation45]) is a trait correlated with higher virulence, both in vivo [Citation46] and in clinical studies [Citation47,Citation48]. However, this characteristic alone does not define a strain as hypervirulent: not all hvKp strains are hypermucoviscous and, viceversa, some hypermucoviscous isolates do not carry virulence genes [Citation49,Citation50]. This misconception led to uncertainty in the scientific literature, especially when this phenotypic trait is used alone to categorize a strain as belonging to the hvKp pathotype [Citation51].
Similarly, the presence of genes encoding for the siderophores-salmochelin (iro), aerobactin (iuc), yersiniabactin (ybt) – the iron-foraging molecules which are associated with systemic infections, is not enough to define an isolate as hvKp [Citation33,Citation52–55]. This is also true for the presence of the K1 or K2 capsule types: though being generally associated with hypervirulence and particularly resistant to phagocytosis and serum [Citation56–58] they cannot be considered as discerning traits. In addition to siderophores, colibactin (clb), a genotoxin first described in Escherichia coli [Citation59] which causes cross-links in the DNA and induces double-strand DNA breaks [Citation60,Citation61] is diffused in K. pneumoniae too. Its prevalence ranges from 3.5% to 4% in the general population [Citation62] to 17–25% in the endemic East Asia region [Citation63]
As a matter of fact, hypervirulence should be considered as a multifactorial phenotype conferred by the presence of multiple virulence genes carried on large virulence plasmids (e.g. pK2044 [Citation64] and pLVPK [Citation65]) and within chromosomally inserted Integrative Conjugative Elements (ICEs).
Taken into consideration the limits associated with the prediction of a phenotype based on the genotype, a valid approach to the evaluation of virulence in a specific strain has been implemented by the creators of Kleborate [Citation66]. This tool does not return a dichotomic answer ‘virulent’, ‘not virulent’, rather it gives a ‘virulence score’, ranging from 0 to 5 . Specifically, the virulence score is calculated as follows: 0 if the isolate is negative for all of ybt, clb, and iuc, 1 if the isolate carries ybt only, 2 if the isolate carries ybt and clb or clb only, 3 if the isolate carries iuc (typically associated with the rmpADC and iro loci) only, 4 if the isolate carries ybt and iuc but not clb and 5 if the isolate carries clb, ybt and iuc. A similar modus operandi is applied to antimicrobial resistance, each isolate is given a score ranging from 0 to 3. Specifically, the score is 0 if no ESBLs nor carbapenemases are detected, 1 if there is one ESBL alone, 2 if there is a carbapenemase and 3 if the carbapenemase is associated with colistin resistance. On top of that, Kleborate also quantifies how many drug classes have at least one resistance gene detected. These approaches, despite the above-mentioned limits, are a valid starting point not only for considering hypervirulence and multidrug-resistance as nuances, but also for the detection and analysis of strains displaying simultaneously both hypervirulence and multidrug-resistance.
Convergence
Compared to the vast literature of MDR- and hvKp, there are not many reports describing isolates characterized by both pathotypes. Owing to the danger of clones possessing both these characteristics, surveillance systems are being implemented [Citation67–69].
Most cases of K. pneumoniae isolates carrying both MDR and virulence traits have been described in Asian countries (China leading this chart, with more than 2/3 of the reports), and this is coherent with the prevalence of isolates displaying these characteristics alone [Citation70]. Yet, this kind of phenomenon is being reported more and more frequently worldwide.
Under an evolutionary light, it is possible to identify three main patterns which can lead to a convergence of the two pathotypes [Citation9,Citation71]: i) hvKp isolates acquire MDR genes [Citation72,Citation73], ii) MDR-Kp isolates acquire hypervirulence plasmids [Citation74–76], and iii) acquisition of hybrid plasmids carrying both virulence and resistance genes [Citation30,Citation77,Citation78].
Research methodology
In January 2022, we searched the literature on PubMed for the keywords ‘Klebsiella pneumoniae’ and one of the selected CGs (GC258, CG147, CG101, CG23, CG65 or CG86), and ‘either associated with MDR or hv’. The STs comprised within each CG were searched on the BIGSdb database hosting the public Klebsiella pneumoniae database (https://bigsdb.web.pasteur.fr/klebsiella/) by using the ‘Search by locus combination’ option to retrieve every isolate belonging to a specific ST. Albeit being a biased search method, since not every isolate reported in literature is sequenced and not all sequenced isolates are uploaded on that database, this methodology allows for standardized and reproducible results ().
Figure 1. Alluvial diagram depicting the distribution of the various clonal groups (CGs) (left) in the sequence types (STs) that compose them (center), sorted by country (right).
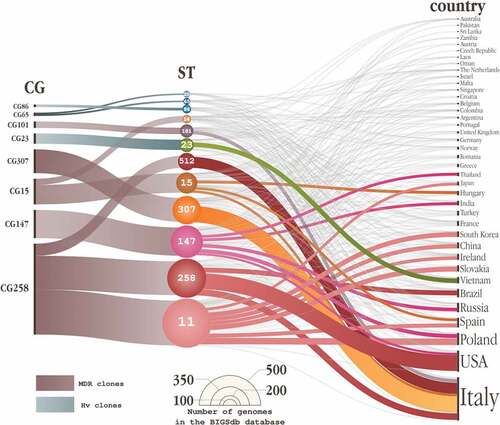
Figure 2. Minimum spanning tree representing one of the potentially many relationship which link the isolates belonging to the different high-risk clones. Numbers on the branches indicate the Single Nucleotide Polymorphisms differentiating the connected Sequence Types.
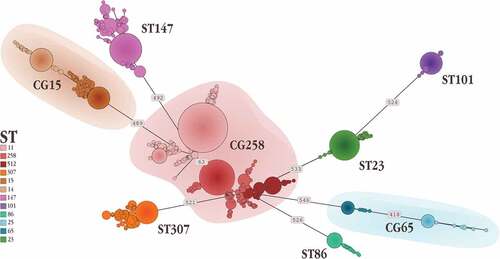
In the interest of simplification and intelligibility, we chose to only illustrate well-documented cases for each CG where strong evidence demonstrated the convergence of virulence and resistance genotypes, reporting about their evolution, spread, and genetic characteristics.
Clonal group 258 (ST11, 258, and ST512)
Three of the globally epidemic STs, namely ST11, ST258 and ST512, belong to the CG258, making this CG the most diffused MDR-Kp group [Citation79,Citation80] in the BIGSdb database, 965 isolates belonged to this CG (n = 476, ST11; n = 363, ST258; n = 126, ST512) ().
CG258 is a frequent carrier of the blaKPC carbapenemase gene, which can be found in most cases on plasmids with a pKpQIL backbone; even if this association is very high, it is neither absolute nor mutual, since this backbone can be found in other clonal groups [Citation81–83]. Furthermore, members of the CG258 can also carry numerous other acquired AMR genes [Citation84,Citation85].
ST11 is by far the dominant clone of KPC-producing K. pneumoniae in China, with the blaKPC-2 gene usually found on plasmids of the IncF type [Citation86–88]. Its endemicity in this country is so high that more than one clone belonging to this ST can cause simultaneous outbreaks in the same hospital [Citation89]. In 2015, two reports describing carbapenem-resistant K. pneumoniae belonging to ST11 with a hypervirulent phenotype were issued from the Beijing region [Citation90,Citation91], alerting the nation for a threat bigger than just carbapenem-resistance. The isolates carried the blaKPC-2 carbapenemase and the capsular type was defined as ‘non typeable’ by the capsular polysaccharide synthesis PCR method [Citation89,Citation90]. A retrospective study demonstrated how, up until 2016, the main carbapenem-resistant K. pneumoniae ST11 lineage of a hospital in the Zhejiang region harbored a K47 capsule which, over time, was replaced by the ST11-K64 sub-clone [Citation92]. In this specific case, the edge of the ST11-K64 was the co-occurrence of a pLVPK-like virulence plasmid interplaying with several other plasmids. Other studies describing outbreaks of carbapenem-resistant and hypervirulent ST11 in China detected K64 as the most common capsular type (up to 25% of the infections caused by carbapenem-resistant K. pneumoniae in certain regions) [Citation93], and highlighted how the fecal carriage of these isolates played a key role in their spread [Citation94].
