ABSTRACT
The fast growing evidences have shown that the plant-derived compound honokiol is a promising candidate for treating multiple human diseases, such as inflammation and cancer. However, the mode-of-action (MoA) of honokiol remains largely unclear. Here, we studied the antifungal activity of honokiol in fission yeast model, with the goal of understanding the honokiol’s mechanism of action from the molecular level. We found that honokiol can inhibit the yeast growth at a dose-dependent way. Microarray analysis showed that honokiol has wide impacts on the fission yeast transcription levels (in total, 512 genes are up-regulated, and 42 genes are down-regulated). Gene set enrichment analysis indicated that over 45% up-regulated genes belong to the core environmental stress responses category. Moreover, network analysis suggested that there are extensive gene–gene interactions amongst the co-expression gene lists, which can assemble several biofunctionally important modules. It is noteworthy that several key components of central carbon metabolism, such as glucose transporters and metabolic enzymes of glycolysis, are involved in honokiol’s MoA. The complexity of the honokiol’s MoA displayed in previous studies and this work demonstrates that multiple omics approaches and bioinformatics tools should be applied together to achieve the complete scenario of honokiol’s antifungal function.
Introduction
Although the first characterisation of honokiol from Magnolia obovata was reported in 1972 (Maruyama and Kuribara Citation2006), this natural product actually has been widely used in traditional medicine in China, Japan and Korea for a long time (Maruyama and Kuribara Citation2006; Lee et al. Citation2011). Honokiol started to capture attention in recent 20 years mainly because of the finding of its promising therapeutic potential to treat multiple human diseases (especially for tumour and thrombus) (Fukuyama et al. Citation2002; Hu et al. Citation2005; Arora et al. Citation2012). Compared with the gradually accumulated knowledge from clinical applications, nevertheless, the understanding of the honokiol’s mode-of-action (MoA) at the molecular levels still remains largely unclear.
Earlier studies indicated that honokiol can target to multiple intracellular pathways depending on the specific disease model used (Fried and Arbiser Citation2009). For instance, honokiol displayed clear pro-apoptotic activity against sarcoma, melanoma, leukaemia, myeloma and colon cancer cell lines, etc. (Bai et al. Citation2003; Battle et al. Citation2005; Ishitsuka et al. Citation2005; Chang et al. Citation2013). There is report of honokiol-mediated inhibition of PI3K/mTOR pathway as a promising strategy to surmount immunoresistance in glioma, breast and prostate cancers (Crane et al. Citation2009). Meanwhile, honokiol has a significant impact on prostacyclin metabolism. Since prostacyclin is well known for its inhibition role of platelet aggregation, above observation may explain the antithrombotic activity of honokiol (Hu et al. Citation2005).
The above-mentioned discoveries suggest the complexity of honokiol’s MoA. Based on our previous study of natural product resveratrol (Wang et al. Citation2016), here, we took advantage of a simple unicellular model, Schizosaccharomyces pombe (S. pombe), and tried to use transcriptomics and bioinformatics analysis to reduce the complexity and understand honokiol’s MoA comprehensively. Fission yeast is an excellent model for studying fundamental cell behaviours, such as cell growth and proliferation. At the same time, fission yeast displays cancer cell-like metabolic behaviour, called Crabtree effect (similar to the Warburg effect of cancer cell) (Piškur et al. Citation2006; Vander Heiden et al. Citation2009), means it prefers to use fermentation rather than respiration for the energy production. Because of above merits, the mechanism discovery from fission yeast system could be immigrated to clinical medicine feasibly. In addition, benefiting from many scientists’ contributions, multiple valuable bioinformatics tools and databases of fission yeast model are available, which directly facilitates the exploration of honokiol’s MoA in a systematic view (Simon and Bedalov Citation2004).
In the present research, with the goal of understanding the common MoA of the anti-proliferative activity of honokiol, we thus investigated the honokiol’s impacts to fission yeast both on cellular level and whole-genome transcriptional level. The experimental procedures, results and discussions are reported as follows.
Materials and methods
Yeast cell culture and honokiol treatment
Fission yeast S. pombe strain 972 h- was used in this study. Honokiol was purchased from Institute of Chinese Materia Medica, Shanghai University of Traditional Chinese Medicine (Shanghai, China). The experimental procedure basically follows previous description (Wang et al. Citation2016). Briefly, for the drug activity experiment, a 10 ml culture of YE liquid medium (0.5% yeast extract, 3% glucose) was inoculated from single colony and was grown overnight at 30°C to the late log phase (OD600 = 2.0–3.0). The yeast culture was next diluted to OD600 = 0.05 and treated with a series of honokiol doses (0, 1, 2, 3, 4, 6 and 8 µg/ml) in 50 ml of fresh YE liquid culture. The optical density was measured at 600 nm at different time points (0, 4, 8, 12, 16, 20, 24, 28 and 32 h), and eventually the IC50 value was computed based on the readout at 20 h after drug treatment.
