?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Aspergillus
is a genus of filamentous and cosmopolitan fungi that includes important species for medical mycology, food, basic research and agro-industry areas. Aspergillus section Nigri are efficient producers of hydrolytic enzymes such as cellulases that are employed in the cellulose conversion. Hence, the search of new cellulolytic isolates and their correct identification is important for carrying out safe biotechnological processes. This study aimed to characterise the cellulolytic potential of Aspergillus sp. LBM 134, isolated from the Paranaense rainforest (Argentina) and to identify the isolate through a polyphasic approach. The fungus was identified as Aspergillus niger and its cellulolytic potential was evaluated by using Congo red technique and fluorescence plate assays for carboxymethyl cellulase, β-glucosidase and cellobiohydrolase, respectively. All three cellulase activities were positive; this bio-prospective positioned A. niger LBM 134 as a promising alternative for industries that require organisms capable of carrying out cellulosic biomass processing.
Introduction
Aspergillus is a diverse group of filamentous and cosmopolitan fungi that grow in almost every natural and artificial substrate. These fungi have a significant impact in modern society and are very studied due to their importance in medical and industrial mycology (Samson et al. Citation2014; Park et al. Citation2017). Raper and Fennell (Citation1965) recognised 150 species and currently, more than 340 species of Aspergillus are recognised in 4 subgenera and 19 sections (Houbraken et al. Citation2014; Samson et al. Citation2014; Park et al. Citation2017).
Black aspergillus is one of the most studied groups since they have huge industrial potential and biotechnological relevance (de Vries et al. Citation2017). They belong to subgenus Circumdati which comprises 26 species distributed into the clades A. niger, A. carbonarius, A. heteromorphus, A. homomorphus and A. acualeatus (Varga et al. Citation2011).
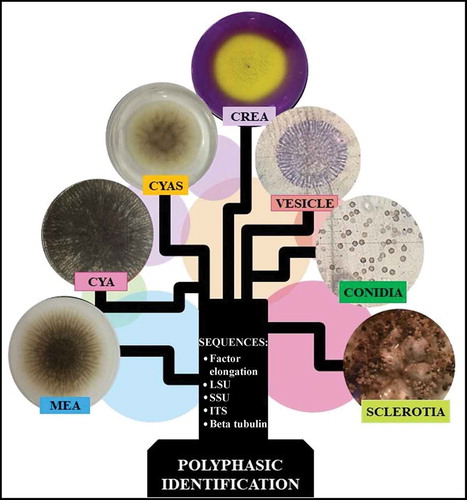
Previously, fungal identification was based almost entirely on macro- and micro-morphological criteria (Raper and Fennell Citation1965; Klich and Pitt Citation1988). However, identification of taxa in the genus Aspergillus is difficult (Abarca Citation2000; Samson et al. Citation2007a) and has now been complemented with molecular identification that compares specific genes or partial sequences named molecular markers (Bennett Citation2010). To identify Aspergillus species a multiple locus identification has been proposed that employs the use of ITS and also beta-tubulin (Bt), calmoduline (CMD) and transcriptional elongation factor 1α (Tef) molecular markers (Samson et al. Citation2014; Palumbo and O’keeffe Citation2015). Unfortunately, this multiple locus identification is often insufficient to identify species belonging to section Nigri. Therefore, a polyphasic approach has been recommended to delimit, identify and describe species of Aspergillus (Frisvad and Samson Citation2004; Samson et al. Citation2007b; Rodrigues et al. Citation2009; Simões et al. Citation2013; Silva et al. Citation2015; Decontardi et al. Citation2018). The polyphasic identification applies different types of data (DNA sequences, morphological, physiological and ecological data and extrolite analysis) in order to obtain a consensus result of higher fidelity and robustness. The polyphasic focus also avoids errors that arise when applying only morphological or/and molecular methods (Samson et al. Citation2004; Silva et al. Citation2011). Reference protocols of polyphasic identification have been proposed for different fungal groups adapting to more significant characters and several prestige laboratories offer this type of identification. The Westerdikj Fungal Biodiversity Institute (formerly CBS) has an interesting tool that is available free online, Polyphasic Identification for the identification of Aspergillus, Penicillium and yeast species comparing user data with its database, CBS-KNAW, to identify a fungal isolate through a polyphasic approach.
Species within Aspergillus section Nigri have been used in different industrial bioprocess with enzymatic production being one of the most popular applications of these fungi (Bennett Citation2010; Park et al. Citation2017). Cellulose degrading enzymes that attack cellulose have been intensely studied due to their wide industrial application (Martínez et al. Citation2005; Kuhad et al. Citation2011; Sohail et al. Citation2016; Park et al. Citation2017). The main cellulases are endo-β-1,4-glucanases (EG, EC 3.2.1.4), cellobiohydrolases (CBH, EC 3.2.1.91) and β-glucosidases (BGL, EC 3.2.1.21) (Singhania et al. Citation2017). EGs act on β-1,4-glucosidic bonds of cellulose to generate reducing and non-reducing ends that are attached by CBHI and CBHII, respectively, releasing cellobioses, which are then hydrolysed by BGLs (Singhania et al. Citation2017).
Cellulosic biomass processing is an attractive source of research that involves cellulases and cellulolytic organisms. Knowing more about new fungal isolates through a biotechnological focus would make available efficient enzymes for industry applications (Kuhad et al. Citation2011; Bertrand et al. Citation2017).
The present work addressed the identification of an Aspergillus isolate from Paranaense rainforest through a polyphasic approach, and investigated its biotechnological prospects by determining its cellulolytic potential.
Materials and methods
Isolate and morphological analysis
The isolate Aspergillus sp. LBM 134 belonging to the section Nigri was used in this study. It was selected from a previous study of screening and obtained from a sample of rotten wood in a natural zone of Misiones rainforest (Latitude 27°09ʹ05” S/Longitude 54°86ʹ77” W) during winter 2015 (Díaz et al. Citation2019a). The fungus was isolated by scrapping the material surface and inoculated on 39 g*L−1 potato dextrose agar (PDA) with 0.5% (p/v) chloramphenicol to inhibit bacterial growth (Benbow and Sugar Citation1999) and identified as Aspergillus sp. based on its macroscopic and microscopic characteristics using the keys of Carrillo (Citation2003). The fungus was deposited in the Culture Collection of the Laboratory of Molecular Biotechnology (LBM) of the Institute of Biotechnology Misiones and was maintained on PDA medium at 4°C and incubated on the same medium at 28°C for its mycelial growth.