Other reports describe how ST11-K47 has also been able to acquire hypervirulence traits. In several cases, the acquisition of a non-conjugative pLVPK-like plasmid has been held responsible for this evolutionary convergence [Citation95]. This type of HGT might have happened with the help of the conjugative IncF plasmid harboring blaKPC-2, which has an 11 kb region homologous to that of pLVPK [Citation96]. Interestingly, in another case, the co-existence of the two pathotypes in a ST11 isolate was possible thanks to the acquisition of a 141 kb IncFII plasmid (pR16-Hv-CRKp1, which does not show high homology with the known virulent plasmids), which harbored both resistance (blaKPC-2, blaCTX-M-65 and rmtB among the others) and putative virulence genes (such as R16_5486) [Citation73].
The acquisition of a virulence plasmid plays a key role in the spread of ST11 carbapenem-resistant hvKp strains [Citation21], and points to a worrisome evolutionary trajectory. This should elicit increased surveillance of the dissemination of these strains not only in nosocomial but also in community settings [Citation97]. Hypervirulent isolates can spread at a faster pace, given the limited fitness costs of virulence plasmids [Citation98], and their longer environment survival [Citation92].
With regard to the treatment, even if the carbapenem-resistant hypervirulent ST11-KL64 sub-clone has been capable of evolving resistance toward tigecycline and colistin during therapy [Citation99], for now ceftazidime-avibactam seems to be a valid therapeutic option against infections caused by KPC-2-producing hvKp [Citation100].
ST11 is also the ancestor of ST258, a hybrid clone composed by a replacement event between ST11, which supplied about 80% of the genetic material, and a single locus variant of ST258 called ST442 [Citation101]. Though there is a tight association between with the blaKPC carbapenemase gene, ST258 and its most common derivative ST512, no description about these two STs being hypervirulent has been outlined.
Clonal group 147 (ST147, ST273 and ST392)
Between 2008 and 2014, outbreaks caused by ST147 were reported in Germany and Hungary, with isolates producing OXA-48, KPC-2 and NDM-1 carbapenemases [Citation102,Citation103]. In 2016–17, three strains carrying the blaOXA-181 located on a 6 kb small ColKP3 plasmid were identified. These strains also carried a 110 kb IncFIB-like plasmid, that encoded tellurite/colicin resistance determinant, and phage-related genes (CP084394.1, CP074089.1) [Citation104]. In 2017, K. pneumoniae ST147 was described in the Middle East and India [Citation103,Citation104]. These isolates produced chromosomally encoded OXA-181, and the majority also produced the NDM-5 carbapenemase. The blaNDM-5 gene was located within an IncFII plasmid [Citation105,Citation106]. In Algeria in 2017, ST147 carried the blaNDM-1 gene located on an IncR plasmid [Citation107]. The evolutionary origin of ST147 and the two closely related ST273 and ST392 (grouped into CG147) was recently investigated. A time-scaled phylogeny reconstructed from this CG was structured into three main branches, each corresponding to ST147, ST273, and ST392, respectively. Within ST147, two main clades were observed, each characterized by distinct KL type and liposaccharide O antigen loci: clade KL64-O2 from Europe, and clade KL10-O3a from Asia, which emerged in 1994 and 2002, respectively. ST147 represents the paradigm of a clone in which antimicrobial resistance and virulence determinants converged thanks to the acquisition and exchange of mosaic plasmids. Different ybt/ICEKp subtypes (ybt16/ICEKp12, ybt10/ICEKp4) were observed within the ST147-KL64 clade, further discerning it in subclades 1 and 2 [Citation108]. Differently, virulence genes were rare among genomes of ST392 and ST273.
In 2018–2019, a large outbreak of NDM-1 producing ST147 K. pneumoniae occurred in Tuscany, Italy, with a total of 1,645 cases [Citation109]. The outbreak clone was resistant to all beta-lactam antibiotics, including carbapenems, and aminoglycosides, but susceptible to colistin, tigecycline, cefiderocol, and the aztreonam – avibactam combination. Reports of ST147 strains in Tuscany continued in 2020 and through 2021 [Citation110,Citation111].
Genomic studies performed on NDM-producing ST147 from Italy revealed that they belonged to the KL64 clade and that they were related with clinical isolates from the Middle East, the USA, Thailand, Myanmar, Egypt, Lebanon, the UK, Denmark, Germany, and Hungary [Citation111]. In Italian ST147, virulence genes encoding aerobactin (iutA-iucABCD), regulators of the mucoid phenotype (rmpADC and rmpA2, the latter often carrying a frameshift mutation), proteins involved in iron metabolism (cobW), and hemin and lysine transport system (shiF) were located on a large hybrid virulence/resistance plasmid carrying the HIB-FIB (Mar) replicons. This also carried blaCTX-M-15 and the 16S methyltransferase armA genes. The origin of the hybrid virulence/resistance plasmid can be traced back to the pNDM-Mar plasmid, carrying the same IncFIB/IncHI1 replicons (IncHI1B_pNDM-Mar; IncFIB_pNDM-Mar JN420336), identified in 2011 in K. pneumoniae ST15 from Morocco. pNDM-Mar was positive for the blaNDM-1, blaCTX-M-15 and qnrB1 genes, but negative for virulence genes [Citation112]. The virulence content probably originates from the pK2044 and pLVpK virulence plasmids of hvKp that are characterized by the replicon IncHI1B_pNDM-Mar but carry a different FIB-like replicon (repB_KLEB_VIR_AP006726).
The blaNDM-1 gene has been located mostly on plasmids of the FIB(pKpQIL)-type, showing 99% identity and 51% coverage with the backbone of plasmid pKpQIL (NC_014016), which massively contributed to the dissemination of KPC-type carbapenemases in CG258 [Citation110]
Virulence/resistance genes were also identified in the chromosome of ST147-KL64, with the most relevant being the yersiniabactin-encoding genes (ybtSXQPA, irp1, irp2, ybtUTE, and fyuA) associated with an ICEKp3, the mrkA-H genes encoding a type 3 fimbriae, as well as two additional blaCTX-15 genes located in the chromosome [Citation110,Citation111,Citation113]
ST147 strains from Italy did not exhibit a maximal hypervirulent phenotype compared with canonical hypervirulent isolate hv-Kp2. This was demonstrated using both a subcutaneous model of infection in immunocompetent CD1 mice but also in the Galleria mellonella infection model [Citation110,Citation111]
Despite the G. mellonella model was shown to not accurately differentiate hypervirulent from less virulent K. pneumoniae strains some strains in the model showed an overall enhanced virulence potential compared with a representative of the ST258 high-risk clone, with LD50 comparable or lower than those of reference hvKp strain NTUH-K2044. Furthermore, in serum bactericidal assays, the Italian ST147 isolates exhibited different grades of serum resistance associated with a different status of pal, csrD, and ramR chromosomal genes. In particular, the inactivation of CsrD has been associated with increased serum fitness by promoting capsule production and thickness, suggesting that ST147 virulence grade could be dependent by the status or function of bacterial surface components that can be variable during the clonal expansion [Citation110].
In May 2020, a new variant of the Italian ST147 was reported in a hospital in Tuscany. A shift in the NDM variant from NDM-1 to NDM-9 was observed, and this strain evolved to extreme-drug resistance by acquisition of resistance to colistin, tigecycline, and fosfomycin. The new clone emerged from highly related strains identified in 2019, showing mutations in RamR and MgrB proteins, implicated in tigecycline and colistin resistance, respectively. The fosfomycin resistance was associated with a defect in the glycerol-3-phosphate transporter [Citation114].
Clonal group 101 (ST101, ST1685, ST2016, ST2017, and ST2502)
The emergence of CG101 is estimated at the beginning of the 1990s [Citation115], yet it only needed a few years to become one of the most well-represented carbapenemase-producing K. pneumoniae in Europe [Citation116,Citation117]. Albeit strongly associated with the yersiniabactin locus [Citation5], there is only one report describing CG101 strains as having acquired hypervirulence trait [Citation78]. These isolates were sampled in 2018 in the United Kingdom and carried the blaOXA-48 carbapenemase (on the IncL/M plasmid CP031374.2), the 16S rRNA-methyltransferase armA and the macrolide resistance genes msr(E) and mph(E) (on the FIB/HI1B pNDM-Mar plasmid CP031372.2) and the quinolone resistance gene qnrS1 (on the IncX3 plasmid CP031373.2). A IncFII(K)/IncFIB(K) virulence plasmid (CP031369.2), carrying rmpA, rmpA2, iutA, iucABD, (with a truncation in both iucB and iucD), was retrieved alongside the resistance ones.