Cell phenotypic and FACS analysis
Cell staining, microscopic and fluorescence-activated cell sorting (FACS) analysis were basically performed as previous described (Wang et al. Citation2016). The septum staining by calcofluor was conducted based on the Dr. Paul Nurse’s Lab Fission Yeast Handbook (Cortés et al. Citation2012). In brief, the yeast cells from late log phase culture (OD600 = 2.0–3.0) was diluted to OD600 = 0.05, 3 μg/ml honokiol was next added to the culture, and finally 107 cells was collected at different time points by centrifugation at 2500 rpm for 5 min. The cell pellets were washed once with cold ddH2O and were re-suspended in 1 ml of cold 70% ethanol for fixation. For calcofluor staining, 30 µl of fixed cells were washed with 1 ml of water, and then mixed with 2× calcofluor stain (50 µg/ml calcofluor, 0.3 mg/ml p-phenylenediamine 50% glycerol). The samples were observed under fluorescence microscopy (DM2500, Leica). For FACS analysis, 0.3 ml of above fixed cells was washed with 50 mM sodium citrate twice. Then, RNA was firstly digested by 0.1 mg/ml RNase A at 37°C for 2 h. After that, we added propidium iodide to final concentration at 2 µg/ml. Just before processing the cells, an ultrasonic treatment of 45 s was applied to preventing the conglutination of the cells. Eventually the DNA contents were detected by FACScan (Becton Dickinson). The raw data were analysed and visualised by Flowjo software.
Microarray analysis
In general, the microarray analysis was performed as previous described (Wang et al. Citation2016). Honokiol (3 µg/ml) and vehicle control were added to YE cultured yeast at OD600 = 0.2. Next, the cells were incubated for 4 h, harvested by centrifugation, and washed once with 25 ml of cold ddH2O. The hot phenol method was used to extract the total RNA, and the RNA was purified using Qiagen RNeasy columns. The three replicated total RNA samples were sent to Shanghai Biochip Co. Ltd. for Affymetrix Yeast 2.0 microarray analysis. After passing the standard microarray quality check, the differential expressed genes were defined as (ratio >2.0, p-value <0.05).
Bioinformatics analysis
The gene expression data was first imported into Gene Cluster 3.0 to perform the hierarchical analysis using the centroid linkage clustering method (de Hoon et al. Citation2004), and the heat-map was visualised by Java Treeview 1.0. Gene Set Enrichment Analysis (GSEA) was conducted by the online tool AnGeLi (Bitton et al. Citation2015). The enriched gene ontology category was selected based on biological process and molecular function using list frequency >5% and p-value <0.01 as the selection criterion. The protein–protein interaction network was constructed by the online tool Pint (Pombe Interactome) based on the support vector machine and random forest algorithm (Pancaldi et al. Citation2012).
Results
Natural product honokiol inhibits fission yeast growth
Previous studies have shown that the natural product honokiol (3ʹ,5-di-2-progenyl-1, 1ʹ-biphenyl-2,4ʹ-diol) structurally belongs to the polyphenol family [)] (Maruyama and Kuribara Citation2006), and it was originally extracted from the genus Magnolia. Here, to explore the feasibility of using the fission yeast S. pombe as a model to study its antifungal activity, we arranged a series of honokiol concentrations (0–8 μg/ml) to treat wild-type fission yeast and quantitated the growth inhibition effect by monitoring the cell densities at OD600. The results showed that honokiol inhibits cell growth in a dose-dependent way [)], with an IC50 value at 3 μg/ml.
Figure 1. Honokiol can inhibit the cell growth of fission yeast. (a) The yeast growth inhibition curve under different doses of honokiol (0, 1, 2, 3, 4, 6 and 8 μg/ml). Cell growth rates were measured as described in “Materials and methods” section. (b) Phase contrast microscopic analysis of yeast cell shape: 3 μg/ml honokiol and mock reagent (ethanol) were used to treat the yeast cells for 4 h. The white bar represents the length of 10 μm. The white arrow shows the cell with abnormal phenotype. (c) Calcofluor staining to visualise the cell septum: 3 μg/ml honokiol and mock reagent (ethanol) were used to treat the yeast cells for 4 h. The white bar represents the length of 10 μm. The white arrow shows the cell with abnormal phenotype. (d) FACS analysis to check honokiol’s effect on cell cycle: 3 μg/ml honokiol was used to treat the cells for 0, 2 and 4 h.
