ABSTRACT
Introduction
Carbapenemase-producing Gram-negative bacilli have been major culprits in hospital-associated infections (HAIs), particularly in critically ill patients suffering device-associated infections (DAIs). The current study aimed to investigate the performance of the modified Hodge test (MHT) as a phenotypic confirmatory method for the detection of carbapenemase-producing Gram-negative bacilli and to compare it to the gold standard PCR for the detection of carbapenemase production in both non-susceptible and phenotypically susceptible isolates. The latter were expected to harbor silent carbapenemase genes, as suspected from the inappropriate response to carbapenem therapy.
Methods
Ninety-five bacterial isolates from 75 critically ill patients were collected over 6 months from several ICUs at Cairo University Hospitals. The isolates were subjected to antibiotic susceptibility testing (AST) for carbapenems and were further screened by MHT, followed by genotypic analysis via multiplex PCR.
Results
Enterobacteriaceae were the most commonly isolated pathogens (55.8% of the total isolates), followed by Acinetobacter spp. (24%). Lower respiratory tract infections were the most common HAIs (42.11%), followed by surgical site infections (27.37%). All isolates demonstrating carbapenem resistance by AST were found to harbor at least one of the following carbapenemase genes: blaKPC, blaOXA-48, blaIPM, blaVIM, and blaNDM-1. Alarmingly, 97.8% of the isolates which exhibited carbapenem-susceptible profile and negative MHT were harboring carbapenemase genes as confirmed by multiplex PCR. With the exception of one isolate (E. coli) which was not harboring any carbapenemase gene, the remaining 94 bacterial isolates were found to carry either a single or multiple carbapenemase genes.
Conclusion
The silent dissemination of different classes of carbapenemases even in isolates with negative MHT is a daunting challenge. It necessitates the implementation of strict antibiotic stewardship along with updated and actionable approach to detect non-expressed carbapenemase genes in phenotypically susceptible isolates.
1. Introduction
Throughout the last decades, hospital-acquired infections have become a grave problem in the environment of intensive care units (ICUs). The advancement in human medicine has been ensued by extended hospitalization, with unwarranted and long-term antibiotic treatment. In a setting of immune-deficient patients with invasive procedures, this could cause serious repercussions on mortality rates and treatment consequences [Citation1].
In the meantime, due to the surging rates of resistance witnessed to commonly prescribed antibiotics, carbapenems have become the most substantial therapeutic option for handling infections caused by Gram-negative bacilli (GNB) [Citation2]. Nonetheless, an alarming concern is the global spread and the cumulative prevalence of anti-microbial resistance to β-lactam antibiotics (including carbapenems) among Pseudomonas aeruginosa, Acinetobacter baumannii as well as members of the Enterobacteriaceae family, which represents a life-threatening medical and public health issue [Citation3].
Prompt molecular identification of carbapenemase genes in GNB is crucial for infection control and prevention, surveillance, and epidemiological studies. Furthermore, it would have a major influence upon determining an appropriate initial treatment, which will positively impact critically ill patients [Citation4]. Several diagnostic modalities have been recognized based on the detection of carbapenem-hydrolyzing activity. Additionally, several phenotypic confirmatory tests have also been implemented, such as NG-test CARBA 5, Xpert Carba-R, mCIM, and eCIM [Citation5].
This study was performed in several ICUs at Kasr Al-Ainy University Hospitals to detect carbapenemase genes among Gram-negative bacteria; both resistant and phenotypically susceptible strains, and to compare the modified Hodge test (MHT) as a phenotypic confirmatory test with multiplex PCR as a gold standard genotypic method.
2. Materials and methods
2.1. Ethical consideration
Before commencing this study, it was approved via the Research Ethics Committee of the Institutional Review Board, Faculty of Medicine, Cairo University. An informed consent was procured from each participant.