Nonetheless, several studies highlight the resistance of this clone not only to first-class antibiotics, such as carbapenems [Citation118], but also to colistin [Citation119–121], which is associated with a higher in-hospital mortality [Citation119], and tigecycline [Citation122]. Furthermore, in high-endemic settings for KPC-producing isolates, such as Italy, ST101 is developing several variants of this specific carbapenemase in order to resist to ceftazidime-avibactam (e.g. blaKPC-39 and blaKPC-68 [Citation38] blaKPC-46 [Citation123] or blaKPC-31 [Citation124].
Meanwhile, this ST is giving rise to its own CG by generating single locus variants (e.g. ST2502 [Citation124,Citation125], ST1685 [Citation115] or ST2016 and 2017 [Citation126]).
Clonal group 23 (ST23, ST26, ST57, and ST163)
Strictly associated with K1 capsular type [Citation49,Citation58], CG23 is highly diffused in the East Asia area [Citation127] () and it has been described as a hvKp with ‘moderate virulence’ in the G. mellonella model [Citation128]. The spread of the main clade of this lineage (CG23-I [Citation17]) is rapidly and steadily involving Western countries through multiple independent transmissions, embedding AMR genes along its way. Since 2018 there have been several reports of isolates belonging to ST23, which developed resistance toward carbapenems, mainly due to the acquisition of the IncL plasmid harboring blaOXA-48 [Citation129,Citation130].
Historically, the first traces of the encounter of this hvKp lineage with a plasmid conferring carbapenem resistance can be dated back to 2012. The strains were isolated from Russia [Citation131] and Germany [Citation103].
Following the report of several hvKp belonging to the ST23 in Ireland in March 2019, the European Center for Disease Prevention and Control (ECDC) performed a Rapid Risk Assessment (RRA), published in March 2021 [Citation67]. The ECDC asked 37 National Reference Laboratories to submit WGS data from ST23, receiving 5 representative isolates from Ireland, 5 from France and 1 from both Finland and Sweden. The isolates were split into two clades, a main one characterized by the K1 capsule loci, and one unrelated one with a K57 capsule. Despite all the Irish ST23 isolates belonged to the K1 clade, and most of them harbored the blaOXA-48 carbapenemase gene, a SNPs analysis on the core genome revealed that they were not related between each other. Differently, 2 out of 5 of the France isolates had a K1 capsule cluster and harbored blaOXA-48, while the other three had a K57 capsule, two of which carried blaOXA-48 and one blaNDM-1. The Swedish isolate had a K1 capsule and did not carry any carbapenemase gene, while the Finnish one had a K57 capsule and carried blaOXA-48. This peculiar spread and the comparison of the European isolates with the most related ones from the public domain show how the acquisition of blaOXA-48 from ST23 is rather recent and happened several times independently across the continent [Citation67].
The spread of ST23 isolates that acquired carbapenemase genes in Europe is not limited to the cases reported by the RRA [Citation132].
An eXtensively Drug Resistant (XDR [Citation39]) K. pneumoniae isolate harboring blaOXA-48 and the 16S rRNA methyltransferase armA on a plasmid has been described in Spain [Citation133]. The more relevant, and worrisome, difference with the RRA isolates is the fact that the virulence plasmid was fused within a large hybrid virulence/resistance plasmid carrying a copy of the blaCTX-M-15, while another copy of blaCTX-M-15, was in the chromosome [Citation133].
Despite more and more studies about hvKp in Europe are being published [Citation134–136], the scattered diffusion of ST23 in this region is a sign of the unnoticed spread of hvKp in the continent. An additional issue is that the convergence of the hypervirulent ST23 with carbapenem resistance in Europe seems to be strictly associated with the blaOXA-48 gene, for which there is a lack of guidelines for treatment [Citation137].
Clonal group 65 (ST25 and ST65)
CG65 is characterized by a K2 capsular serotype [Citation18,Citation138], and it is considered to be hypervirulent [Citation71] due to the carriage of pLVPK-like virulence plasmids and of the pks gene cluster (which is necessary for the synthesis of colibactin) [Citation63]. It comprises two STs, which differ in their epidemiology.
ST65 is spread worldwide, and it has the ability to acquire several plasmids harboring various carbapenemase genes. The epicenter of reports of carbapenem-resistant ST65 is China where, starting from 2015, several isolates manifesting both characteristics have been described. Two isolates have been reported carrying blaKPC-2 in the 2010–2014 period [Citation91], suggesting that this convergence happened prior to 2010, and one isolate in 2017–2018 period causing a bloodstream infection (BSI) [Citation95]. Furthermore, a case blaNDM-5 on a ‘canonical’ IncX3 plasmid [Citation139] was described [Citation140].
With regard to less diffused carbapenemases, two isolates belonging to ST65 carrying blaIMP-4 on IncU and IncN plasmids, in the latter case also coupled with tigecycline resistance [Citation141] were described.
While epidemiological studies in China report a high level of genetic diversity and an absence of MDR in hvKp [Citation142,Citation143], they only provide us with a glimpse of the tip of the iceberg (i.e. strains with peculiar phenotypes): the high plasticity of the ST65 genome, which can accept different resistance plasmids maintaining hypervirulence traits, poses a great danger.
There have been several reports of carbapenem resistance in ST65 also in other countries, such as Japan, where a study reported that out of 104 IMP-producing K. pneumoniae 12 were ST65-K2 [Citation144], or in Argentina, where blaKPC-2 has been described carried by IncM plasmid [Citation145].
ST25, instead, is the second most common carbapenemase-producing K. pneumoniae in Argentina, where in recent years there has been a shift from primarily ST258 [Citation146,Citation147] to a number of STs [Citation148,Citation149]. A study undertaken to characterize the isolates showing both MDR and hypervirulent phenotype collected during a 6-month period reported how, out of 35 isolates, 13 were blaKPC-2 harboring-ST25-K2 [Citation150]. Furthermore, this clone is spreading in the South American continent, particularly in Colombia [Citation151] and in Ecuador [Citation152].
Clonal group 86 (ST86 and ST3994)
Most of the isolates belonging to ST86 are characterized by a K2 capsular serotype and by the presence of a pLVPK-like plasmid backbone, which confer the hypervirulent phenotype [Citation58]. This ST is an international clone with global distribution, and a cause of community acquired infections in most continents, with a slight predisposition for animal infection.
In Australia, it is the cause of well-documented community-acquired infections since 1977, demonstrating a high PFGE similarity level throughout the years [Citation153]. Starting from the 2001–2003 breeding seasons, ST86 caused several outbreaks in New Zealand sea lion pups, eventually becoming endemic in the population in this country [Citation154].
The American continent has been affected from north to south by ST86. In North America, a carbapenemase-producing (KPC-2) ST86 isolate has been described as the cause of urine infections in Canada [Citation155], while in the Caribbean, a case of meningitis sustained by an hvKp has been reported [Citation156]. In South America, specifically in Brazil in 2013 [Citation157], an isolate belonging to the ST3994-K2 (a single locus variant of ST86, belonging to the same CG) was reported as the cause of a bloodstream infection [Citation157], while in the same nation in 2019, a ST86 clone was the cause of sudden death in 11 captive marmosets [Citation158].
Several reports have been provided by Asian countries, mainly from China, where the first report of ST86 can be traced back to two fatal infections caused by a susceptible isolate [Citation159], and it is considered an important cause of asymptomatic bacteriuria, which may lead to a BSI [Citation160].
Over time, ST86 has been described as associated with the blaKPC-2 carbapenemase gene. Alone, on an atypical IncX6 plasmid [Citation161], or in association with blaNDM-1 [Citation162]. In Japan, it has been held up as cause of severe infections, from community-acquired pneumonia [Citation163,Citation164] to infections in the veterinary field, particularly in captive-bred ruffed lemurs [Citation165].
In Europe, ST86 has been described as a cause of BSIs in a teaching hospital in Spain [Citation166] and there have been multiple descriptions of this ST in France [Citation167]. In this country, the first reports date back to 2011, when two cases of fatal infection in patients who recently returned from international travels were ascribed to this clone [Citation168]. The number of cases grew subsequently and in a 5-year period (2011–2016). Five community acquired infections (mainly pneumonia) that required ICU hospitalization were attributed to this ST [Citation167].
Despite all the previously mentioned strains were susceptible, this is not always the case: in the same nation two isolates belonging to this ST, both harboring several virulence genes, have been detected as MDR, the first one because of the presence of the blaOXA-48 on a IncL plasmid, the latter because of blaCTX-M-15 on an IncN plasmid coupled with deletions in the ompK36 and ramR genes [Citation169].