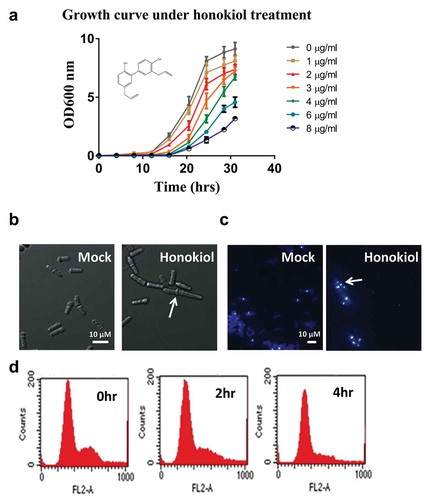
Honokiol inhibits a small group of cell’s separation mildly
When using the IC50 concentration (3 μg/ml) of honokiol to challenge the yeast for 4 h, microscopic analysis showed that there is no obvious phenotypic change amongst most of the cells, although we observed elongated cell occasionally [less than 5% in the whole cell population, shown in )]. We also utilised the chemical calcofluor white to probe the cell septum, as shown in ), the septum of several cells became noticeably thicker in contrast to the mock-treated samples, and we rarely found some cells hold more than one septum. This suggests that this group of cell’s separation is delayed. However the delaying effect is very mild (or only take effect on a small portion of the whole cell population), because there is no significant cell cycling difference between the mock and drug treated groups, based on the FACS results [)].
Honokiol-induced transcriptomic change is closely related to core environmental stress response genes
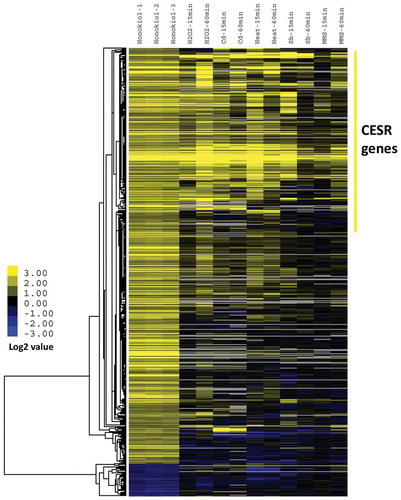
For the purpose of understanding the molecular mechanism of honokiol, we utilised microarray analysis to identify the gene transcriptional consequences after honokiol treatment. After adding 3 μg/ml honokiol to fission yeast culture at the early time point (4 h), we characterised total 554 genes (512 genes with increased expression levels and 42 genes with decreased expression levels) whose expression levels are significantly affected by honokiol treatment (fold >2, p-value < 0.05, shown in Supplementary Table 1). We next performed the GSEA via the web-tool AnGeLi for comprehensively analysing the honokiol-modulated gene lists (Bitton et al. Citation2015). As shown in , honokiol treatment can significantly trigger the stress-related genes’ expression. For example, over 45% up-regulated genes (232 genes in total, p-value = 3.09 E-133) are recognised as the “Core Environmental Stress Response (CESR) Induced” category, which is largely overlapped with the secondary highest hit “Oxidative Stress Cluster4” (182 genes in total, p-value = 2.18E-102). It is worth noting that previous studies have proven that the b-ZIP transcription factor Atf1 is the key component of stress-dependent gene transcription (Zhou et al. Citation2012). Consistently, we found that “Atf1 activated” categories are over-represented in the statistical test (52 genes in total, p-value = 1.20E-43). We, therefore, did the combinational and comparative analysis between our results and the previous published microarray data of fission yeast responding to various environmental stresses (oxidative stress, heavy metal stress, osmotic stress, heat shock and DNA damage) (Dongrong Chen et al., Citation2003). As shown in , the result of hierarchical analysis indicates that honokiol-caused transcriptomic modulation is significantly consistent with stress-caused changes, especially for H2O2, Cd and heat-induced stresses. Compared to over 500 honokiol up-regulated genes, there are only 42 are down-regulated by the drug, and which cannot be clustered into any meaningful category. However, it is interesting to see that pyruvate decarboxylase (SPAC13A11.06) has the obviously decreased expression level, which implies a potential modulation of metabolic fluxes.
Table 1. The gene set enrichment analysis of the honokiol up-regulated genes.