DNA isolation, amplification and analyses
Total genomic DNA was extracted from axenic isolates grown for 3 d on 15 g*L−1 yeast extract 30 g*L−1 sucrose (YES) medium, following the protocols of Fonseca et al. (Citation2015). Partial gene sequences were determined for the ITS1-5,8S gene-ITS2 region, using primers ITS1 and ITS4 (White et al. Citation1990), the large subunit 28S of rDNA gene (D1/D2) using primers NL1 and NL4 (Kurtzman and Robnett Citation1997), β-tubulin gene (Bt) using primers Bt2a and Bt2b (Glass and Donaldson Citation1995), calmodulin gene (CMD) using primers CMD5 and CMD6 (Hong et al. Citation2005) and the translation elongation factor 1-alpha gene (Tef) using primers EF1S and Tef1R (Yergeau et al. Citation2005; Samuels and Ismaiel Citation2009). Amplicons were sequenced in both directions and consensus sequences were determined using the software Chromas v2.6.5 and BioEdit Sequence Alignment Editor v7.0.5. Subsequent alignments were carried out using ITS, Bt and CMD sequences belonging to type strains (less Aspergillus sp. LBM 134) of culture collection CBS-KNAW referenced by Varga et al. (Citation2011). Alignments were generated for each individual locus and then assembled using MEGA v.7.0 and manually corrected if necessary. Concatenated sequences were included in the phylogenetic inference based on maximum likelihood (ML) and neighbour joining (NJ) applying substitution Kimura’s model and a bootstrap with 1 000 replicates. A. flavus CBS 100927 was used as external group. The phylogenetic inference was carried out with software MEGA version 7.0.
Polyphasic identification
Polyphasic identification was carried out through the online tool Polyphasic Identification of the website http://www.westerdijkinstitute.nl/Aspergillus/DefaultInfo.aspx?Page=Home of the Westerdijk Fungal Biodiversity Institute of Royal Netherlands Academy of Arts and Sciences. Polyphasic identification required the following information of Aspergillus sp. LBM 134: macro-morphology, micro-morphology, acid production tests and DNA sequences of ITS, D1/D2, Bt, CMD and Tef.
Macro-morphology
Aspergillus sp. LBM 134 was grown on different media as recommended by the International Commission for Penicillium and Aspergillus for polyphasic identification. Aspergillus sp. LBM 134 growth was measured and macro-morphological characteristics were observed. The culture media were Czapek yeast extract agar (CYA; 3 g*L−1 NaNO3, 5 g*L−1 yeast extract, 30 g*L−1 sucrose, 1.3 g*L−1 K2HPO4.3H2O, 0.5 g*L−1 MgSO4.7H2O, 0.5 g*L−1 KCl, 0.01 g*L−1 FeSO4.7H2O and 15 g*L−1 agar, pH 6.3 ± 0.2 (Pitt Citation1979); malt extract agar (MEA; 30 g*L−1 malt extract, 1 g*L−1 bacteriological peptone, 20 g*L−1 glucose and 20 g*L−1 agar, pH 5.3 ± 0.3) and, Czapek Yeast Extract with salt (CYAS; 3 g*L−1 NaNO3, 50 g*L−1 NaCl, 5 g*L−1 yeast extract, 30 g*L−1 sucrose, 1.3 g*L−1 K2HPO4.3H2O, 0.5 g*L−1 MgSO4.7H2O, 0.5 g*L−1 KCl, 0.01 g*L−1 FeSO4.7H2O and 15 g*L−1 agar, pH 6.3 ± 0.2) (Pitt Citation1979). All media were autoclaved at 121°C for 20 min to sterilise. Aspergillus sp. LBM 134 was inoculated in the centre of 90 mm Petri plate containing the media outlined above and incubated at 25°C in the dark. The fungus was also incubated at 30°C and 37°C in CYA medium. Cultures were examined each day and measured at 7 d of incubation.
Micro-morphology
Aspergillus sp. LBM 134 was grown on MEA and PDA media at 28°C for 3 d to observe microscopic characteristics of mycelium. Young mycelium was stained by cotton blue-lactophenol technique. Micro-characteristics of stipes, conidiophores, vesicles, phialides, conidia, cleiostothecia/sclerotia, ascospores were photographed by a digital Canon Power Shot camera G10 and measured using the software ImageJ.
Growth and acid production on creatine sucrose agar (CREA) culture medium
The capability of growth and production of acid by the fungus was tested on CREA medium: 3.0 g*L−1 creatine, 30 g*L−1 sucrose, 0.5 g*L−1 KCl, 0.5 g*L−1 MgSO4.7H2O, 0.5 g*L−1 FeSO4.7H2O, 1.3 g*L−1 K2HPO4.3H2O, 0.05 g*L−1 Bromocresol purple and 15.0 g*L−1 agar (Frivsad Citation1985). The pH of the medium was adjusted to 8 with Tris buffer and autoclaved at 121°C for 20. The fungus was inoculated at the centre of Petri plates containing CREA medium and incubated at 25°C in the dark for 7 d. If violet CREA medium turns yellow after incubation, then the fungus produces acids on this medium.
Test of Ehrlich to evaluate the cyclopiazonic acid production
Aspergillus sp. LBM 134 was examined for the production of cyclopiazonic acid and other alkaloids reacting with Ehrlich reagent (2 g 4-dimethylaminobenzaldehyde in 85 mL of 96% ethanol and 15 mL of 10 N HCl) (Lund Citation1995) using the filter paper method. Firstly, Aspergillus sp. LBM 134 was grown on CYA medium and incubated at 25°C for 7 d. Plugs of 5 mm diameter covered with fungal mycelium were cut out of the centre of the colony and a 1 cm x 1 cm square of filter paper (Whatman No. 1) wetted with Ehrlich reagent was placed on the mycelial side of the plug. The presence of violet, yellow or pink/red ring around the plug after 2 to 6 min, means that the fungus produces cyclopiazonic acid or related alkaloids (Frisvad and Samson Citation2004).