2.2. Population of study and disease condition
This cross-sectional analytical study was conducted on 75 critically ill patients with healthcare-associated infections (HAIs), comprising device-associated infections (catheter-associated urinary tract infections, CAUTI; and ventilator-associated events, VAE), during a 6-month period in several ICUs at Kasr Al-Ainy Hospitals, Cairo University.
2.2.1. Inclusion criteria
Patients with HAIs after 2 calendar days of admission according to the CDC [Citation6].
Patients with CAUTI having a UTI where an indwelling urinary catheter (IUC) was in place for >2 calendar days on the date of event, with theday of device placement being day 1, and an indwelling urinary catheter was in place on the date of event or the day before [Citation6].
Patients with VAE/Ventilator-Associated Pneumonia (VAP): According to the CDC definition [Citation7], the patient must have fulfilled the criteria of the three stages of VAE following a baseline period of stability or improvement on the ventilator.
2.3. Samples
During the study period, 95 bacterial isolates were collected from 75 critically ill patients with HAIs. These isolates were obtained by cultivating the following clinical specimens: sputum, bronchoalveolar lavage (BAL), endotracheal tube (ETT) aspirate, urine, wound swab, and drainage of chest tube. The isolation was done using blood agar and MacConkey’s agar plates incubated aerobically at 37°C for 24–48 hours. Identification of the isolates was done according to the conventional microbiological standard tests: Gram’s stain, glucose fermentation, lactose fermentation, and oxidase test [Citation8]. Identification up to the species level was done using Microbact-12A, Oxoid, UK, and API-20E, bioMérieux, France, identification systems.
2.4. Antimicrobial susceptibility testing and carbapenem resistance screening by disk diffusion
The antimicrobial susceptibility profiles were performed by employing the following antibiotics (Oxoid, UK): Ertapenem (10 µg), Imipenem (10 µg), Meropenem (10 µg), Ceftazidime (30 µg), Cefepime (30 µg), Aztreonam (30 µg), Amikacin (30 µg), Ciprofloxacin (5 µg), and Tigecycline (15 µg). Zone of inhibition for each antibiotic was measured as per the standard CLSI and EUCAST guidelines and interpreted as susceptible, resistant, or intermediate if applicable [Citation9,Citation10].
2.5. Phenotypic confirmatory test for carbapenemase production using the modified Hodge test (MHT)
All isolates were subjected to phenotypic confirmatory tests using the MHT. The standard suspension (E. coli ATCC 25,922 at a turbidity of 0.5 McFarland) was diluted as 1:10 in sterile saline. Thereafter, it was inoculated on Mueller-Hinton agar plate that was supplemented with zinc sulfate. For each plate, the tested colonies were streaked out from the edge of meropenem disk (10 µg) toward the plate periphery. After overnight incubation, indentation of the inhibition zone(s) indicated that the test strain attacks carbapenems. This test was considered negative for carbapenemase production in case there was no enhanced growth [Citation11].
2.6. Genotypic detection of carbapenemase genes
Discrete colonies were carefully isolated from each plate and hence sub-cultivated to obtain non-contaminated pure isolates.
Genotypic analysis procedures were performed in a well-equipped molecular laboratory at the Medical Biochemistry Department, Faculty of Medicine, Cairo University.
Multiplex PCR was operated to detect carbapenem resistance genes (blaKPC, blaOXA-48, blaIPM, blaVIM, and blaNDM-1) using specific primers (Invitrogen by life technologies, Thermo Fisher Scientific Inc. USA) to amplify internal fragments with sizes from 181 bp to 744 bp [Citation12] ().
DNA was extracted using a commercially available kit (Qiagen DNeasy Blood & Tissue Kit; Qiagen GmbH, Hilden, Germany) as per the manufacturer’s instructions. The extracted DNA was subjected to enzymatic amplification via a commercially available kit (Bio Basic. Canada INC) in a final volume of 50 μl using a thermal cycler (Applied Biosystems). The PCR products were resolved by electrophoresis on 1.5% agarose gel.