Figure 2. Honokiol-induced transcriptomic change is closely related to core environmental stress response. IC50 value honokiol (3 µg/ml) and vehicle control were added to YE medium cultured yeast at OD600 = 0.2. Next, the cells were incubated for 4 h. Afterwards, the cells were collected and mRNA was extracted for the microarray analysis. The detailed data analysis is described in “Materials and methods” section. Basically, the data used in clustering analysis were selected by both the fold change (>2 folds) and p-value (<0.05). Honokiol-1, -2, -3 represent three biological replicates. Other stress treatment conditions are reported before (Dongrong et al. Citation2003). Briefly, these stress conditions include: oxidative stress (0.5 mM H2O2, treat 15 and 60 min), heavy metal stress (0.5 mM CdSO4, treat 15 and 60 min), heat stress (39°C, treat 15 and 60 min), osmotic stress (1 M sorbitol, treat 15 and 60 min) and alkylating agent (0.2%, w/v methylmethane sulfonate, treat 15 and 60 min).
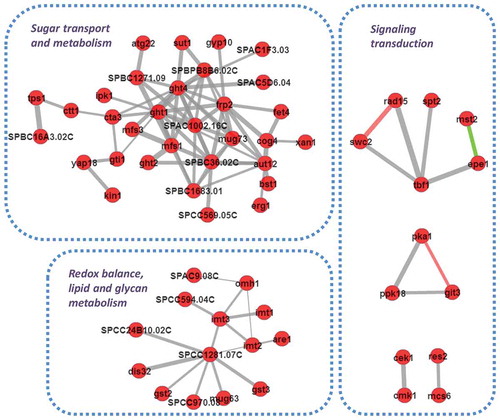
Network analysis of honokiol up-regulated gene sets indicates extensive protein–protein interactions are existed and form biofunctionally important modules
To keep dissecting the honokiol-caused transcriptomic modulation, we next constructed the protein interaction networks using the honokiol up-regulated gene sets. As shown in , there are highly frequent protein–protein interaction events in the prediction. Interestingly, several biofunctionally important modules are emerged based on those interaction networks. The first noteworthy module is related to sugar transport and metabolism, because this module includes at least three hexose transmembrane transporters (Ght1, Ght2 and Ght4) (Heiland et al. Citation2000), gluconate transmembrane transporter inducer (Gti1) (Caspari Citation1997) and glucoside transmembrane transporter (Sut1) (Reinders and Ward Citation2001), which suggests the glycolysis activation is a part of honokiol’s MoA. Another interesting module is related to redox balance, lipid and glycan metabolism. It includes three glutathione S-transferase genes (Gst2, Gst3 and SPCC1281.07C) (Kim et al. Citation2004), one NAD+/NADH kinase (SPCC24B10.02C), four mannosyltransferase (Imt1, Imt2, Imt3 and Omh1) (Nakase et al. Citation2010), and one acyl-CoA-sterol acyltransferase (Are1). We also observed several modules which are related to signalling transduction. For examples, there is a predicted transcriptional regulation complex including DNA binding factor (Tbf1 and Spt2) (Cockell et al. Citation2009), transcription factor TFIIH complex subunit (Rad15) (Murray et al. Citation1992), histone acetyltransferase (Mst2) (Gómez et al. Citation2005) and Jmjc domain chromatin-associated protein (Epe1) (Trewick et al. Citation2007). There is also a potential signalling module associated with protein kinase and secondary messenger cAMP, which consists of cAMP-dependent protein kinase catalytic subunit (Pka1) (Matsuo et al. Citation2008), serine/threonine protein kinase Ppk18 (Beltrao et al. Citation2009) and a G-protein coupled receptor, Git3, which can activate the adenylate cyclase activity (Wang et al. Citation2005).
Figure 3. Network analysis of honokiol up-regulated gene sets indicates extensive protein–protein interactions are existed and form biofunctionally important modules. The predicted interactions are retrieved with scores >0.5 based on the support vector machine and random forest algorithm. The red circles represent the candidate proteins, the grey lines represent the predicted interaction, the green lines represent the interactions has experimental evidences. The line’s thickness represents the relative possibilities of the interaction event.
The honokiol’s transcriptional regulation on central carbon metabolism
We next analysed the honokiol’s impact on central carbon metabolism (CCM) to answer if its antifungal activity is related to dysregulation of crabtree effect or not. The carbtree effect-related metabolic enzyme’s expression levels were investigated. As shown in , Fbp1 (Fructose-1,6-bisphosphatase) is strongly up-regulated by honokiol (increased to 28 folds). In glycolysis, Fbp1 is responsible for converting fructose-6-phosphate to fructose-1,6-bisphosphate, for which the reactions need ATP donating one phosphate group. However, we also observed that pyruvate kinase (Pyk1), pyruvate dehydrogenase E1 component alpha subunit (Pda1), pyruvate dehydrogenase E1 component beta subunit (Pdb1), and especially pyruvate decarboxylase (Pdc) are down-regulated obviously, suggesting that the synthesis of acetyl-CoA is impaired although its upstream metabolites, such as fructose-1,6-bisphosphate and may be phosphoenolpyruvate, are accumulated, which becomes a waste for both carbon scaffolds and ATP molecule.