Though mostly being caused by susceptible isolates, the impact of outbreaks happening in veterinary settings should not be underestimated. The presence of hvKp in reservoirs close to human is of concern, and the application of a One Health approach is mandatory to control and contrast it [Citation170].
Conclusions
The identification of carbapenem resistance genes is nowadays routine in most clinical microbiology laboratories, but the detection of virulence genes is not. Phenotypic methods, such as the string test, cannot be considered reliable, since they lack sensitivity [Citation49]. Apart from the vast symptoms range to which clinicians may not be used to, as of today it is mainly the simultaneous presence of carbapenem resistance and hypervirulence that leads to the identification of the latter.
To tackle these issues and to be prepared to face the spread of hvKp, there are two strategies, which are not mutually exclusive: worthwhile tests to identify virulence genes/lineages (e.g. MLST screening of clinical isolates of K. pneumoniae [Citation171] or multiplex PCR assay for identification of clones with capsular serotype K2 [Citation172)], and a wider worldwide Whole Genome Sequencing and analysis coverage.
This identification is of the uttermost importance due to the fact that for a long time, the two K. pneumoniae MDR-Kp and hvKp pathotypes have been considered as two distinct, non-intersecting phenotypes, the first mostly associated with infections in immunocompromised patients, the second causing infections in healthy patients. As of today, oversimplified line we drew to keep MDR-Kp and hvKp apart is fading away since the two pathotypes are converging. We are not also allowed to conceive the risk of acquisition of hvKp and MDR-Kp strains restricted to patients linked to Asia or to patients from western countries with weakened immune systems, respectively.
Currently, the boundaries dividing the two pathotypes are less and less marked. Yet, since the intersection of the two pathotypes is something completely new, their description is moving on different tracks, and genomic epidemiology is needed as part of the diagnostic routine. On the one hand, case reports of single isolates and/or outbreaks are published with higher frequency, making a sufficient noise to depict a threat more and more tangible as time passes by, but from a quantitative point of view they are just scratching the surface. On the other hand, epidemiological studies portrait a dire summary of what the situation actually is, and the direction toward which we are heading: the spread of strains with converging pathotypes is rising, and it is just a matter of time before these strains are going to have the upper hand against the single pathotypes.
While focusing on the similarities and the distinctive traits of the global spread of MDR-Kp and of hvKp STs, this review highlights that, in the future years, the definition of these two pathotypes as dichotomous K. pneumoniae manifestations is going to be much harder. Rather, it is clear that, to tackle the diffusion of this global pathogen, the best approach will be to assess spectra of resistance and virulence.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Wareth G, Neubauer H. The animal-foods-environment interface of Klebsiella pneumoniae in Germany: an observational study on pathogenicity, resistance development and the current situation. Vet Res. 2021;52(16). DOI:10.1186/s13567-020-00875-w
- Podschun R, Ullmann U. Klebsiella spp. As nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589–603.
- Conlan S, Kong HH, Segre JA. Species-Level analysis of DNA sequence data from the NIH human microbiome project. PLoS One. 2012;7(10):e47075.
- Martin RM, Bachman MA. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol. 2018;8(4). DOI:10.3389/fcimb.2018.00004
- Wyres KL, Lam MMC, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020;18(6):344–359.
- Martin RM, Cao J, Brisse S, et al. Molecular epidemiology of colonizing and infecting isolates of Klebsiella pneumoniae. mSphere. 2016;1(5): DOI:10.1128/mSphere.00261-16.
- Gorrie CL, Mirceta M, Wick RR, et al. Antimicrobial-Resistant Klebsiella pneumoniae carriage and infection in specialized geriatric care wards linked to acquisition in the referring hospital. Clinl Infect Dis. 2018;67(2):161–170. DOI:10.1093/cid/ciy027
- Xu L, Sun X, Ma X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob. 2017;16(1):18.
- Lan P, Jiang Y, Zhou J, et al. A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J Glob Antimicrob Resist. 2021;25:26–34.
- Rao, K., Patel, A., Sun, Y., et al. Risk factors for Klebsiella Infections among Hospitalized Patients with Preexisting Colonization. Msphere. 2021; 6(3),e00132-21. doi:10.1128/mSphere.00132-21.
- Magill SS, Edwards JR, Bamberg W, et al. Multistate point-prevalence survey of health care–associated infections. N Engl J Med. 2014;370(13):1198–1208. DOI:10.1056/NEJMoa1306801
- Cassini A, Högberg LD, Plachouras D, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European economic area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019;19(1):56–66. DOI:10.1016/S1473-3099(18)30605-4
- ECDC. Outbreak of carbapenemase-producing (NDM-1 and OXA-48) and colistin-resistant Klebsiella pneumoniae ST307, north-east Germany, 2019. (2019).
- ECDC. Regional outbreak of New Delhi metallo-beta-lactamase-producing carbapenem-resistant Enterobacteriaceae, Italy, 2018–2019. 7 (2019).
- ECDC. Combined clonal and plasmid-mediated outbreak of carbapenemase-producing Enterobacterales, Lithuania, 2019–2020. 9 (2020).
- Liu YC, Cheng DL, Lin CL. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch Intern Med. 1986;146(10):1913–1916.
- Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32(3):42.
- Bialek-Davenet S, Criscuolo A, Ailloud F, et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis. 2014;20(11):1812–1820. DOI:10.3201/eid2011.140206
- Diancourt L, Passet V, Verhoef J, et al. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178–4182.
- Murray CJ, Ikuta KS, Sharara F, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. S0140673621027240. DOI:10.1016/S0140-6736(21)02724-0.
- Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. DOI:10.1016/S1473-3099(17)30753-3
- Ah Y-M, Kim A-J, Lee J-Y. Colistin resistance in Klebsiella pneumoniae. Int J Antimicrob Agents. 2014;44(1):8–15.
- Sheng Z-K, Hu F, Wang W, et al. Mechanisms of tigecycline resistance among Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother. 2014;58(11):6982–6985. DOI:10.1128/AAC.03808-14
- Wyres KL, Holt KE. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol. 2016;24(12):944–956.
- Chaves J, Ladona MG, Segura C, et al. SHV-1 β-lactamase is mainly a chromosomally encoded species-specific enzyme in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45(10):2856–2861. DOI:10.1128/AAC.45.10.2856-2861.2001
- Bauernfeind A, Schweighart S, Chong Y. β-Laktamase in Klebsiella pneumoniae mit erweitertem breitspektrum einschliesslich resistenz gegen cephamycine. Infection. 1989;17(5):316–321.
- Jacoby GA, Medeiros AA. More extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1991;35(9):1697–1704.
- Cantón R, González-Alba JM, Galán JC. CTX-M enzymes: origin and diffusion. Front Microbiol. 2012;3:110.
- D’Andrea MM, Arena F, Pallecchi L, et al. CTX-M-Type β-lactamases: a successful story of antibiotic resistance. Int J Med Microbiol. 2013;303(6–7):305–317.
- Lam MMC, Wyres KL, Wick RR, et al. Convergence of virulence and MDR in a single plasmid vector in MDR Klebsiella pneumoniae ST15. J Antimicrob Chemother. 2019;74(5):1218–1222. DOI:10.1093/jac/dkz028
- Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):252–275.
- Yigit H, Queenan AM, Anderson GJ, et al. Novel Carbapenem-Hydrolyzing β-Lactamase, KPC-1, from a Carbapenem-Resistant Strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45(4):1151–1161. DOI:10.1128/AAC.45.4.1151-1161.2001
- Holt KE, Wertheim H, Zadoks RN, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae , an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112(27):E3574–3581. DOI:10.1073/pnas.1501049112
- Ramirez MS, Traglia GM, Lin DL, et al. Plasmid-Mediated Antibiotic Resistance and Virulence in Gram-Negatives: the Klebsiella pneumoniae Paradigm. Microbiol Spectr. 2014;2(5). DOI:10.1128/microbiolspec.PLAS-0016-2013
- Iredell J, Brown J, Tagg K. Antibiotic resistance in Enterobacteriaceae: mechanisms and clinical implications. Bmj. 2016;352:h6420.
- Leavitt A, Chmelnitsky I, Carmeli Y, et al. Complete nucleotide sequence of KPC-3-encoding plasmid pKpqil in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob Agents Chemother. 2010;54(10):4493–4496.