Table 1. List of primers used in the study
2.7. Statistical analysis
Data were coded and entered employing the statistical package SPSS (Statistical Package for the Social Sciences) version 24. For quantitative data, mean, standard deviation, median, minimum, and maximum were employed, while for categorical data, frequency (count) and relative frequency (percentage) were employed. The standard diagnostic indices including sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and diagnostic efficacy were all calculated.
3. Results
The current study was carried out on 95 clinical isolates retrieved from 75 critically ill patients; where some enrolled patients yielded more than one isolate from different sites.
The study population comprised 51 males (68%) and 24 females (32%), with their ages between 16 and 84 years (mean 49.9). The patients were identified as having HAIs according to the CDC criteria, with 3–70 days (mean 12.36) of hospital stay prior to infection onset.
Among the 75 patients, 67 (89%) had inserted urinary catheters and 41 (54.6%) were on mechanical ventilators.
Among the 95 bacterial isolates, Enterobacteriaceae were the most frequent pathogens (55.8% of the total isolates), followed by Acinetobacter spp. 23/95 (24% of the total isolates), and lastly Pseudomonas spp. 15/95 (16%). Thirty-eight (40%) isolates were retrieved from sputum specimens, and 30 (31.5%) isolates from wounds, as depicted in .
Table 2. Isolated bacteria from clinical samples (n = 95)
BAL: Bronchoalveolar lavage; ETT aspirate: endotracheal tube aspirate
Pneumonia was the most common entity of HAIs, with 40 retrieved isolates (42.11%), followed by surgical site infections (SSIs) with 26 isolates (27.37%), then urinary tract infections (UTIs) with 24 isolates (25.26%).
Out of 95 HAIs isolates, 41 isolates were from DAIs (43.2%). VAEs fulfilling the criteria of CDC affected 21% of all critically ill patients, and 48.78% of mechanically ventilated patients, while CAUTIs affected 22.1% of all patients and 31.34% of patients with urinary catheters.
3.1. Susceptibility pattern using disk diffusion and modified Hodge test
The antibiotic susceptibility test was performed using disk diffusion AST for all isolates (n = 95), including tigecycline which was used for Enterobacteriaceae only. Tigecycline zone diameter ≤15 mm was considered resistant, while zone diameter ≥18 mm was considered susceptible according to EUCAST [Citation10].
All three carbapenems (Meropenem, Imipenem, and Ertapenem) were used to test for susceptibility pattern of each isolate of Enterobacteriaceae, while only imipenem and meropenem were used for non-fermentative Gram-negative bacteria.
Each isolate found to be resistant to any of the carbapenems used was considered “carbapenem resistant” (Group A), while isolates which were susceptible to all tested carbapenems were considered “carbapenem susceptible” (Group B). No isolate showed an intermediate result. Antibiotic resistance pattern for all isolates is shown in .
The susceptibility patterns among all 95 isolates using disk diffusion (Kirby–Bauer) method revealed resistance to carbapenems in 49 isolates (Group A). Further phenotypic confirmation for carbapenemase was performed for all isolates using MHT. The prevalence of carbapenemase producers was as follows: out of the 49 carbapenem-resistant isolates by AST, 44 isolates were positive according to MHT as evidenced by the indentation of the inhibition zone of the MEM disc; whereas 3 isolates were negative, and only 2 isolates were indeterminate.
In the remaining 46 isolates which were carbapenem-susceptible via disk diffusion (Group B); 41 isolates were negative according to MHT, while 5 isolates were indeterminate ().