Qualitative determination of cellulolytic potential Aspergillus sp. LBM 134
Determination of CMCase activity
CMCase activity of Aspergillus sp. LBM 134 was carried out on agar plates containing Czapek-agar medium (2 g*L−1 NaNO3, 1 g*L−1 K2HPO4, 0.5 g*L−1 KCl, 0.5 g*L−1 MgSO4.7H2O, 0.01 g*L−1 FeSO4.7H2O, 20 g*L−1 agar) supplemented with 0.1% CMC. The pH of the medium was adjusted to 4.5 ± 0.2 with 100% glacial acetic acid. Plugs (5 mm diameter) were placed in the centre of Petri dishes containing the medium. A non-inoculated plate and an inoculated medium without the substrate (CMC) were used as negative controls. After incubation at 28°C for 4 d, the colony diameter was measured as growth halo and all Petri dishes were incubated at 50°C for 60 min covered with 50 mM sodium acetate buffer at pH 4.8 ± 0.2. Then, plates were stained with an aqueous solution of 0.1% Congo red and shaken at 80 rpm for 15 min. The Congo red solution was then poured off by washing with distillated water and 1 M NaCl solution to reveal the degradation halo which was measured. The cellulose degradation coefficient (CDC) was determined as follows:
where dh is the degradation halo, dc is the colony diameter.
Determination of BGL and CBH activities of Aspergillus sp. LBM 134 by fluorescence plate assays
The fungus was inoculated on agar plates containing 1 g*L−1 CMC, 1 g*L−1 yeast extract and 20 g*L−1 agar supplemented with 0.1 mM 4-methyl umbelliferyl glucoside (Mu-g; Sigma, MO, USA) for BGL determination and 0.1 mM 4-methyl umbelliferyl cellobioside (Mu-c; Sigma, MO, USA) for CBH assay. The pH of the media was adjusted to 4.2 with 100% glacial acetic acid. Non-inoculated plates and inoculated plates without the substrates were used as negative controls. All plates were incubated at 28 ± 2°C for 3 d. After incubation, the plates were exposed under UV light. If the fungus shows a fluorescence halo under UV light, it is considered as positive BGL or CBH producer.
Quantitative determination of Aspergillus sp. LBM 134 cellulolytic potential
Aspergillus sp. LBM 134 was grown according to Díaz et al. (Citation2019b). After incubation, the culture media were centrifuged at 4°C (15 min, 10,000 g) and supernatants were used to determine CMCase, BGL, CBH and Filter Paper activity (FPase).
CMCase and FPase activities were determined according to Ghose (Citation1987) using CMC (Sigma-Aldrich, USA) and Whatman no. 1 filter paper as substrates, respectively. Reducing sugars were measured by 1,3-dinitrosalicylic acid (DNS) assay (Miller Citation1959) using glucose as standard curve. Enzyme activities were expressed as international units (U), defined as the amount of enzyme needed to produce 1 µmol of glucose per min at 50°C.
BGL activity was assayed by the method described by Herr et al. (Citation1978) using nitrophenyl-β-D-glucoside (pNPG; Sigma, MO, USA) as substrate. The CBH activity was assayed following a modified method described by Wu et al. (Citation2006) using nitrophenyl-β-D-cellobioside (pNPC; Sigma, MO, USA) as substrate. In both enzyme assays, the amount of p-nitrophenol released was measured and expressed as international units (U), defined as the amount of enzyme necessary to release 1 μmol of p-nitrophenol per minute at 50°C.
Results
Concatenated sequences trees
Using only ITS sequences, the trees formed did not distinguish Aspergillus sp. LBM 134 from A. lacticoffetaus, A. welwitschiae (A. awamori sensu Perrone), A.foetidus and A. niger (subclade A. niger) (data not shown). Therefore, in this work, we amplified for ITS, Bt, CMD and also for D1-D2 and Tef. The consensus sequences are deposited in GenBank of National Centre for Biotechnology Information (NCBI). The sequence access numbers are MK457457, MK465342, MK465341, MK463630 and MK465340, respectively.
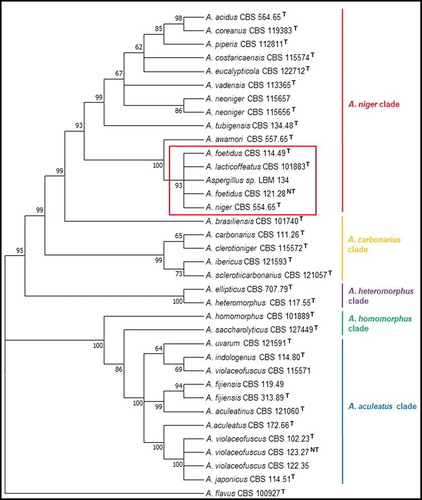
ITS, Bt and CMD sequences were used and they were concatenated for constructing NJ and ML trees. The alignment of the concatenated sequences was deposited in the website TreeBASE (https://treebase.org/) under the submission number: TB2:S24742 (http://purl.org/phylo/treebase/phylows/study/TB2:S24742). These trees grouped the CBS type strains into the expected clades of section Nigri: A. niger, A. carbonarius, A. heteromorphus, A. homomorphus, A.aculeatus. The phylogenetic tree using NJ method showed Aspergillus sp. LBM 134 belonged to the subclade A. niger and grouped with A. niger CBS 554.65, A. lacticoffeatus CBS 101883, A. foetidus CBS 114.49 and A. foetidus CBS 121.28, with a bootstrap value of 96 (). The tree also showed that the subclade A. niger was included into the clade A. niger with the other subclade A. tubigensis with the following type species: A. piperis, A. vadensis, A. costaricaencis, A. acidus and A. neoniger. Similarly, the tree obtained by ML method grouped Aspergillus sp. LBM 134 with A. niger CBS 554.65, A. lacticoffeatus CBS 101883, A. foetidus CBS 114.49 and A. foetidus CBS 121.28, with a bootstrap value of 93 () staying inside the subclade A. niger and the clade A. niger. Moreover, the analysis using concatenated sequences could set aside A. awamori from the cluster formed using only ITS sequences. Therefore, we discarded that the isolate was A. welwitschiae. However, due to the impossibility to appropriately identify our isolate and reach to a species-specific identification, we carried out a polyphasic identification.