- Villa L, García-Fernández A, Fortini D, et al. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother. 2010;65(12):2518–2529.
- Carattoli A, Arcari G, Bibbolino G, et al. Evolutionary trajectories toward ceftazidime-avibactam resistance in Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother. 2021;65(10):e0057421. DOI:10.1128/AAC.00574-21
- Magiorakos A-P, Srinivasan A, Carey RB, et al. Multidrug-Resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. DOI:10.1111/j.1469-0691.2011.03570.x
- Harada S, Doi Y, Diekema DJ. Hypervirulent Klebsiella pneumoniae: a call for consensus definition and international collaboration. J Clin Microbiol. 2018;56(9). DOI:10.1128/JCM.00959-18
- Fang C-T, Chuang Y-P, Shun C-T, et al. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199(5):697–705.
- Bachman MA, Breen P, Deornellas V, et al. Genome-Wide identification of Klebsiella pneumoniae fitness genes during lung infection. Mbio. 2015;6(3):e00775. DOI:10.1128/mBio.00775-15
- Mike LA, Stark AJ, Forsyth VS, et al. A systematic analysis of hypermucoviscosity and capsule reveals distinct and overlapping genes that impact Klebsiella pneumoniae fitness. PLoS Pathog. 2021;17(3):e1009376. DOI:10.1371/journal.ppat.1009376
- Lee C-R, Lee JH, Park KS, et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017;7:483. DOI:10.3389/fcimb.2017.00483
- Walker KA, Treat LP, Sepúlveda VE, et al. The small protein RmpD drives hypermucoviscosity in Klebsiella pneumoniae. Mbio. 2020;11(5):20–e01750.
- Yoshida K, Uchida K, Tsujimoto S, et al. Role of bacterial capsule in local and systemic inflammatory responses of mice during pulmonary infection with Klebsiella pneumoniae. J Med Microbiol. 2000;49(11):1003–1010. DOI:10.1099/0022-1317-49-11-1003
- Lee C-H, Liu J-W, Su L-H, et al. Hypermucoviscosity associated with Klebsiella pneumoniae-mediated invasive syndrome: a prospective cross-sectional study in Taiwan. Int J Infect Dis. 2010;14(8):e688–692. DOI:10.1016/j.ijid.2010.01.007
- Kim D, Park BY, Choi MH, et al. Antimicrobial resistance and virulence factors of Klebsiella pneumoniae affecting 30 day mortality in patients with bloodstream infection. J Antimicrob Chemother. 2019;74(1):190–199. DOI:10.1093/jac/dky397
- Catalán-Nájera JC, Garza-Ramos U, Barrios-Camacho H. Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence. 2017;8(7):1111–1123.
- Russo TA, Olson R, Fang C-T, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2018;56(9):18–e00776. DOI:10.1128/JCM.00776-18
- Gonzalez-Ferrer S, Peñaloza HF, Budnick JA, et al. Finding order in the chaos: outstanding questions in Klebsiella pneumoniae pathogenesis. Infect Immun. 2021;89(4):20–e00693. DOI:10.1128/IAI.00693-20
- Shon AS, Bajwa RPS, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–118.
- Bachman MA, Oyler JE, Burns SH, et al. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect Immun. 2011;79(8):3309–3316. DOI:10.1128/IAI.05114-11
- Behnsen J, Raffatellu M. Siderophores: more than stealing iron. Mbio. 2016;7(6):e01906–16.
- Holden VI, Breen P, Houle S, et al. Klebsiella pneumoniae siderophores induce inflammation, bacterial dissemination, and HIF-1α stabilization during pneumonia. Mbio. 2016;7(5):16–e01397.
- Lee IR, Molton JS, Wyres KL, et al. Differential host susceptibility and bacterial virulence factors driving Klebsiella liver abscess in an ethnically diverse population. Sci Rep. 2016;6(1):29316. DOI:10.1038/srep29316
- Kabha K, Nissimov L, Athamna A, et al. Relationships among capsular structure, phagocytosis, and mouse virulence in Klebsiella pneumoniae. Infect Immun. 1995;63(3):847–852. DOI:10.1128/iai.63.3.847-852.1995
- Struve C, Roe CC, Stegger M, et al. Mapping the evolution of hypervirulent Klebsiella pneumoniae. Mbio. 2015;6(4): DOI:10.1128/mBio.00630-15.
- Nougayrède J-P, Homburg S, Taieb F, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313(5788):848–851. DOI:10.1126/science.1127059
- Lai Y-C, Lin A-C, Chiang M-K, et al. Genotoxic Klebsiella pneumoniae in Taiwan. PLoS One. 2014;9(5):e96292. DOI:10.1371/journal.pone.0096292
- Vizcaino MI, Crawford JM. The colibactin warhead crosslinks DNA. Nat Chem. 2015;7(5):411–417.
- Putze J, Hennequin C, Nougayréde J-P, et al. Genetic structure and distribution of the colibactin genomic island among members of the family enterobacteriaceae. Infect Immun. 2009;77(11):4696–4703. DOI:10.1128/IAI.00522-09
- Chen Y-T, Lai Y-C, Tan M-C, et al. Prevalence and characteristics of pks genotoxin gene cluster-positive clinical Klebsiella pneumoniae isolates in Taiwan. Sci Rep. 2017;7(1):43120. DOI:10.1038/srep43120
- Wu K-M, Li L-H, Yan J-J, et al. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol. 2009;191(14):4492–4501. DOI:10.1128/JB.00315-09
- Chen Y-T, et al. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene. 2004;337:189–198. DOI:10.1016/j.gene.2004.05.008
- Lam MMC, Wick RR, Watts SC, et al. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12(1):4188. DOI:10.1038/s41467-021-24448-3
- European Centre for Disease Prevention and Control. Emergence of hypervirulent Klebsiella pneumoniae ST23 carrying carbapenemase genes in EU/EEA countries (Stockholm: ECDC)17 March 2021 . 14.
- Wyres KL, Nguyen TNT, Lam MMC, et al. Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from South and Southeast Asia. Genome Med. 2020;12(1):11. DOI:10.1186/s13073-019-0706-y
- Silvester R, Madhavan A, Kokkat A, et al. Global surveillance of antimicrobial resistance and hypervirulence in Klebsiella pneumoniae from LMICs: an in-silico approach. Sci Total Environ. 2022;802:149859. DOI:10.1016/j.scitotenv.2021.149859
- Tang M, Kong X, Hao J, et al. Epidemiological characteristics and formation mechanisms of multidrug-resistant hypervirulent Klebsiella pneumoniae. Front Microbiol. 2020;11:581543.
- Wyres KL, Wick RR, Judd LM, et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019;15(4):e1008114. DOI:10.1371/journal.pgen.1008114
- Xie Y, Tian L, Li G, et al. Emergence of the third-generation cephalosporin-resistant hypervirulent Klebsiella pneumoniae due to the acquisition of a self-transferable bla dha-1 -carrying plasmid by an ST23 strain. Virulence. 2018;9(1):838–844. DOI:10.1080/21505594.2018.1456229
- Yang Q, Jia X, Zhou M, et al. Emergence of ST11-K47 and ST11-K64 hypervirulent carbapenem-resistant Klebsiella pneumoniae in bacterial liver abscesses from China: a molecular, biological, and epidemiological study. Emerging Microbes Infect. 2020;9(1):320–331. DOI:10.1080/22221751.2020.1721334
- Xu Y, Zhang J, Wang M, et al. Mobilization of the nonconjugative virulence plasmid from hypervirulent Klebsiella pneumoniae. Genome Med. 2021;13(1):119. DOI:10.1186/s13073-021-00936-5
- Tian D, Wang W, Li M, et al. Acquisition of the conjugative virulence plasmid from a CG23 hypervirulent Klebsiella pneumoniae strain enhances bacterial virulence. Front Cell Infect Microbiol. 2021;11:752011. DOI:10.3389/fcimb.2021.752011
- Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46. DOI:10.1016/S1473-3099(17)30489-9
- Zhang R, Lin D, Chan E-W-C, et al. Emergence oF carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother. 2016;60(1):709–711. DOI:10.1128/AAC.02173-15
- Turton J, Davies F, Turton J, et al. Hybrid resistance and virulence plasmids in “high-risk” clones of Klebsiella pneumoniae, including those carrying blaNDM-5. Microorganisms. 2019;7(9):326. DOI:10.3390/microorganisms7090326
- Munoz-Price LS, Poirel L, Bonomo RA, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13(9):785–796. DOI:10.1016/S1473-3099(13)70190-7
- Pitout JDD, Nordmann P, Poirel L. Carbapenemase-Producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59(10):5873–5884.