Figure 1. MHT showing indentation (clover-leaf appearance) indicating carbapenemase production by the tested isolates (A), and isolates with no indentation indicate negative MHT (B), while isolates with inhibited E. coli ATCC 25,922 growth indicate indeterminate result (C). Meropenem (MEM) disc (10 µg) was used
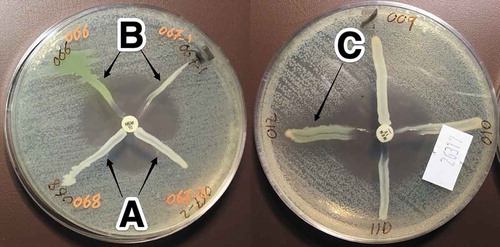
Table 3. Resistance pattern for both groups: carbapenem resistant (A) and susceptible (B)
3.2. Genotypic analysis
All 95 isolates of the study were subjected to genotypic analysis via multiplex PCR as a gold standard method.
All 49 suspected carbapenem-resistant isolates (Group A) were found to harbor at least one of the following carbapenemase genes: blaKPC, blaOXA-48, blaIPM, blaVIM, and blaNDM-Of note, only 44 isolates out of 49 were MHT positive; thus in Group A only, the sensitivity of MHT when compared to PCR as the gold standard method was 89.8% and the accuracy was also 89.8%.
Although apparently sensitive, PCR was performed for Group B because it was expected that they harbor silent carbapenemase genes, as suspected from the inappropriate response to carbapenem therapy.
As expected, 45 out of 46 carbapenem-susceptible isolates (Group B) were found to harbor at least one carbapenemase gene. These non-expressed (silent) genes were isolated from different bacterial species among patients with inappropriate clinical response to carbapenem therapy ().
Excluding indeterminate results, the overall sensitivity and specificity of MHT in relation to PCR as a gold standard in all 95 isolates (both Groups A and B) were 50.57% and 100%, respectively, as shown in .
Except for a single isolate (E. coli) that was not harboring any carbapenemase gene, the remaining 94 bacterial isolates were found to carry at least one carbapenemase gene.
The most common carbapenemase gene identified in the current study was KPC in 51/95 (53.7%) of isolates, followed by OXA-48 in 39/95 (41%) then VIM, IPM and NDM-1 in 28/95 (29.5%), 26/95 (27.3%), and 14/95 (14.7%) of isolates, respectively.
Out of 94 bacterial isolates harboring carbapenemase genes in the present study, only 39 isolates were harboring a single gene (41%); 21 were harboring blaKPC gene, 13 isolates were harboring blaIPM, two were harboring blaOXA-48, two were harboring blaVIM, and only one isolate was harboring blaNDM-1 gene. The remaining 55 isolates (57.9%) were harboring multiple (double or triple) carbapenemase genes.
Table 4. Distribution of expressed (Group A) and non-expressed (silent; Group B) carbapenemase genes isolated from different bacterial species
Table 5. Sensitivity and specificity, PPV & NPV of MHT in relation to PCR
4. Discussion
Infections attributed to GNB have been considerably escalating in the ICUs [Citation3]. In the present study, lower respiratory tract infections were the most frequently encountered HAIs (42.11%), while wound infections came next at 32.6%, then urinary tract infections (25.26%). That was partly in accordance with Pradhan et al. [Citation13] and Sileem et al. [Citation14], who reported that respiratory tract infections were the most frequent nosocomial infections (65.8% and 79.5%, respectively) and were followed by urinary tract infections (17.1% and 14.1%, respectively).
In the current study, the prevalence of VAE was 21% in all critically ill patients, and 48.78% in mechanically ventilated patients. The result of the current study was concordant with the earlier results by Mathai et al. [Citation15] who demonstrated a high VAP rate (37.5%) among mechanically ventilated ICU patients. Moreover, a study by the American Thoracic Society [Citation16] encountered VAP in 8–20% of all ICU patients and up to 27% of mechanically ventilated patients.
In the meantime, infections triggered by multi-drug resistant (MDR)-GNB are not only challenging to treat at the individual patient level, but they also contribute to augmented healthcare expenditure, hospital length of stay (LOS), high morbidity, and high mortality. The WHO grades carbapenem-resistant Enterobacteriaceae (CRE) amongst the highest priority pathogens [Citation17,Citation18]. In this study, Enterobacteriaceae were the most common isolates (55.8%), followed by Acinetobacter spp. (24%), and lastly Pseudomonas spp. (16%). On the other hand, a study by Katchanov et al. [Citation18] on 119 patients colonized or infected with MDR-GNB revealed that P. aeruginosa was identified in 55.5% of patients, Enterobacteriaceae in 37%, and A. baumannii in 15.1% [Citation18].