Figure 1. Phylogenetic tree obtained by the neighbour joining method, showing placement of Aspergillus sp. LBM 134 among species of section Nigri and reference species A. flavus (the outgroup species in the analysis) as presented by the bootstrap consensus tree inferred from 1 000 replicates derived from analysis of ITS-Bt-CMD. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1 000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as used for evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using Kimura’s method. All but LBM 134 are type strains
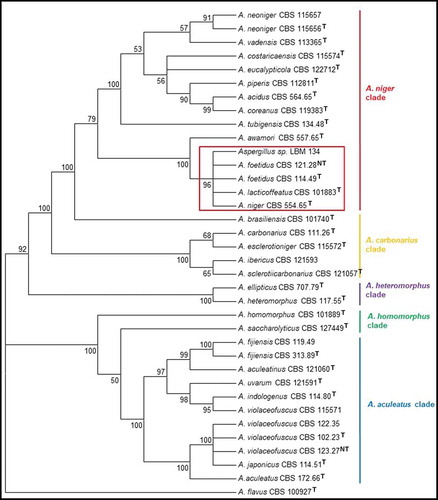
Figure 2. Phylogenetic tree obtained by the maximum likelihood method, showing placement of Aspergillus sp. LBM 134 among species of section Nigri and reference species A. flavus (the outgroup species in the analysis) as presented by the bootstrap consensus tree inferred from 1 000 replicates derived from analysis of ITS-Bt-CMD. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1 000 replicates) are shown next to the branches. The evolutionary distances were computed using Kimura’s method. All but LBM 134 are type strains
Polyphasic identification
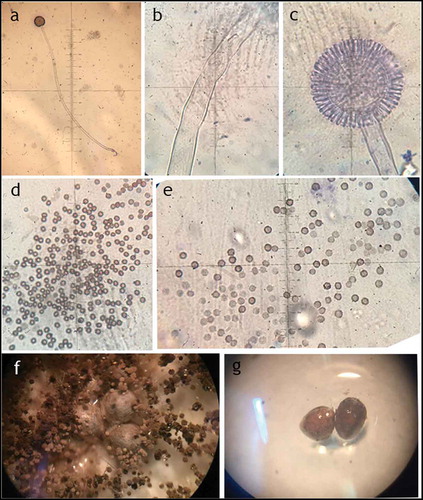
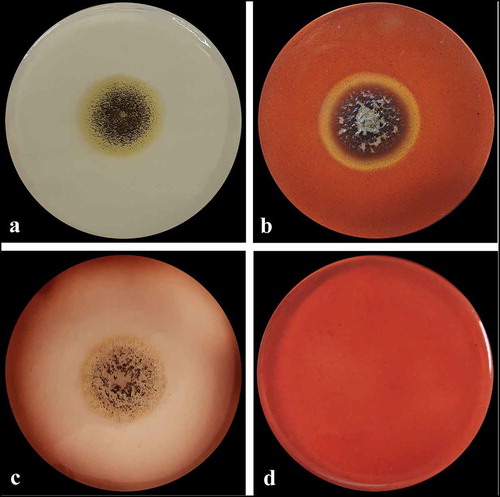
Aspergillus sp. LBM 134 showed differences in its macro-morphology in different media and temperatures (). The colour of the “top view” of the mycelium is due to the fungus conidia colour. Black colouration, characteristic for conidia of black Aspergillus, and cream reverse were observed on MEA medium (). The conidia were greenish browns to dark brown or almost black on CYA media, getting darker as the temperature of incubation increased (from 25°C to 37°C); the colour of the reverse also showed the same pattern, from cream to dark brown as the temperature of incubation increased (). Conidia of paler colour were produced on CYAS medium as well as the colour reverse ). Aspergillus sp. LBM 134 grown on MEA or CYAS media did not cover all the plate during the incubation time. Data are summarised in .
Table 1. Macro-morphological characteristics registered for Aspergillus sp. LBM 134 grown on different media for 7 d of incubation
Figure 3. Macro-morphology and production of acid-base of Aspergillus sp. LBM 134 grown for 7 d on different agar media. On MEA at 25 C: (a) conidia colour and (b) reverse colour; CYA at 25 C: (c) conidia colour and (d) reverse colour; CYA at 30 C: (e) conidia colour and (f) reverse colour; CYA at 37 C: (g) conidia colour and (h) reverse colour; CYAS at 25 C: (i) conidia colour and (j) reverse colour. Acid production on CREA medium: (k) inoculated plate and (l) plate without inoculating
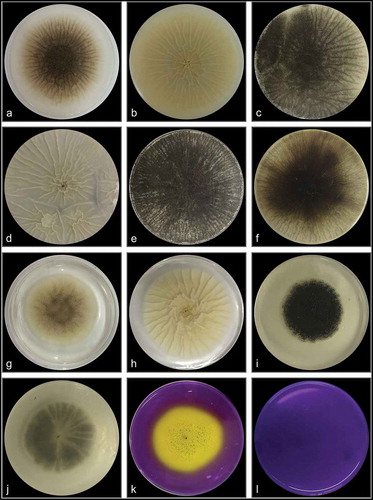
Aspergillus sp. LBM 134 grown on CREA medium registered a moderate production of acids visualised by the turn of violet to yellow of culture medium ). Positive acid production on CREA medium is reported for both A. niger and A. lacticoffeatus. The same occurs for the Ehrlich test; the Ehrlich reagent did not react with the Aspergillus sp. LBM 134 mycelium () suggesting that it does not produce cyclopiazonic acid or any related alkaloid.
Figure 4. Test of Ehrlich reagent using the filter paper method to detect cyclopiazonic acid and related alkaloids. (a) Aspergillus sp. LBM 134 developed on CYA medium where plugs were cut off. (b) Filter paper did not show any colour ring indicating the absence of production of cyclopiazonic acid or any related alkaloid
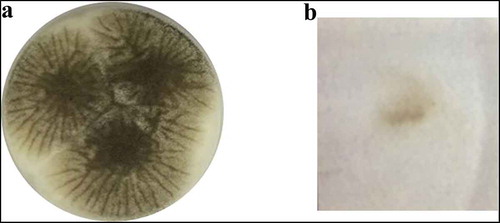
Micro-morphological characteristics of Aspergillus sp. LBM 134 are shown in and . Stipes were wide, hyaline, long without any ornamentation (). A. niger has hyaline conidiophores with long and hyaline stipes, similar to Aspergillus sp. LBM 134; stipes from A. lacticoffeatus are shorter and orange. Vesicles of Aspergillus sp. LBM 134 had almost a spherical shape and metulae () and conidia have apiculate ornamentation (). An interesting characteristic shown by Aspergillus sp. LBM 134 grown on CYA medium for 14 d was the presence of sclerotia (). Sclerotia are asexual, multicellular reproduction structures in fungi (Chet and Henis Citation1975) and there is no study that describes A. lacticoffeatus species as sclerotia producer.