- Adler A, Paikin S, Sterlin Y, et al. A swordless knight: epidemiology and molecular characteristics of the bla KPC -negative sequence type 258 Klebsiella pneumoniae clone. J Clin Microbiol. 2012;50(10):3180–3185. DOI:10.1128/JCM.00987-12
- David S, Cohen V, Reuter S, et al. Integrated chromosomal and plasmid sequence analyses reveal diverse modes of carbapenemase gene spread among Klebsiella pneumoniae. Proc Natl Acad Sci, USA. 2020;117(40):25043–25054. DOI:10.1073/pnas.2003407117
- Hendrickx APA, Landman F, de Haan A, et al. Plasmid diversity among genetically related Klebsiella pneumoniae blaKPC-2 and blaKPC-3 isolates collected in the Dutch national surveillance. Sci Rep. 2020;10(1):16778. DOI:10.1038/s41598-020-73440-2
- Di Pilato V, Arena F, Tascini C, et al. Mcr-1.2 , a new mcr variant carried on a transferable plasmid from a colistin-resistant KPC Carbapenemase-Producing Klebsiella pneumoniae strain of sequence type 512. Antimicrob Agents Chemother. 2016;60(9):5612–5615. DOI:10.1128/AAC.01075-16
- Kocsis B, Kádár B, Tóth Á, et al. MgrB variants in colistin-susceptible and colistin-resistant Klebsiella pneumoniae ST258. J Microbiol Immunol Infect. 2017;50(5):735–736.
- Qi Y, Wei Z, Ji S, et al. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66(2):307–312. DOI:10.1093/jac/dkq431
- Zhao J, Liu C, Liu Y, et al. Genomic characteristics of clinically important ST11 Klebsiella pneumoniae strains worldwide. J Glob Antimicrob Resist. 2020;22:519–526. DOI:10.1016/j.jgar.2020.03.023
- Xiong L, Su L, Tan H, et al. Molecular epidemiological analysis of ST11-K64 extensively drug-resistant Klebsiella pneumoniae infections outbreak in intensive care and neurosurgery units based on whole-genome sequencing. Front Microbiol. 2021;12:709356. DOI:10.3389/fmicb.2021.709356
- Zhang M, Li J, Lu Y, et al. Expanding of ST11 Carbapenemase-Producing Klebsiella pneumoniae subclones in a Chinese hospital, Shenzhen, China. infect Drug Resist. 2021;14:1415–1422. DOI:10.2147/IDR.S299478
- Zhang Y, Zeng J, Liu W, et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect. 2015;71(5):553–560. DOI:10.1016/j.jinf.2015.07.010
- Yao B, Xiao X, Wang F, et al. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Inter J Infect Dis. 2015;37:107–112. DOI:10.1016/j.ijid.2015.06.023
- Zhou K, Xiao T, David S, et al. Novel subclone of carbapenem-resistant Klebsiella pneumoniae sequence type 11 with enhanced virulence and transmissibility, China. Emerg Infect Dis. 2020;26(2):289–297. DOI:10.3201/eid2602.190594
- Zhang R, Li J, Wang Y, et al. Presence of NDM in non-E. coli Enterobacteriaceae in the poultry production environment. J Antimicrob Chemother. 2019;74(8):2209–2213. DOI:10.1093/jac/dkz193
- Zheng B, Xu H, Lv T, et al. Stool samples of acute diarrhea inpatients as a reservoir of ST11 hypervirulent KPC-2-producing Klebsiella pneumoniae. mSystems. 2020;5(3): DOI:10.1128/mSystems.00498-20.
- Du F, Huang Q-S, Wei D-D, et al. Prevalence of carbapenem-resistant Klebsiella pneumoniae co-harboring blaKPC-carrying plasmid and pLVPK-like virulence plasmid in bloodstream infections. Front Cell Infect Microbiol. 2021;10:556654. DOI:10.3389/fcimb.2020.556654
- Dong N, Yang X, Zhang R, et al. Tracking microevolution events among ST11 carbapenemase-producing hypervirulent Klebsiella pneumoniae outbreak strains. Emerging Microbes Infect. 2018;7(1):1–8.
- Xie M, Yang X, Xu Q, et al. Clinical evolution of ST11 carbapenem resistant and hypervirulent Klebsiella pneumoniae. Commun Biol. 2021;4(1):650. DOI:10.1038/s42003-021-02148-4
- Zhang Y, Jin L, Ouyang P, et al. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother. 2020;75(2):327–336. DOI:10.1093/jac/dkz446
- Jin X, Chen Q, Shen F, et al. Resistance evolution of hypervirulent carbapenem-resistant Klebsiella pneumoniae ST11 during treatment with tigecycline and polymyxin. Emerging Microbes Infect. 2021;10(1):1129–1136. DOI:10.1080/22221751.2021.1937327
- Yu F, Lv J, Niu S, et al. In vitro activity of ceftazidime-avibactam against carbapenem-resistant and hypervirulent Klebsiella pneumoniae isolates. Antimicrob Agents Chemother. 2018;62(8): DOI:10.1128/AAC.01031-18.
- Chen L, Mathema B, Pitout JDD, et al. Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. Mbio. 2014;5(3). DOI:10.1128/mBio.01355-14
- Szilágyi E, Füzi M, Böröcz K, et al. Risk factors and outcomes for bloodstream infections with extended-spectrum β-lactamase-producing Klebsiella pneumoniae; findings of the nosocomial surveillance system in Hungary. Acta Microbiol Immunol Hung. 2009;56(3):251–262. DOI:10.1556/AMicr.56.2009.3.5
- Becker L, Kaase M, Pfeifer Y, et al. Genome-Based analysis of carbapenemase-producing Klebsiella pneumoniae isolates from German hospital patients, 2008-2014. Antimicrob Resist Infect Control. 2018;7(1):62. DOI:10.1186/s13756-018-0352-y
- Xanthopoulou K, Carattoli A, Wille J, et al. Antibiotic resistance and mobile genetic elements in extensively drug-resistant Klebsiella pneumoniae sequence type 147 recovered from Germany. Antibiotics (Basel). 2020;9(10):E675. DOI:10.3390/antibiotics9100675
- Sonnevend Á, Ghazawi A, Hashmey R, et al. Multihospital occurrence of pan-resistant Klebsiella pneumoniae sequence type 147 with an is Ecp1 -directed bla oxa-181 Insertion in the mgrB gene in the United Arab Emirates. Antimicrob Agents Chemother. 2017;61(7): DOI:10.1128/AAC.00418-17.
- Rojas LJ, Hujer AM, Rudin SD, et al. NDM-5 and OXA-181 beta-lactamases, a significant threat continues to spread in the Americas. Antimicrob Agents Chemother. 2017;61(7): DOI:10.1128/AAC.00454-17.
- Abderrahim A, Djahmi N, Pujol C, et al. First case of NDM-1-producing Klebsiella pneumoniae in Annaba University Hospital, Algeria. Microbial Drug Resist. 2017;23(7):895–900. DOI:10.1089/mdr.2016.0213
- Rodrigues C, Desai S, Passet V, et al. Genomic evolution of the globally disseminated multidrug-resistant Klebsiella pneumoniae clonal group 147. Microb Genom. 2022;8(1). DOI:10.1099/mgen.0.000737
- Tavoschi L, Forni S, Porretta A, et al. Prolonged outbreak of New Delhi metallo-beta-lactamase-producing carbapenem-resistant enterobacterales (NDM-CRE), Tuscany, Italy, 2018 to 2019. Euro Surveill. 2020;25(6): DOI:10.2807/1560-7917.ES.2020.25.6.2000085.
- Di Pilato V., De Angelis L.H, and Aiezza N., et al. Resistome and virulome accretion in an NDM-1-producing ST147 sublineage of Klebsiella pneumoniae associated with an outbreak in Tuscany, Italy: a genotypic and phenotypic characterisation. The Lancet Microbe. 2022:3(3),e224-e234.doi:10.1016/S2666-5247(21)00268-8 .
- Martin MJ, Corey BW, Sannio F, et al. Anatomy of an extensively drug-resistant Klebsiella pneumoniae outbreak in Tuscany, Italy. Proc Natl Acad Sci, USA. 2021;118(48):e2110227118. DOI:10.1073/pnas.2110227118
- Villa L, Poirel L, Nordmann P, et al. Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15 and qnrB1 genes. J Antimicrob Chemother. 2012;67(7):1645–1650.