For the treatment of critically ill patients having life-threatening infections caused by MDR-GNB, the carbapenems, such as meropenem and imipenem, are currently considered the last resort [Citation19]. Resistance to carbapenems is chiefly due to the production of the hydrolyzing enzymes; carbapenemases. Moreover, other β-lactamase-like AmpCs (either chromosomal or acquired); and ESBLs joined with other mechanisms such as porin mutations, increased expression of efflux systems or penicillin-binding protein alterations can also result in carbapenem resistance [Citation20,Citation21]. Consequently, it is vital to comprehend carbapenem resistance mechanisms and their identifying methods. In this context, the application of a simple and precise laboratory method to detect carbapenemase production in GNB would be of a prominent benefit.
In the present study, we attempted to determine the presence and the prevalence of five carbapenemase genes (blaKPC, blaOXA-48, blaIPM, blaVIM, and blaNDM-1), among clinical isolates of Enterobacteriaceae, Acinetobacter spp., and Pseudomonas spp., preliminary by AST followed by the phenotypic confirmatory test (MHT), and to compare the result of the latter with genotypic detection of these genes via multiplex PCR as the gold standard method.
Out of the 95 studied isolates; 49 isolates were non-susceptible to carbapenems according to AST (Group A), and 46 were susceptible to carbapenems (Group B). MHT was positive in 44 out of 49 carbapenem non-susceptible (Group A) isolates (89.8%). This was in accordance with the result reported by Anwar et al. [Citation22] who revealed that 83.3% of the meropenem resistant strains screened by MHT tested positive.
Employing MHT, three isolates tested negative although they were carbapenem non-susceptible by AST. Meanwhile, none of the isolates in the carbapenem-susceptible group (Group B) yielded false-positive results via MHT. On the other hand, the two indeterminate results by MHT could correspond to isolates producing inhibitory substances, such as colicin, that inhibited the growth of E. coli ATCC 25,922 in this study [Citation23].
Although PCR is not suitable for daily testing principally due to its high cost, PCR analysis remains the gold standard method for the detection of carbapenemase producers [Citation24]. In the present study, all 49 suspected carbapenem-resistant isolates by AST, underwent genotypic confirmation by multiplex PCR, and all of them were found to harbor at least one carbapenemase gene. Of note, 44 out of these 49 isolates were MHT-positive; hence, the sensitivity of MHT compared to PCR as the gold standard method was 89.8%. In the meantime, all of the MHT-positive isolates were carbapenemase producers as confirmed by PCR. Nonetheless, these results differ from those by Takayama et al. [Citation25] who reported that only 58.3% of the Enterobacteriaceae isolates showing positive MHT (using meropenem disc) had carbapenemase genes detected by PCR.
It is worth addressing that numerous studies evaluated MHT performance in the detection of carbapenemase producers; however, to our knowledge, only a few of them studied the genotypic profile of negative-MHT isolates for non-expressed (silent) genes. In the present study, 45 out of 46 isolates in Group B which showed phenotypically susceptible carbapenem profile by AST, and 41 of which showed negative MHT were harborers of carbapenemase genes as confirmed by multiplex PCR. This, if considered, renders the overall sensitivity and specificity of MHT in relation to PCR in all 95 isolates (both Groups A and B) 50.57% and 100%, respectively. This study findings mirror those of Doyle et al. [Citation26] and Solanki et al. [Citation27]; both of which documented that the sensitivity and specificity for MHT were 58% and 93%, respectively. In contrast, according to one Egyptian study on 100 Klebsiella pneumoniae isolates, MHT had sensitivity and specificity of 100% and 47%, respectively [Citation28].