Figure 4. The honokiol’s transcriptional regulation on fermentation module. The metabolic structure was drawn based on the KEGG (http://www.genome.jp/kegg/). For the metabolic enzymes (brown fonts, italics), their relative expression level after honokiol treatment are shown as numbers (red fonts represent up-regulated genes, which changes >20%; green fonts represent down-regulated genes, which changes >20%) in parallel. The abbreviations of these metabolic enzymes represent: PGI1 (glucose-6-phosphate isomerase), FBA1 (fructose-bisphosphate aldolase), FBP1 (fructose-1,6-bisphosphatase), TDH1 (glyceraldehyde-3-phosphate dehydrogenase), PGK1 (phosphoglycerate kinase), ENO1 (enolase), PYK1 (pyruvate kinase), PDA1 (pyruvate dehydrogenase E1 component alpha subunit), PDB1 (pyruvate dehydrogenase E1 component beta subunit), PDC (pyruvate decarboxylase), CIT1 (citrate synthase), ACO1 (aconitate hydratase), IDH1 (isocitrate dehydrogenase), IDH2 (isocitrate dehydrogenase), KGD1 (2-oxoglutarate dehydrogenase), SDH1 (succinate dehydrogenase), SDH2 (succinate dehydrogenase), FUM1 (fumarate hydratase), MDH1 (malate dehydrogenase). For the metabolites (red fonts), these abbreviations represent: glucose-6-P (glucose 6-phosphate), fructose-6-P (fructose 6-phosphate), FBP (fructose 1,6-bisphosphate), G3P (glyceraldehyde 3-phosphate), 1,3 BPG (glycerate 1,3-diphosphate), PG (phosphoglycerate), PEP (phosphoenolpyruvate), PYR (pyruvate), Acetyl-CoA (acetyl coenzyme A), CIT (citrate), ISOCIT (iso-citrate), α-KG (alpha-ketoglutarate), SUC (succinate), FUM (fumarate), MAL (malate), OAA (oxaloacetate).
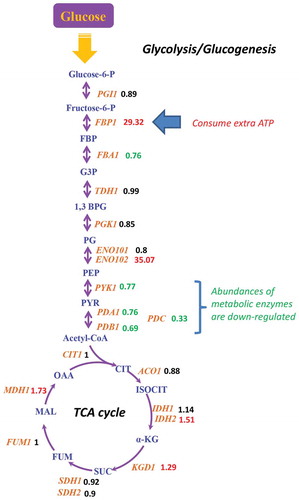
Although the production of acetyl-CoA (the precursor of TCA cycle) is slowed down, we did not find significantly changed gene expression levels in the metabolic enzymes of TCA cycle () and the components of mitochondrial respiration chain (Supplementary Figure 1). Above results imply that the transcriptional modulation in fermentation, rather than respiration, is part of honokiol’s MoA.
Summary of honokiol’s MoA
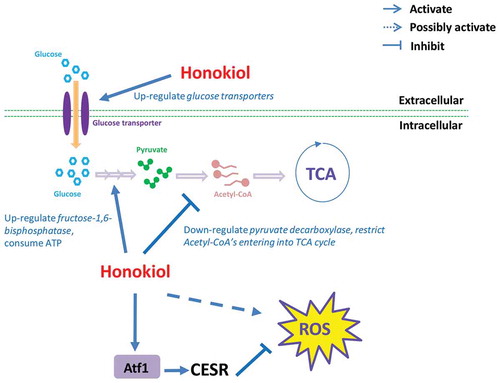
Based on above results, we have summarised the honokiol’s antifungal MoA. As shown in , upon honokiol penetrating into yeast cell, it rapidly up-regulate glucose uptake by enhancing hexose transmembrane transporters’ expression. Honokiol will keep improving the converting from fructose-6-phosphate to fructose-1,6-bisphosphate by up-regulating fructose-1,6-bisphosphatase’s expression, which becomes a huge energy consuming/wasting step because meanwhile the metabolic flux of downstream glycolysis is shrinked due to the down-regulated expression of pyruvate decarboxylase. At the same time, honokiol extensively triggers the expression of CESR/oxidative stress-related genes. Many of those genes are downstream target of transcription faction Atf1, which is well-known regulator of self-adaptation for resisting the accumulated ROS. However, long-termed or high-dosed honokiol treatment may disturb the redox homeostasis eventually, which will contribute to honokiol’s cidal effect, similarly as the mechanism of many known antifungals and antibiotics.