Table 2. Micro-morphological characteristics of Aspergillus sp. LBM 134 grown on CYA and MEA media for 3 d of incubation
Figure 5. Micro-morphology of Aspergillus sp. LBM 134. (a) Conidiophore, 10X; (b) stipe, 400X; (c) vesicle, 400X; (d) and (e) conidia, 1000X; (f) and (g) sclerotia, 4X
After analysing all this information (macro, micro-morphological, acid and alkaloid production) about Aspergillus sp. LBM 134 and the sequence information (ITS, D1-D2, Bt, CMD and Tef) we introduced this information into the online tool Polyphasic Identification and identified the isolate LBM 134 as A. phoenicis, currently, A. niger.
Cellulolytic bioprospection of A. niger LBM 134
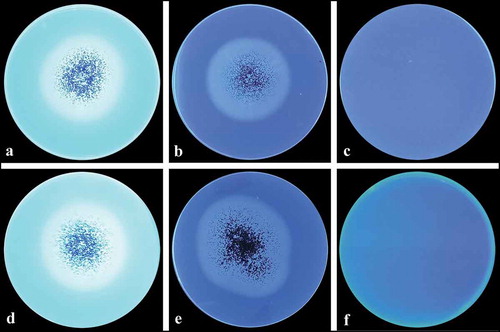
For CMCase activity determination, A. niger LBM 134 was grown on Czapek medium supplemented with CMC for 4 d (). Degradation of CMC in agar-plates, which may be considered a semiquantitative technique, revealed that A. niger LBM 134 had a value of CDC of 1.2 ± 0.01, indicating that the degradation halo is greater than the growth halo.
Figure 6. CMCase activity of Aspergillus sp. LBM 134 determined by the Congo red assay. (a) Developed mycelium for 4 d without revealing; (b) developed mycelium on medium with the substrate (CMC) and revealed showing a degraded halo around the mycelium; (c) control plate, developed mycelium on medium without substrate; (d) plate control, medium without inoculating
On the other hand, screening for BGL and CBH enzymes in agar plates may be considered exclusively qualitative and A. niger LBM 134 was classified as positive (). Furthermore, EG, BGL, CBH and PFase were quantitatively determined in two optimised media () showing high levels on these enzymatic enzymes.
Table 3. CMCase, BGL, CBH and FPase activities of Aspergillus niger LBM 134 grown on two optimised media supplemented with sugarcane bagasse (SCB) and cassava bagasse (CB)
Figure 7. BGL and CBH activities of Aspergillus sp. LBM 134 determined by fluorescence plate assay. (a) Developed mycelium for 4 d on medium containing the substrate (Mu-g) and revealed; (b) plate control, developed mycelium for 4 d on medium without the substrate; (c) control plate, medium containing the substrate without inoculating. (d) Developed mycelium for 4 d on medium containing the substrate (Mu-c) and revealed; (e) plate control, developed mycelium for 4 d on medium without the substrate; (f) control plate, medium containing the substrate without inoculating
Discussion
The taxonomy of filamentous fungi is complicated and classifying the species of Aspergillus is not an exception (Samson et al. Citation2007a). Identifying Aspergillus species of section Nigri is not easy due to their phenotypic similarities (macro and microscopic) and therefore, it is necessary to use several strategies for identifying them. DNA sequence information is increasingly being used for species identification and diagnosis. Although ITS is known as the fungal barcode (Schoch et al. Citation2012), this molecular marker does not allow to identify closely related species. Some authors (Citation2017; Samson et al. Citation2014) recommended the use of a second or even a third molecular marker for identifying black aspergillus. In this sense, the use of Bt, CMD, RPB2 and Tef gene sequences in addition of ITS would achieve a species-specific identification.
While RPB2 is not easy to amplify, Tef is not reported for all Aspergillus species in the database. Bt is the easiest gene to amplify and almost all species of section Nigri can be identified using this gene sequence although it is impossible to discriminate between Bt sequences belonging to A. niger and A. lacticoffeatus. Moreover, there is a risk of amplifying Bt paralog genes (Koonin Citation2005) because of the variations inside the gene introns and their number (Peterson Citation2008; Hubka and Kolarik Citation2012; Samson et al. Citation2014). The same phenomenon occurs with CMD gene; it is easy to amplify and has the capacity to distinguish most species of Aspergillus; so is the second temporal marker proposed for Aspergillus (Samson et al. Citation2014).
In this work, the trees formed using the NJ and ML methods grouped the type species CBS into the expected clades of section Nigri, reproducing the phylogeny for the fungi group described by Varga et al. (Citation2011) and Samson et al. (Citation2014). Both trees showed that Aspergillus sp. LBM 134 belonged to the subclade A. niger. The use of international renowned species such as CBS type strains ensures that the trees were constructed with correct sequences and taxonomic names (Fungaro et al. Citation2017). We used that criteria and decided to not use any sequence submitted in GenBank since it is a public, archival database and accepts all sequences submitted and cannot always verify the taxonomy. Hence, results from BLAST search may give hits to misidentified sequences in the database (Fungaro et al. Citation2017).
A. niger LBM 134 has been grouped within the subclade A. niger with the species A. welwitschiae, A. foetidus and A. lacticoffeatus by using only molecular techniques. The trees using concatenated sequences separated A. welwitschiae from the A. niger clade. For a long time, A. welwitschiae was considered as a synonym of A. niger. Currently, both species are considered as cryptic species (Perrone et al. Citation2011) in speciation process; hence, it is necessary to use more than one molecular marker to distinguish them (Samson et al. Citation2014; von Hertwig et al. Citation2018). Regarding A. foetidus is not currently use anymore and the correct name is A. niger (Samson et al. Citation2004; Varga et al. Citation2011). A. lacticoffeatus was reported for the first time in 2004 by Frisvad and Samson (Citation2004), is in the list of accepted species of genus Aspergillus. However, taxonomic studies by Varga et al. (Citation2011), de Vries et al. (Citation2017) and Vesth et al. (Citation2018) and the study of growth and hydrolytic profiles by Meijer et al. (Citation2011) considered the species A. lacticoffeatus as a synonym of A. niger. A study of A. lacticoffeatus strains isolated from coffee grains in Venezuela and Indonesia (Frisvad and Samson Citation2004) suggested that A. lacticoffeatus could be a mutant strain of A. niger.