- Falcone M, Tiseo G, Arcari G, et al. Spread of hypervirulent multidrug-resistant ST147 Klebsiella pneumoniae in patients with severe COVID-19: an observational study from Italy, 2020–21. J Antimicrob Chemother. 2022;77(4):1140–1145. DOI:10.1093/jac/dkab495
- Falcone M, Giordano C, Barnini S, et al. Extremely drug-resistant NDM-9-producing ST147 Klebsiella pneumoniae causing infections in Italy, may 2020. Euro Surveill. 2020;25(48): DOI:10.2807/1560-7917.ES.2020.25.48.2001779.
- Roe CC, Vazquez AJ, Esposito EP, et al. Diversity, virulence, and antimicrobial resistance in isolates from the newly emerging Klebsiella pneumoniae ST101 lineage. Front Microbiol. 2019;10:542.
- David S, Reuter S, Harris SR, et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4(11):1919–1929. DOI:10.1038/s41564-019-0492-8
- Di Pilato V, Errico G, Monaco M, et al. The changing epidemiology of carbapenemase-producing Klebsiella pneumoniae in Italy: toward polyclonal evolution with emergence of high-risk lineages. J Antimicrob Chemother. 2021;76(2):355–361. DOI:10.1093/jac/dkaa431
- Avgoulea K, Di Pilato V, Zarkotou O, et al. Characterization of extensively drug-resistant or pandrug-resistant sequence type 147 and 101 OXA-48-producing Klebsiella pneumoniae causing bloodstream infections in patients in an intensive care unit. Antimicrob Agents Chemother. 2018;62(7): DOI:10.1128/AAC.02457-17.
- Can F, Menekse S, Ispir P, et al. Impact of the ST101 clone on fatality among patients with colistin-resistant Klebsiella pneumoniae infection. J Antimicrob Chemother. 2018;73(5):1235–1241. DOI:10.1093/jac/dkx532
- Esposito EP, Cervoni M, Bernardo M, et al. Molecular epidemiology and virulence profiles of colistin-resistant klebsiella pneumoniae blood isolates from the hospital agency “Ospedale dei Colli,” Naples, Italy. Front Microbiol. 2018;9:1463. DOI:10.3389/fmicb.2018.01463
- Nirwan PK, Chatterjee N, Panwar R, et al. Mutations in two component system (PhoPQ and PmrAB) in colistin resistant Klebsiella pneumoniae from North Indian tertiary care hospital. J Antibiot. 2021;74(7):450–457.
- Pena I, Picazo JJ, Rodríguez-Avial C, et al. Carbapenemase-Producing enterobacteriaceae in a tertiary hospital in Madrid, Spain: high percentage of colistin resistance among VIM-1-producing Klebsiella pneumoniae ST11 isolates. Int J Antimicrob Agents. 2014;43(5):460–464.
- Venditti C, Butera O, Meledandri M, et al. Molecular analysis of clinical isolates of ceftazidime-avibactam-resistant Klebsiella pneumoniae. Clin Microbiol Infect. 2021;27(7):1040.e1–1040.e6. DOI:10.1016/j.cmi.2021.03.001
- Arcari G, Oliva A, Sacco F, et al. Interplay between Klebsiella pneumoniae producing KPC-31 and KPC-3 under treatment with high dosage meropenem: a case report. Eur J Clin Microbiol Infect Dis. 2022;41(3):495–500. DOI:10.1007/s10096-021-04388-y
- Arena F, Di Pilato V, Vannetti F, et al. Population structure of KPC carbapenemase-producing Klebsiella pneumoniae in a long-term acute-care rehabilitation facility: identification of a new lineage of clonal group 101, associated with local hyperendemicity. Microb Genom. 2020;6(1): DOI:10.1099/mgen.0.000308.
- Pedersen T, Sekyere JO, Govinden U, et al. Spread of plasmid-encoded NDM-1 and GES-5 carbapenemases among extensively drug-resistant and pandrug-resistant clinical enterobacteriaceae in Durban, South Africa. Antimicrob Agents Chemother. 2018;62(5): DOI:10.1128/AAC.02178-17.
- Lam MMC, Wyres KL, Duchêne S, et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun. 2018;9(1):2703. DOI:10.1038/s41467-018-05114-7
- Shi Q, Lan P, Huang D, et al. Diversity of virulence level phenotype of hypervirulent Klebsiella pneumoniae from different sequence type lineage. BMC Microbiol. 2018;18(1):94. DOI:10.1186/s12866-018-1236-2
- Lev AI, Astashkin EI, Kislichkina AA, et al. Comparative analysis of Klebsiella pneumoniae strains isolated in 2012–2016 that differ by antibiotic resistance genes and virulence genes profiles. Pathog Glob Health. 2018;112(3):142–151. DOI:10.1080/20477724.2018.1460949
- Pitout JDD, Peirano G, Kock MM, et al. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev. 2019;33(1). DOI:10.1128/CMR.00102-19
- Volozhantsev NV, Kislichkina AA, Mukhina TN, et al. Draft genome sequences of clinical K1-Type Klebsiella pneumoniae strains isolated in Russia. Microbiol Resour Announc. 2020;9(1). DOI:10.1128/MRA.01250-19
- Blanc DS, Poirel L, Van Singer M, et al. Hypervirulent Klebsiella pneumoniae ST23 producing OXA-48 in Switzerland. Int J Antimicrob Agents. 2021;58(6):106457.
- Hernández M, López-Urrutia L, Abad D, et al. First report of an extensively drug-resistant ST23 Klebsiella pneumoniae of capsular serotype K1 co-producing CTX-M-15, OXA-48 and ArmA in Spain. Antibiotics. 2021;10(2):157. DOI:10.3390/antibiotics10020157
- Turton JF, Payne Z, Coward A, et al. Virulence genes in isolates of Klebsiella pneumoniae from the UK during 2016, including among carbapenemase gene-positive hypervirulent K1-ST23 and ‘non-hypervirulent’ types ST147, ST15 and ST383. J Med Microbiol. 2018;67(1):118–128. DOI:10.1099/jmm.0.000653
- Sánchez-López J, García-Caballero A, Navarro-San Francisco C, et al. Hypermucoviscous Klebsiella pneumoniae: a challenge in community acquired infection. Idcases. 2019;17:e00547. DOI:10.1016/j.idcr.2019.e00547
- Baron SA, Pascale L-M, Million M, et al. Whole genome sequencing to decipher the virulence phenotype of hypervirulent Klebsiella pneumoniae responsible for liver abscess, Marseille, France. Eur J Clin Microbiol Infect Dis. 2021;40(5):1073–1077. DOI:10.1007/s10096-020-04080-7
- Castanheira M, Doyle TB, Collingsworth TD, et al. Increasing frequency of OXA-48-producing Enterobacterales worldwide and activity of ceftazidime/avibactam, meropenem/vaborbactam and comparators against these isolates. J Antimicrob Chemother. 2021;76(12):3125–3134.
- Liao CH, Huang YT, Chang CY, et al. Capsular serotypes and multilocus sequence types of bacteremic Klebsiella pneumoniae isolates associated with different types of infections. Eur J Clin Microbiol Infect Dis. 2014;33(3):365–369.
- Wu W, Feng Y, Tang G, et al. NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin Microbiol Rev. 2019;32(2):e00115–18. DOI:10.1128/CMR.00115-18
- Zhao J, Zhang Y, Fan Y, et al. Characterization of an NDM-5-producing hypervirulent Klebsiella pneumoniae sequence type 65 clone from a lung transplant recipient. Emerging Microbes Infect. 2021;10(1):396–399. DOI:10.1080/22221751.2021.1889932
- Zhang Y, Wang X, Wang Q, et al. Emergence of tigecycline nonsusceptible and IMP-4 carbapenemase-producing K2-ST65 hypervirulent Klebsiella pneumoniae in China. Microbiol Spectr. 2021;9(2):e01305–21. DOI:10.1128/Spectrum.01305-21
- Zhao J, Chen J, Zhao M, et al. Multilocus sequence types and virulence determinants of hypermucoviscosity-positive Klebsiella pneumoniae isolated from community-acquired infection cases in Harbin, North China. Jpn J Infect Dis. 2016;69(5):357–360. DOI:10.7883/yoken.JJID.2015.321
- Ma Y, Bao C, Liu J, et al. Microbiological characterisation of Klebsiella pneumoniae isolates causing bloodstream infections from five tertiary hospitals in Beijing, China. J Glob Antimicrob Resist. 2018;12:162–166. DOI:10.1016/j.jgar.2017.10.002
- Yonekawa S, Mizuno T, Nakano R, et al. Molecular and epidemiological characteristics of carbapenemase-producing Klebsiella pneumoniae clinical isolates in Japan. mSphere. 2020;5(5): DOI:10.1128/mSphere.00490-20.