Figure 5. The mode-of-action of honokiol caused antifungal activity in fission yeast. As shown in this figure, honokiol can up-regulate a series of glucose transmembrane transporters, thus increase the glucose uptake rate. Honokiol can greatly up-regulated fructose 1,6-bisphosphate, which consumes much more ATP to produce fructose 1,6-bisphosphate. However, honokiol also significantly down-regulates pyruvate decarboxylase, which restricts the flux entering into TCA cycle, and removes waste of the previous ATP molecules as well. At the same time, honokiol can widely trigger the overexpression of many CESR genes. Although above event may represent cell’s self-adaptation to the ROS stress (many are mediated by transcription factor Atf1), but the long-termed dysregulation of CESR may also disrupt the redox homeostasis, which is harmful to the viability of cell.
Discussion
Despite the widely recognised pharmacological potential of the plant-derived compound honokiol, its underlying MoA still remains largely incomplete. Our studies elucidated that widespread transcriptional reprogramming and subsequent modulation of CCM are the major MoA of honokiol in response to its antifungal function in fission yeast.
Honokiol is normally extracted from Magnolia, and it chemically belongs to polyphenol, which has been well known as the capability against oxidative stress via its metal-chelating properties. However, sometimes things go to the opposite side, means polyphenol can also increase the generation of reactive oxygen species, such as the recent findings that red wine polyphenols and resveratrol can increase the ROS in cancer cells (Heiss et al. Citation2007; Sharif et al. Citation2010). Here, we proposed a similar model that honokiol may inhibit the cell growth also partly by stimulating the production of ROS ( and and ). The strong evidences come from the microarray analysis-both GSEA and hierarchical analysis indicates that the application of honokiol can trigger a similar transcriptional signature such as the ones from environmental stress stimuli, especially for H2O2 treatment. It is worth to mention that the drug dose used for microarray analysis is sublethal (IC50 concentration), since we tried to avoid to see the dead effect. Correspondingly, there are some of the transcriptional modulations of CESR or oxidative stress responsive genes that belong to cell’s self-adaptation or detoxification program. They are mainly controlled by the well-known stress regulator Atf1 (its transcript increases three folds by honokiol treatment), because there is a large proportion overlapping between Atf1 targets and honokiol regulated genes. We, therefore, proposed that b-ZIP transcription factor Atf1 plays a key role in honokiol’s MoA as well.
Although there is significant growth inhibition effect, microscopic analysis indicated that most of cells still hold the normal phenotype. Only a small proportion of cells (less than 5% in the whole population) showed elongated cell shape and delayed sister cell separation. The GSEA suggested that some meiotic genes are dysregulated by honokiol, but it is hard to make conclusion, since over 95% cells without cell cycle defects. Although the mechanism of those “outlier cells” is not fully understood, it may be related to epigenetics modulation. However, this possibility still remains for further investigation, such as using single cell transcriptomic approach.
Recent study shows that honokiol is a promising lead to bind the peroxisome proliferator-activated receptor gamma (Atanasov et al. Citation2013) and stimulate basal glucose uptake in differentiated adipocytes. Interestingly, at least three glucose transmembrane transporters (Ght1, Ght2 and Ght4) are up-regulated by honokiol treatment in fission yeast (), which implies that the improvement of sugar uptake and potential faster glycolysis rate is a common honokiol’s MoA within both yeast and mammalian cells. We kept questioning if honokiol’s antifungal activity comes from sugar-induced cell death (SICD) or not. The transcriptional analysis of crabtree effect-related genes indicated that there is no significant regulation on mitochondrial respiration chain (Supplementary Figure 1), but in glycolysis module, fructose-1,6-bisphosphatase is strongly up-regulated while pyruvate decarboxylase is obviously down-regulated. The above-mentioned modulation caused greatly increased ATP consuming, but it eventually becomes energy and carbon scaffolds wasteful since the flux of downstream glycolysis is shrinked. These results suggested that SICD may not be the MoA of honokiol, because it normally characterised with dysregulation of mitochondrial respiration.