A polyphasic approach was used to distinguish if the isolate LBM 134 was A. niger or A. lacticoffeatus and LBM 134 was correctly identified as A. niger. The colour of conidia (greenish brown-black) developed by A. niger LBM 134 on MEA and CYA which effectively coincides with conidia from A. niger (Abarca Citation2000) and contrasts with soft brown conidia from A. lacticoffeatus (Samson et al. Citation2004; Cabañes and Bragulat Citation2018). Another characteristic feature was the ornamentation of conidia of A. niger LBM 134; these had their surface less echinulate than A. lacticoffeatus conidia (Frisvad and Samson Citation2004). Also, A. niger LBM 134 showed sclerotia production, characteristic for A. niger grown on CYA medium supplemented with fruit pieces and rice as reported by Frisvad et al. (Citation2014). The Polyphasic identification online tool showed that the isolate LBM 134 was A. phoenicis, this name is antique and correspond to A. niger (as we call it henceforth) (Abarca et al. Citation2004). The employment of online software to identify species allows to verify the obtained results in the laboratory and, at the same time, eliminates the subjectivity when putting weight on the criteria used in identification. Although the software solved that the isolate LBM 134 was A. phoenisis, this denomination is rejected over the conserved species A. niger (Frisvad et al. Citation1990; Kozakiewicz et al. Citation1992). This fact emphasises the importance of a critical analysis carried out by the researcher on the available bioinformatic tools.
Black Aspergillus are efficient producers of hydrolytic enzymes and A. niger has been employed for enzyme production to be used in different biotechnological processes. Among hydrolytic enzymes, we focused on cellulase production by A. niger LBM 134 since they are essential enzymes for the bioprocessing of cellulosic biomass. As A. niger LBM 134 was isolated from rotten wood, we understood that this fungus was a potential cellulolytic producer due to it grew on cellulosic biomass and probably used cellulose as carbon and energy. For carrying out the bio-prospective studies of A. niger LBM 134, we employed qualitative methods for detection of cellulase enzymes. In first place, we evaluated EG activity on plate assay using CMC as substrate using Congo red, a colourant that interacts with β-1,4-glucosidic bonds (Sazci et al. Citation1986), contrasting with the discoloured halo that represents the degraded substrate by CMCase activity. We measured the CMCase activity by determining the CDC which is the most used parameter for evaluating the cellulase production by microorganisms on solid media. There is a direct correlation between the degradative diameter and the degradation capacity (Lin et al. Citation1991). Since CMC can be degraded by other enzymes in addition to EG, talking about CMCase activity is correct. In this sense, we can say that A. niger LBM 134 has positive CMCase activity which coincides with the literature (Castrillo et al. Citation2012; Martínez et al. Citation2005; Panda et al. Citation2012; Singhania et al. Citation2017; Maia and Fraga Citation2017). Respect on BGL and CBH activities, the fluorescence was intense after a few days of incubation; data agreed with the literature that describes species of A. niger as BGLs and CBHs producer and the technique of fluorescence plate contributed more significant results in the study of the potential cellulolytic of A. niger LBM 134. These qualitative, novelty, specific and simple assays provide key information when investigating new isolates not yet characterised that could have a big biotechnological potential, for example, in the cellulosic bioconversion (Coniglio et al. Citation2017). To our knowledge, this is the first time the substrates Mu-g and Mu-c were used for qualitative determination of BGL and CBH activities in ascomycetes.
Cellulolytic potential of A. niger LBM 134 was reinforced by quantitative determination of CMCase, BGL, CBH and PFase. Levels of these cellulolytic enzymes of A. niger LBM 134 were higher than values found in other species of Aspergillus grown on CMC (El-Hadi et al. Citation2014; Sohail et al. Citation2016). Furthermore, the cellulolytic ability of A. niger LBM 134 was confirmed by the successful bioconversion of two agroindustrial wastes: sugarcane bagasse and cassava bagasse (paper in press).
Acknowledgements
This work has been possible thanks to the Consejo Nacional de Investigaciones Científicas & Técnicas (CONICET) and Mycology Chatedra of the Facultad de Ciencias Exactas, Químicas y Naturales, of the Universidad Nacional de Misiones.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Abarca M, Accensi F, Cano J, Cabañes FJ. 2004. Taxonomy and significance of black aspergilli. Anton Leeuw Int J G 86(1):33–49
- Abarca ML. 2000. Taxonomy and identification of the species involved in nosocomial aspergillosis. Rev Iberoam Micol. 17:S79–S84.
- Benbow JM, Sugar D. 1999. Fruit surface colonization and biological control of postharvest diseases of pear by preharvest yeast applications. Plant Dis. 83(9):839–844. doi:https://doi.org/10.1094/PDIS.1999.83.9.839.
- Bennett JW. 2010. An overview of the genus Aspergillus. Aspergillus: molecular biology and genomics 1-17.
- Bertrand B, Martínez‐Morales F, Trejo‐Hernández MR. 2017. Upgrading laccase production and biochemical properties: strategies and challenges. Biotechnol Progr. 33(44):1015–1034. doi:https://doi.org/10.1002/btpr.2482.
- Cabañes FJ, Bragulat MR. 2018. Black aspergilli and ochratoxin A-producing species in foods. Curr Opin Food Sci. 23:1–10. doi:https://doi.org/10.1016/j.cofs.2018.01.006.
- Carrillo L. 2003. Aspergillus: morfología. Identificación. Cultivos. Ambiente. Micotoxinas: aflatoxinas, ocratoxina A, esterigmatocistina, ácido ciclopiazónico, neurotoxinas, otras toxinas. In: Carrillo, editor. Los hongos de alimentos y forrajes. 1st ed. Argentina: Universidad Nacional de Salta; p. 44–61.
- Castrillo ML, Fonseca MI, Bich GA, Jerke G, Horianski MA, Zapata PD. 2012. Taxonomy and phylogenetic analysis of Aspergillus section nigri isolated from yerba mate in Misiones (Argentina). J Basic Appl Genet. 23:2.
- Chet I, Henis Y. 1975. Sclerotial morphogenesis in fungi. Annu Rev Phytopathol. 13(1):169–192. doi:https://doi.org/10.1146/annurev.py.13.090175.001125.
- Coniglio RO, Fonseca MI, Villalba LL, Zapata PD. 2017. Screening of new secretory cellulases from different supernatants of white rot fungi from Misiones, Argentina. Mycology. 8:1–10. doi:https://doi.org/10.1080/21501203.2016.1267047.
- de Vries RP, Riley R, Wiebenga A, Aguilar-Osorio G, Amillis S, Uchima CA, …, Battaglia E. 2017. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 18(1):1–45
- Decontardi S, Soares C, Lima N, Battilani P. 2018. Polyphasic identification of Penicillia and Aspergilli isolated from Italian grana cheese. Food Microbiol. 73:137–149. doi:https://doi.org/10.1016/j.fm.2018.01.012.