Notably, while MHT is useful for the detection of carbapenemases, it possesses low sensitivity and low specificity for metalloenzymes [Citation23,Citation27]. In addition, MHT may yield false-positive results chiefly due to CTX-M-producing strains with reduced outer membrane permeability and high-level AmpC producers [Citation29,Citation30]. Consequently, the diminished expression levels of carbapenemase-encoding genes are thought to contribute to silent dissemination within hospital settings because carbapenem MICs remain low and phenotypic tests may test as negative [Citation31].
On the other hand, the most common carbapenemase gene identified in the current study was blaKPC in 53.7% of the isolates, followed by blaOXA-48 in 41% of the isolates, then blaVIM, blaIPM, and blaNDM-1 in 29.5%, 27.3%, and 14.7% of the isolates, respectively. This was discordant with a previous study from Egypt, demonstrating that 28.57% of Klebsiella pneumoniae isolates produced carbapenemase class D, 25.2% produced class A enzyme, and 16.66% produced class B [Citation28]. However, AlTamimi et al. [Citation30] revealed a high prevalence of blaOXA-48 carbapenemase producers in 38.33% of isolates, followed by blaNDM in 8.3%, while blaVIM was detected in one isolate only of K. pneumoniae.
Of note, the combined carbapenemases in a single isolate render its overall hydrolytic spectrum wider and the available antibiotics even fewer. For example, isolates that once harbored an MBL and subsequently acquired a KPC, become fully resistant even to aztreonam, which could have remained active without the presence of KPC [Citation31].
Out of 94 bacterial isolates harboring carbapenemase genes in the present study, only 39 isolates were carrying a single gene (41%). The remaining 55 isolates (57.9%) were carrying multiple (double or triple) carbapenemase genes. In a study by Kazi et al. [Citation12] 18.5% of the studied isolates possessed dual carbapenemase genes. On the other hand, Baran and Aksu [Citation20] identified one carbapenemase-encoding gene in 90 out of 181 (49.7%) of the carbapenem non-susceptible isolates, and in only four (2.2%) isolates, multiple carbapenemase genes were identified.
The prompt identification of carbapenem-resistant pathogens is crucial not only for the commencement of accurate antimicrobial regimen but also for discontinuing their spread. Phenotypic methods are growth-dependent and time-consuming as they take 18–24 h for completion; hence, they are not clinically useful and results are also subjective [Citation30].
Therefore, identification by molecular methods such as real time-PCR has proven to be sensitive and more accurate. Multiplex PCR will also help in simultaneous detection of various genes, hence decreasing the required materials, manpower, and expenses. This assists in defining the epidemiology of these genes and is of significant infection control concern [Citation27].
The strength of this study is based on the fact that few studies had performed the genotypic analysis of phenotypically sensitive isolates, which revealed the harboring of silent carbapenemase genes among different species of Gram-negative bacteria. This study may raise questions about the molecular methods of carbapenemase gene expression in vivo, despite being phenotypically sensitive in vitro. In addition, highlighting the problem of the DAIs in healthcare settings in Egypt, especially VAE and CAUTIs, gives an alarming data for improving the infection control measures. On the other hand, this study which reveals the extent of carbapenem resistance spread may guide to implement an effective antibiotic stewardship in Cairo University Hospitals.
The limitation of this study could be the need for the whole genome sequencing to confirm the presence of carbapenemase genes. Moreover, the complementary step to detect mRNA expression is also important to reveal the extent of carbapenemase gene expression. Noteworthy, the use of MHT as a confirmatory test for carbapenem resistance may be replaced by more recent tests in the last few years.
5. Conclusion
Multiplex real-time PCR is a robust, reliable, and rapid method for detection of the most prevalent carbapenemase genes. In the meantime, our study results provide evidence of the diminished expression levels of carbapenemase-encoding genes which contribute to silent dissemination within hospital settings where carbapenem AST may well yield negative results.