Conclusion
In summary, we elucidated that the natural product honokiol can inhibit the cell growth of fission yeast, primarily through the dysregulation of glycolysis and may be also caused by disrupting the redox homeostasis. At the same time, sublethal dose of honokiol can trigger the extensive regulation of stress responsive genes, which is controlled by the well-known transcription factor Atf1. The complexity of the honokiol’s MoA demonstrates that it is essential to apply multiple omics approaches to obtain a complete picture of its antifungal activity.
Supplementary_material.docx
Download MS Word (690.3 KB)Acknowledgments
This work is supported by National Natural Science Foundation of China (No.30900018), National Science Foundation for Postdoctoral Scientists of China (No.20090450651), Shanghai Science Foundation for Postdoctoral Scientists (No.08R214105) to Zhe Wang.
Disclosure statement
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
Supplemental data
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Arora S, Singh S, Piazza GA, Contreras CM, Panyam J, Singh AP. 2012. Honokiol: a novel natural agent for cancer prevention and therapy. Curr Mol Med. 12:1244–1252.
- Atanasov AG, Wang JN, Gu SP, Bu J, Kramer MP, Baumgartner L, Fakhrudin N, Ladurner A, Malainer C, Vuorinen A, et al. 2013. Honokiol: a non-adipogenic PPARγ agonist from nature. Biochim Biophys Acta. 1830:4813–4819.
- Bai X, Cerimele F, Ushio-Fukai M, Waqas M, Campbell PM, Govindarajan B, Der CJ, Battle T, Frank DA, Ye K, et al. 2003. Honokiol, a small molecular weight natural product, inhibits angiogenesis in vitro and tumor growth in vivo. J Biol Chem. 278:35501–35507.
- Battle TE, Arbiser J, Frank DA. 2005. The natural product honokiol induces caspase-dependent apoptosis in B-cell chronic lymphocytic leukemia (B-CLL) cells. Blood. 106:690–697.
- Beltrao P, Trinidad JC, Fiedler D, Roguev A, Lim WA, Shokat KM, Burlingame AL, Krogan NJ. 2009. Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 7:1–12.
- Bitton DA, Schubert F, Dey S, Okoniewski M, Smith GC, Khadayate S, Pancaldi V, Wood V, Bähler J. 2015. AnGeLi: a tool for the analysis of gene lists from fission yeast. Front Genet. 6:1–9.
- Caspari T. 1997. Onset of gluconate-H+ symport in Schizosaccharomyces pombe is regulated by the kinases Wis1 and Pka1, and requires the gti1+ gene product. J Cell Sci. 110:2599–2608.
- Chang KH, Yan MD, Yao CJ, Lin PC, Lai GM. 2013. Honokiol-induced apoptosis and autophagy in glioblastoma multiforme cells. Oncol Lett. 6:1435–1438.
- Cockell MM, Lo Presti L, Cerutti L, Del Rosario EC, Hauser PM, Simanis V. 2009. Functional differentiation of tbf1 orthologues in fission and budding yeasts. Eukaryot Cell. 8:207–216.
- Cortés JC, Sato M, Muñoz J, Belén Moreno M, Clemente-Ramos JA, Ramos M, Okada H, Osumi M, Durán A, Ribas JC. 2012. Fission yeast Ags1 confers the essential septum strength needed for safe gradual cell abscission. J Cell Biol. 198:637–656.
- Crane C, Panner A, Pieper RO, Arbiser J, Parsa AT. 2009. Honokiol-mediated inhibition of PI3K/mTOR pathway: a potential strategy to overcome immunoresistance in glioma, breast, and prostate carcinoma without impacting T cell function. J Immunother. 32:585–592.
- De Hoon MJ, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics. 20:1453–1454.
- Dongrong C, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma A, Jones N, Bähler J. 2003. Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell. 14:2559–2569.
- Fried LE, Arbiser JL. 2009. Honokiol, a multifunctional antiangiogenic and antitumor agent. Antioxid Redox Signal. 11:1139–1148.
- Fukuyama Y, Nakade K, Minoshima Y, Yokoyama R, Zhai H, Mitsumoto Y. 2002. Neurotrophic activity of honokiol on the cultures of fetal rat cortical neurons. Bioorganic Med Chem Lett. 12:1163–1166.
- Gómez EB, Espinosa JM, Forsburg SL. 2005. Schizosaccharomyces pombe mst2+ encodes a MYST family histone acetyltransferase that negatively regulates telomere silencing. Mol Cell Biol. 25:8887–8903.
- Heiland S, Radovanovic N, Hofer M, Winderickx J, Lichtenberg H. 2000. Multiple hexose transporter of Schizosaccharomyces pombe. J Bacteriol. 182:2153–2162.