- Jure MA, Castillo ME, Musa HE, et al. Novel patterns in the molecular epidemiology of KPC-producing Klebsiella pneumoniae in Tucumán, Argentina. J Glob Antimicrob Resist. 2019;19:183–187. DOI:10.1016/j.jgar.2019.02.015
- Gomez SA, Pasteran FG, Faccone D, et al. Clonal dissemination of Klebsiella pneumoniae ST258 harbouring KPC-2 in Argentina. Clin Microbiol Infect. 2011;17(10):1520–1524. DOI:10.1111/j.1469-0691.2011.03600.x
- Cejas D, Fernandez Canigia L, Nastro M, et al. Hyperendemic clone of KPC producing Klebsiella pneumoniae ST 258 in Buenos Aires hospitals. Infect Genet Evol. 2012;12(3):499–501. DOI:10.1016/j.meegid.2011.09.018
- Cejas D, Elena A, Guevara Nuñez D, et al. Changing epidemiology of KPC-producing Klebsiella pneumoniae in Argentina: emergence of hypermucoviscous ST25 and high-risk clone ST307. J Glob Antimicrob Resist. 2019;18:238–242. DOI:10.1016/j.jgar.2019.06.005
- Jure MA, Albarracin L, Vargas JM, et al. Draft genome sequences of two hypermucoviscous carbapenem-resistant ST25 Klebsiella pneumoniae strains causing respiratory and systemic infections. J Glob Antimicrob Resist. 2021;26:174–176. DOI:10.1016/j.jgar.2021.05.018
- Vargas JM, et al. Virulence factors and clinical patterns of multiple-clone hypermucoviscous KPC-2 producing K. pneumoniae. Heliyon. 2019;5:e01829.
- Cienfuegos-Gallet AV, Ocampo de Los Ríos AM, Sierra Viana P, et al. Risk factors and survival of patients infected with carbapenem-resistant Klebsiella pneumoniae in a KPC endemic setting: a case-control and cohort study. BMC Infect Dis. 2019;19(1):830. DOI:10.1186/s12879-019-4461-x
- Reyes J, Cárdenas P, Tamayo R, et al. Characterization of bla kpc-2 -harboring Klebsiella pneumoniae isolates and mobile genetic elements from outbreaks in a hospital in Ecuador. Microbial Drug Resist. 2021;27(6):752–759. DOI:10.1089/mdr.2019.0433
- Hall JM, Ingram PR, O’Reilly LC, et al. Temporal flux in β-lactam resistance among Klebsiella pneumoniae in Western Australia. J Med Microbiol. 2016;65(5):429–437.
- Pinpimai, K., Roe, W.D, and Biggs, et al. DrafT whole-genome sequences of seven isolates of Klebsiella pneumoniae from New Zealand sea lions. Microbiol Resour Announcements. 2018;7(20),e01270-18.doi:10.1128/MRA.01270-18.
- Mataseje LF, Boyd DA, Mulvey MR, et al. Two hypervirulent Klebsiella pneumoniae isolates producing a bla kpc-2 carbapenemase from a Canadian patient. Antimicrob Agents Chemother. 2019;63(7). DOI:10.1128/AAC.00517-19
- Melot B, Brisse S, Breurec S, et al. Community-Acquired meningitis caused by a CG86 hypervirulent Klebsiella pneumoniae strain: first case report in the Caribbean. BMC Infect Dis. 2016;16(1):736. DOI:10.1186/s12879-016-2065-2
- Cerdeira L, Nakamura-Silva R, Oliveira-Silva M, et al. A novel hypermucoviscous Klebsiella pneumoniae ST3994-K2 clone belonging to clonal group 86. Pathog Dis. 2021;79(8):ftab047. DOI:10.1093/femspd/ftab047
- Guerra JM, Fernandes NCCDA, Morales dos Santos AL, et al. Hypervirulent Klebsiella pneumoniae as unexpected cause of fatal outbreak in captive Marmosets, Brazil. Emerg Infect Dis. 2020;26(12):3039–3043. DOI:10.3201/eid2612.191562
- Zhang Y, Sun J, Mi C, et al. First report of two rapid-onset fatal infections caused by a newly emerging hypervirulent K. Pneumonia ST86 strain of serotype K2 in China. Front Microbiol. 2015;6. DOI:10.3389/fmicb.2015.00721.
- Li J, Li Y, Tang M, et al. Distribution, characterization, and antibiotic resistance of hypervirulent Klebsiella pneumoniae isolates in a Chinese population with asymptomatic bacteriuria. BMC Microbiol. 2022;22(1):29. DOI:10.1186/s12866-021-02413-w
- Liu Z, Chu W, Li X, et al. Genomic features and virulence characteristics of a community-acquired bloodstream infection-causing hypervirulent Klebsiella pneumoniae ST86 strain harboring KPC-2-encoding IncX6 plasmid. Microbial Drug Resist. 2021;27(3):360–368. DOI:10.1089/mdr.2019.0394
- Liu Y, Long D, Xiang T-X, et al. Whole genome assembly and functional portrait of hypervirulent extensively drug-resistant NDM-1 and KPC-2 co-producing Klebsiella pneumoniae of capsular serotype K2 and ST86. J Antimicrob Chemother. 2019;74(5):1233–1240. DOI:10.1093/jac/dkz023
- Hirai J, Sakanashi D, Kinjo T, et al. The first case of community-acquired pneumonia due to capsular genotype K2-ST86 hypervirulent Klebsiella pneumoniae in Okinawa, Japan: a case report and literature review. Idr. 2020;13:2237–2243.
- Yamamoto H, Iijima A, Kawamura K, et al. Fatal fulminant community-acquired pneumonia caused by hypervirulent Klebsiella pneumoniae K2-ST86: case report. Medicine (Baltimore). 2020;99(21):e20360. DOI:10.1097/MD.0000000000020360
- Sasaki E, Tokiwa T, Tsugo K, et al. Peracute bacterial meningitis due to infection with Klebsiella pneumoniae in captive-bred ruffed lemurs (Varecia variegate). J Comp Pathol. 2017;156(2–3):281–285. DOI:10.1016/j.jcpa.2016.12.003
- Cubero M, Grau I, Tubau F, et al. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007–2013). Clin Microbiol Infect. 2016;22(2):154–160. DOI:10.1016/j.cmi.2015.09.025
- Rafat C, Messika J, Barnaud G, et al. Hypervirulent Klebsiella pneumoniae, a 5-year study in a French ICU. J Med Microbiol. 2018;67(8):1083–1089. DOI:10.1099/jmm.0.000788
- Decré D, Verdet C, Emirian A, et al. Emerging severe and fatal infections due to Klebsiella pneumoniae in two university hospitals in France. J Clin Microbiol. 2011;49(8):3012–3014. DOI:10.1128/JCM.00676-11
- Beyrouthy R, Dalmasso G, Birer A, et al. Carbapenem resistance conferred by OXA-48 in K2-ST86 hypervirulent Klebsiella pneumoniae , France. Emerg Infect Dis. 2020;26(7):1529–1533.
- Butaye P, Stegger M, Moodley A, et al. One health genomic study of human and animal Klebsiella pneumoniae isolated at diagnostic laboratories on a small Caribbean Island. Antibiotics. 2021;11(1):42. DOI:10.3390/antibiotics11010042
- Yu F, Lv J, Niu S, et al. Multiplex PCR analysis for rapid detection of Klebsiella pneumoniae carbapenem-resistant (Sequence type 258 [ST258] and ST11) and hypervirulent (ST23, ST65, ST86, and ST375) strains. J Clin Microbiol. 2018;56(9): DOI:10.1128/JCM.00731-18.
- Bialek-Davenet S, Criscuolo A, Ailloud F, et al. Development of a multiplex PCR assay for identification of Klebsiella pneumoniae hypervirulent clones of capsular serotype K2. J Med Microbiol. 2014;63(12):1608–1614. DOI:10.1099/jmm.0.081448-0