Future studies should include a multiplex PCR assay confirmed by whole genome sequencing together with mRNA expression assay that is capable of differentiating between expressed and silent genes to provide timely and accurate detection of potential carbapenem resistance among susceptible isolates.
Acknowledgments
We acknowledge Prof. Dr. Abd El-Fattah Attia, the late Professor of Medical Microbiology and Immunology, Faculty of Medicine, Cairo University, for his sincere help, persistent effort, and generous advice throughout this work.
Disclosure statement
The authors declare there are no conflicts of interest.
Data availability
All datasets generated or analyzed during this study are included in the manuscript.
Additional information
Notes on contributors
Ahmed Salah Emira
Ahmed Salah Emira M.Sc. Medical Microbiology and Immunology, Faculty of Medicine, Cairo University.
Lamiaa Abd El-Fattah Madkour
Lamiaa Abd El-Fattah Madkour M.D. in Medical Microbiology and Immunology, Associate Professor at the Medical Microbiology and Immunology Department, Faculty of Medicine, Cairo University.
Nazmy Edward Seif
Nazmy Edward Seif M.D. in Anesthesia and Intensive Care Medicine, Anesthesia and Intensive Care Department, Faculty of Medicine, Cairo University.
Reham Ali Dwedar
Reham Ali Dwedar M.D. in Medical Microbiology and Immunology, Professor at the Medical Microbiology and Immunology Department, Faculty of Medicine, Cairo University.
References
- Kani RKS, Srinivasan B, Glesby MJ, et al. Current state of the art in rapid diagnostics for antimicrobial resistance. Lab Chip 2020; 20(15), 2607–2625.
- Zohar I, Schwartz O, Yossepowitch O, et al. Aminoglycoside versus carbapenem or piperacillin/tazobactam treatment for bloodstream infections of urinary source caused by Gram-negative ESBL-producing Enterobacteriaceae. J Antimicrob Chemother 2020; 75(2), 458–465.
- Morris S, Trends CE, Epidemiology, and management of multi-drug resistant gram-negative bacterial infections in the hospitalized setting. Antibiotics 2020; 9(4), 196.
- Braun SD, Monecke S, Thürmer A, et al. Rapid identification of carbapenemase genes in gram-negative bacteria with an oligonucleotide microarray-based assay. PLoS One 2014; 9(7): e102232.
- Jenkins S, Ledeboer NA, Westblade LF, et al. Evaluation of NG-test carba 5 for rapid phenotypic detection and differentiation of five common carbapenemase families: results of a multicenter clinical evaluation. J Clin Microbiol 2020; 58(7), e00344-20. doi:10.1128/JCM.00344-20.
- Centers for Disease Control and Prevention (CDC). Identifying healthcare-associated infections (HAI) for NHSN surveillance. Atlanta, 2018. Centers for Disease Control and Prevention.
- Centers for Disease Control and Prevention (CDC). Device-associated Module VAE, 2018.
- Collee JG, Marr W Specimen collection, culture containers and media. Mackie & McCartney practical medical microbiology. In: Collee JG, Fraser AG, Marmion BP, et al., editors. New York: Churchill Livingstone; 1996, p. 85–111.
- Clinical Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. 29th ed. Wayne (PA): CLSI Supplement M100; 2019.
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Version 7.1, 2017. [cited 2020 Oct 26]. http://www.eucast.org
- Clinical Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. 27th ed. Wayne (PA): CLSI Supplement M100; 2017.
- Kazi M, Drego L, Nikam C, et al. Molecular characterization of carbapenem-resistant Enterobacteriaceae at a tertiary care laboratory in Mumbai. Eur J Clin Microbiol Infect Dis 2015; 34(3), 467–472.
- Pradhan NP, Bhat SM, Ghadage DP Nosocomial infections in the medical ICU: a retrospective study highlighting their prevalence, microbiological profile and impact on ICU stay and mortality. J Assoc Physicians India 2014; 62(10), 18–21.