- Heiss EH, Schilder YD, Dirsch VM. 2007. Chronic treatment with resveratrol induces redox stress- and ataxia telangiectasia-mutated (ATM)-dependent senescence in p53-positive cancer cells. J Biol Chem. 282:26759–26766.
- Hu H, Zhang XX, Wang YY, Chen SZ. 2005. Honokiol inhibits arterial thrombosis through endothelial cell protection and stimulation of prostacyclin. Acta Pharmacol Sin. 26:1063–1068.
- Ishitsuka K, Hideshima T, Hamasaki M, Raje N, Kumar S, Hideshima H, Shiraishi N, Yasui H, Roccaro AM, Richardson P, et al. 2005. Honokiol overcomes conventional drug resistance in human multiple myeloma by induction of caspase-dependent and -independent apoptosis. Blood. 106:1794–1800.
- Kim HG, Kim BC, Park EH, Ahn K, Lim C. 2004. Differential regulation of three genes encoding glutathione S-transferases in Schizosaccharomyces pombe. Mol Cells. 18:332–339.
- Lee YJ, Lee YM, Lee CK, Jung JK, Han SB, Hong JT. 2011. Therapeutic applications of compounds in the Magnolia family. Pharmacol Ther. 130:157–176.
- Maruyama Y, Kuribara H. 2006. Overview of the Pharmacological Features of Honokiol. CNS Drug Rev. 6:35–44.
- Matsuo Y, McInnis B, Marcus S. 2008. Regulation of the subcellular localization of cyclic AMP-dependent protein kinase in response to physiological stresses and sexual differentiation in the fission yeast Schizosaccharomyces pombe. Eukaryot Cell. 7:1450–1459.
- Murray JM, Doe CL, Schenk P, Carr AM, Lehmann AR, Watts FZ. 1992. Cloning and characterisation of the S. pombe rad15 gene, a homologue to the S. cerevisiae RAD3 and human ERCC2 genes. Nucleic Acids Res. 20:2673–2678.
- Nakase M, Tani M, Morita T, Kitamoto HK, Kashiwazaki J, Nakamura T, Hosomi A, Tanaka N, Takegawa K. 2010. Mannosylinositol phosphorylceramide is a major sphingolipid component and is required for proper localization of plasma-membrane proteins in Schizosaccharomyces pombe. J Cell Sci. 123:1578–1587.
- Pancaldi V, Saraç OS, Rallis C, McLean JR, Převorovský M, Gould K, Beyer A, Bähler J. 2012. Predicting the fission yeast protein interaction network. G3 (Bethesda). 2:453–467.
- Piškur J, Rozpedowska E, Polakova S, Merico A, Compagno C. 2006. How did Saccharomyces evolve to become a good brewer? Trends Genet. 22:183–186.
- Reinders A, Ward JM. 2001. Functional characterization of the alpha-glucoside transporter Sut1p from Schizosaccharomyces pombe, the first fungal homologue of plant sucrose transporters. Mol Microbiol. 39:445–454.
- Sharif T, Auger C, Alhosin M, Ebel C, Achour M, Étienne-Selloum N, Fuhrmann G, Bronner C, Schini-Kerth VB. 2010. Red wine polyphenols cause growth inhibition and apoptosis in acute lymphoblastic leukaemia cells by inducing a redox-sensitive up-regulation of p73 and down-regulation of UHRF1. Eur J Cancer. 46:983–994.
- Simon JA, Bedalov A. 2004. Opinion: yeast as a model system for anticancer drug discovery. Nat Rev Cancer. 4:1–8.
- Trewick SC, Minc E, Antonelli R, Urano T, Allshire RC. 2007. The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. Embo J. 26:4670–4682.
- Vander Heiden MG, Cantley LC, Thompson CB. 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 324:1029–1033.
- Wang L, Griffiths K, Zhang TH, Ivey FD, Hoffman CS. 2005. Schizosaccharomyces pombe adenylate cyclase suppressor mutations suggest a role for cAMP phosphodiesterase regulation in feedback control of glucose/cAMP signaling. Genetics. 171:1523–1533.
- Wang Z, Gu Z, Shen Y, Wang Y, Li J, Lv H, Huo K. 2016. The natural product resveratrol inhibits yeast cell separation by extensively modulating the transcriptional landscape and reprogramming the intracellular metabolome. PLoS One. 11:1–20.
- Zhou X, Yan M, Kato T, Kuno T. 2012. A measurable activation of the bZIP transcription factor Atf1 in a fission yeast strain devoid of stress-activated and cell integrity mitogen-activated protein kinase (MAPK) activities. J Biol Chem. 287:23434–23439.