- Díaz GV, Coniglio RO, Velazquez JE, Zapata PD, Villalba L, Fonseca MI. 2019b. Adding value to lignocellulosic wastes via their use for endoxylanase production by Aspergillus fungi. Mycologia. 111(2):195–205. doi:https://doi.org/10.1080/00275514.2018.1556557.
- Díaz GV, Zapata PD, Villalba LL, Fonseca MI. 2019a. Evaluation of new xylanolytic-producing isolates of Aspergillus from Misiones subtropical rainforest using sugarcane bagasse. Arab J Basic Appl Sci. 26(1):292–301. doi:https://doi.org/10.1080/25765299.2019.1622922.
- El-Hadi AA, El-Nour SA, Hammad A, Kamel Z, Anwar M. 2014. Optimization of cultural and nutritional conditions for carboxymethyl cellulase production by Aspergillus hortai. J Radiat Res Appl Sci. 7(1):23–28. doi:https://doi.org/10.1016/j.jrras.2013.11.003.
- Fonseca MI, Fariña JI, Sadanoski MA, D’Errico R, Villalba LL, Zapata PD. 2015. Decolorization of kraft liquor effluents and biochemical characterization of laccases from Phlebia brevispora BAFC 633. Int Biodeter Biodegr. 104:443–451. doi:https://doi.org/10.1016/j.ibiod.2015.07.014.
- Frisvad JC, Hawksworth DL, Kozakiewicz Z, Pitt JI, Samson RA, Stolk AC. 1990. Proposals to conserve important species names in Aspergillus and Penicillium. In: Frisvad JC, Hawksworth DL, Kozakiewicz Z, Pitt JI, Samson RA, Stolk AC, editors. Modern concepts in Penicillium and Aspergillus classification. 1st edn ed. Boston (MA): Springer; p. 83–89.
- Frisvad JC, Petersen LM, Lyhne EK, Larsen TO, McCluskey K. 2014. Formation of sclerotia and production of indoloterpenes by Aspergillus niger and other species in section Nigri. PLoS One. 9:e94857. doi:https://doi.org/10.1371/journal.pone.0094857.
- Frisvad JC, Samson RA. 2004. Polyphasic taxonomy of Penicillium subgenus Penicillium. ** Stud Mycol. 49:1–174.
- Frivsad JC. 1985. Creatine sucrose agar, a differential medium for mycotoxin producing terverticillate Penicillium species. Lett Appl Microbiol. 1:109–113. doi:https://doi.org/10.1111/j.1472-765X.1985.tb01500.x.
- Fungaro MHP, Ferranti LS, Massi FP, da Silva JJ, Sartori D, Taniwaki MH, Frisvad JC, Iamanaka BT. 2017. Aspergillus labruscus sp. nov., a new species of Aspergillus section Nigri discovered in Brazil. Sci Rep. 7: 6203
- Ghose TK. 1987. Measurement of cellulase activities. Pure Appl Chem. 59(2):257–268. doi:https://doi.org/10.1351/pac198759020257.
- Glass NL, Donaldson G. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 61:1323–1330. doi:https://doi.org/10.1128/AEM.61.4.1323-1330.1995.
- Herr D, Baumer F, Dellweg H. 1978. Purification and properties of an extraeellular endo-l,4-β-glucanase from Lenzites trabea. Appl Microbiol Biotechnol. 5:29–36. doi:https://doi.org/10.1007/BF00515684.
- Hong SB, Go SJ, Shin HD, Frisvad JC, Samson RA. 2005. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia. 97:1316–1329. doi:https://doi.org/10.1080/15572536.2006.11832738.
- Houbraken J, de Vries RP, Samson RA. 2014. Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Adv Appl Microbiol. 86:199–249.
- Hubka V, Kolarik M. 2012. β-tubulin paralogue tubC is frequently misidentified as the benA gene in Aspergillus section Nigri taxonomy: primer specificity testing and taxonomic consequences. Persoonia. 29:1–10. doi:https://doi.org/10.3767/003158512X658123.
- Klich MA, Pitt JI. 1988. A laboratory guide to common Aspergillus species and their teleomorphs. North Ryde: CSIRO Division of Food Processing.
- Koonin EV. 2005. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 39:309–338. doi:https://doi.org/10.1146/annurev.genet.39.073003.114725.
- Kozakiewicz Z, Frisvad JC, Hawksworth DL, Pitt JI, Samson RA, Stolk AC. 1992. Proposals for nomina specifica conservanda and rejicienda in Aspergillus and Penicillium (Fungi). Taxon. 41:109–113. doi:https://doi.org/10.2307/1222500.
- Kuhad RC, Gupta R, Singh A. 2011. Microbial Cellulases and Their Industrial Applications. Enzyme Research 2011 ID 280396 2011 doi:https://doi.org/10.4061/2011/280696
- Kurtzman CP, Robnett CJ. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5ʹend of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol. 35:1216–1223. doi:https://doi.org/10.1128/JCM.35.5.1216-1223.1997.
- Lin JE, Chang DC, Shen GJ, Wang HY. 1991. Correlations among several screening methods used for identifying wood-decay fungi that can degrade toxic chemicals. Biotechnol Tech. 5:275–280. doi:https://doi.org/10.1007/BF02438662.
- Lund F. 1995. Differentiating Penicillium species by detection of indole metabolites using a filter paper method. Lett Appl Microbiol. 20:228–231. doi:https://doi.org/10.1111/j.1472-765X.1995.tb00434.x.
- Maia TF, Fraga ME. 2017. Bioprospecting Aspergillus section Nigri in Atlantic Forest soil and plant litter. In: Arquivos do Instituto Biológico. p. 84. e0502015 0 doi:https://doi.org/10.1590/1808-1657000502015
- Martínez A, Speranza M, Ruiz Dueñas F, Ferreira P, Camarero S, Guillén F, Martínez M, Gutiérrez A, Del Rio J. 2005. Biodegradation of lignocellulosics microbial, chemical and enzymatic aspects of fungal attack to lignin. Int Microbiol. 8:195–204.
- Meijer M, Houbraken JAMP, Dalhuijsen S, Samson RA, de Vries RP. 2011. Growth and hydrolase profiles can be used as characteristics to distinguish Aspergillus niger and other black aspergilli. Stud Mycol. 69:19–30. doi:https://doi.org/10.3114/sim.2011.69.02.
- Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 31(3):426–428. doi:https://doi.org/10.1021/ac60147a030.
- Palumbo JD, O’keeffe TL. 2015. Detection and discrimination of four Aspergillus section Nigri species by PCR. Lett Appl Microbiol. 60:188–195. doi:https://doi.org/10.1111/lam.12358.
- Panda SS, Sahoo K, Das R, Dhal NK. 2012. Pectinolytic and cellulolytic activity of soil fungal isolates from similipal bioreserve forest. World Environ. 2:1–3. doi:https://doi.org/10.5923/j.env.20120202.01.
- Park HS, Jun SC, Han KH, Hong SB, Yu JH. 2017. Diversity, application, and synthetic biology of industrially important Aspergillus fungi. Adv Appl Microbiol. 100:161–202.
- Perrone G, Stea G, Epifani F, Varga J, Frisvad JC, Samson RA. 2011. Aspergillus niger contains the cryptic phylogenetic species A. Fungal Biol. 115:1138–1150. doi:https://doi.org/10.1016/j.funbio.2011.07.008.
- Peterson S. 2008. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia. 100:205–226. doi:https://doi.org/10.1080/15572536.2008.11832477.
- Pitt JI. 1979. The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. London (UK): Academic Press.
- Raper KB, Fennell DI. 1965. The genus Aspergillus. Baltimore: Willians and Wilkins; p. 736–737.
- Rodrigues P, Venâncio A, Kozakiewicz Z, Lima N. 2009. A polyphasic approach to the identification of aflatoxigenic and non-aflatoxigenic strains of Aspergillus section Flavi isolated from Portuguese almonds. Int J Food Microbiol. 129:187–193. doi:https://doi.org/10.1016/j.ijfoodmicro.2008.11.023.
- Samson RA, Hong S, Peterson SW, Frisvad JC, Varga J. 2007a. Polyphasic taxonomy of Aspergillus section Fumigati and its teleomorph Neosartorya. Stud Mycol. 59:147–203. doi:https://doi.org/10.3114/sim.2007.59.14.
- Samson RA, Houbraken JA, Kuijpers AF, Frank JM. 2004. New ochratoxin A or sclerotium producing species in Aspergillus section Nigri. Stud Mycol. 50:45–61.
- Samson RA, Noonim P, Meijer M, Houbraken JAMP, Frisvad JC, Varga J. 2007b. Diagnostic tools to identify black aspergilli. Stud Mycol. 59:129–145. doi:https://doi.org/10.3114/sim.2007.59.13.
- Samson RA, Visagie CM, Houbraken J, Hong SB, Hubka V, Klaassen CH, Perrone G, Seifert KA, Susca A, Tanney JB, et al. 2014. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol. 78:141–173. doi:https://doi.org/10.1016/j.simyco.2014.07.004.
- Samuels GJ, Ismaiel A. 2009. Trichoderma evansii and T. lieckfeldtiae: two new T. hamatumlike species. Mycologia. 101:142–156.
- Sazci A, Erenler K, Radford A. 1986. Detection of cellulolytic fungi by using Congo red as an indicator: a comparative study with the dinitrosalicyclic acid reagent method. J Appl Bacteriol. 61:559–562. doi:https://doi.org/10.1111/j.1365-2672.1986.tb01729.x.
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Miller AN, Bolchacova E, Voigt K, Crous PW. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. P Natl Acad Sci. 109:6241–6246. doi:https://doi.org/10.1073/pnas.1117018109.
- Silva DM, Batista LR, Rezende EF, Fungaro MHP, Sartori D, Alves E. 2011. Identification of fungi of the genus Aspergillus section nigri using polyphasic taxonomy. Braz J Microbiol. 42:761–773. doi:https://doi.org/10.1590/S1517-83822011000200044.
- Silva FC, Chalfoun SM, Batista LR, Santos C, Lima N. 2015. Use of polyphasic approach including MALDI-TOF MS for identification of Aspergillus section Flavi strains isolated from food commodities in Brazil. Ann Microbiol. 65:2119–2129. doi:https://doi.org/10.1007/s13213-015-1050-0.
- Simões MF, Pereira L, Santos C, Lima N. 2013. Polyphasic identification and preservation of fungal diversity: concepts and applications. In: Malik A, Grohmann E, Alves M, editors. Management of microbial resources in the environment. Dordrecht: Springer; p. 91–117.
- Singhania RR, Adsul M, Pandey A, Patel AK. 2017. Cellulases. Curr Devel Biotechnol Bioeng. 4:73–101.
- Sohail M, Ahmad A, Khan SA. 2016. Production of cellulase from Aspergillus terreus MS105 on crude and commercially purified substrates. 3 Biotech. 6(1):103. doi:https://doi.org/10.1007/s13205-016-0420-z.
- Varga J, Frisvad JC, Kocsubé S, Brankovics B, Tóth B, Szigeti G, Samson RA. 2011. New and revisited species in Aspergillus section Nigri. Stud Mycol. 69:1–17. doi:https://doi.org/10.3114/sim.2011.69.01.
- Vesth TC, Nybo JL, Theobald S, Frisvad JC, Larsen TO, Nielsen KF, Hoof JB, Brandl J, Salamov A, Riley R, et al. 2018. Investigation of inter-and intraspecies variation through genome sequencing of Aspergillus section Nigri. Nat Genet. 50:1688–1695. doi:https://doi.org/10.1038/s41588-018-0246-1.
- von Hertwig AM, Sant’Ana AS, Sartori D, da Silva JJ, Nascimento MS, Iamanaka BT, Pelegrinelli Fungaro MH, Taniwaki MH. 2018. Real-time PCR-based method for rapid detection of Aspergillus niger and Aspergillus welwitschiae isolated from coffee. J Microbiol Meth. 148:87–92. doi:https://doi.org/10.1016/j.mimet.2018.03.010.
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky KK, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; p. 315–322.
- Wu B, Zhao Y, Gao PJ. 2006. Estimation of cellobiohydrolase I activity by numerical differentiation of dynamic ultraviolet spectroscopy. Acta Biochim Biophys. 38:372–378.
- Yergeau E, Filion M, Vujanovic V, St-Arnaud M. 2005. A PCR-denaturing gradient gel electrophoresis approach to assess Fusarium diversity in asparagus. J Microbiol Meth. 60:143–154. doi:https://doi.org/10.1016/j.mimet.2004.09.006.