- Sileem AE, Said AM, Meleha MS Acinetobacter baumannii in ICU patients: A prospective study highlighting their incidence, antibiotic sensitivity pattern and impact on ICU stay and mortality. Egypt J Chest Dis Tuberc, 2017; 66(4), 693–698.
- Mathai AS, Phillips A, Isaac R Ventilator-associated pneumonia: A persistent healthcare problem in Indian intensive care units. Lung India 2016; 33(5), 512–516.
- American Thoracic Society. Infectious diseases society of America. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med 2005; 171: 388–416.
- Avery LM, Nicolau DP Investigational drugs for the treatment of infections caused by multidrug-resistant Gram-negative bacteria. Expert Opin Inv Drug 2018; 27(4): 325–338.
- Katchanov J, Asar L, Klupp EM, et al. Carbapenem-resistant Gram-negative pathogens in a German university medical center: prevalence, clinical implications and the role of novel β-lactam/β-lactamase inhibitor combinations. PLoS One 2018; 13(4), e0195757.
- Bassetti M, Peghin M, Vena A, et al. Treatment of infections due to MDR Gram-negative bacteria. Front Med (Lausanne). 2019; (4) 16; 6:74.
- Baran I, Aksu N Phenotypic and genotypic characteristics of carbapenem-resistant Enterobacteriaceae in a tertiary-level reference hospital in Turkey. Ann Clin Microb Anti 2016; 15(20): 1–11.
- Aguirre-Quiñonero A, Martínez-Martínez L Non-molecular detection of carbapenemases in Enterobacteriaceae clinical isolates. J Infect Chemother 2017; 23(1): 1–11.
- Anwar M, Ejaz H, Zafar A, et al. Phenotypic detection of metallo-beta-lactamases in carbapenem resistant acinetobacter baumannii isolated from pediatric patients in Pakistan. J Pathog 2016; 1–6. https://doi.org/10.1155/2016/8603964
- Girlich D, Poirel L, Nordmann P Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J Clin Microbiol 2011; 50(2): 477–479.
- Zhou M, Wang D, Kudinha T, et al. A comparative evaluation of four phenotypic methods for detection of class A and B carbapenemase-producing Enterobacteriaceae in China. J Clin Microbiol 2018; 56(8): JCM–00395.
- Takayama Y, Adachi Y, Nihonyanagi S, et al. Modified Hodge test using Mueller–Hinton agar supplemented with cloxacillin improves screening for carbapenemase-producing clinical isolates of Enterobacteriaceae. J Med Microbiol 2015; 64(7), 774–777.
- Doyle D, Peirano G, Lascols C, et al. Laboratory detection of enterobacteriaceae that produce carbapenemases. J Clin Microbiol 2012; 50(12): 3877–3880.
- Solanki R, Vanjari L, Sreevidya Subramanian AB, et al. Comparative evaluation of multiplex PCR and routine laboratory phenotypic methods for detection of carbapenemases among Gram negative bacilli. J Clin Diagn Res 2014; 8(12), DC23.
- Morsi SS Comparative evaluation of phenotypic and genotypic methods for detection of carbapenemases in clinically significant Klebsiella pneumoniae Isolates. Egypt J Med Microbiol 2016; 38(87), 1–8.
- Pasteran F, Gonzalez LJ, Albornoz E, et al. Triton Hodge test: improved protocol for modified hodge test for enhanced detection of NDM and other carbapenemase producers. J Clin Microbiol 2015; 54(3), 640–649.
- AlTamimi M, AlSalamah A, AlKhulaifi M, et al. Comparison of phenotypic and PCR methods for detection of carbapenemases production by Enterobacteriaceae. Saudi J Biol Sci 2017; 24(1): 155–161.
- Meletis G, Chatzidimitriou D, Malisiovas N Double- and multi-carbapenemase-producers: the excessively armored bacilli of the current decade. Eur J Clin Microbiol 2015; 34(8), 1487–1493
