ABSTRACT
Transcription factors generally regulate gene expression of multiple targets. In contrast, our recent finding suggests that the zinc finger protein Ouija board controls steroid hormone biosynthesis through specific regulation of only one gene spookier in Drosophila. It sheds light on a specialized but essential factor that evolved for one target.
Introduction
In the context of developmental programs, a variety of gene cascades are regulated by a number of transcription factors (TFs). To accomplish the precise morphogenesis and tissue organization, it is important to regulate gene expression profiles appropriately in space and time. In this sense, the expression of TFs should be also tightly controlled in spatiotemporal-dependent manners.Citation1
Some TFs are known as the “master” regulators that are critical for tissue morphogenesis and pattern formation. For example, Hox genes control a set of downstream genes that produce tissues and organs along the body axes.Citation2 Another striking example is Eyeless/Pax6, which serves as a master TF for eye morphogenesis.Citation2 Such master TFs are usually highly conserved among animal phyla, and the transcription of multiple components for a particular developmental process can be cooperatively regulated in a wide variety of animals.
While most TFs, including the master TFs, usually bind multiple sites in the genome to regulate a large number of target genes, less is known about the specific TFs that regulate only few or even one specific target(s). It also remains to be determined whether such specialized TFs are essential in development.
We have recently identified a new TF designated Ouija board (Ouib) essential for steroid hormone biosynthesis in the fruit fly Drosophila melanogaster.Citation3 It is predominantly expressed in the steroidogenic organs and, as far as we know, represents the first example of a TF that appears to be specialized for inducing the expression of only one steroidogenic enzyme gene. In this review, we discuss a role of Ouib and its biological significance for development and evolution.
Transcriptional regulation of the steroid hormone biosynthesis pathway
Steroid hormones have pleiotropic actions on cells, controlling development, homeostasis, and reproduction. They are derivatives of cholesterol and other suitable sterols, being biosynthesized in specialized endocrine organs, such as the adrenal gland and gonads in vertebrates. In the biosynthesis pathway, a battery of steroidogenic enzymes, including cytochrome P450 family, is required to convert suitable sterols into steroid hormones through several intermediate multiple steps. These enzyme genes are specifically expressed in the steroidogenic organs. Therefore, the high activity of steroid hormone biosynthesis is achieved by the high transcriptional activity of these steroidogenic genes.
Vertebrate steroidogenic genes are coordinately regulated by a TF called NR5R1/Steroidodogenic factor-1 (SF-1).Citation4 SF-1 is predominantly expressed in steroidogenic organs. SF-1 knockout mice lose gonads and adrenal glands, and die shortly after birth.Citation5,6 Moreover, the expression of SF-1 is sufficient for the differentiation of non-steroidogenic stem cells into steroidogenic cells by inducing the expression of steroidogenic enzyme genes.Citation7 In this sense, vertebrate steroidogenesis depends on the transcriptional activity of SF-1, and as such, SF-1 is a master regulator for the development of gonads and adrenal glands as well as for steroidogenesis following organogenesis.
SF-1 genes are well conserved in animal evolution. Nevertheless, it seems that the function of SF-1 may differ between vertebrates and invertebrates, as an insect ortholog of SF-1, Ftz-F1, is highly expressed not only in steroidogenic organs but also in many other tissues.Citation8 A previous study has argued that the steroid hormone biosynthesis pathway and its transcriptional regulatory mechanism have separately evolved in insects,Citation9 and how to transcriptionally regulate steroidogenic enzyme genes has not been known until recently.
The biosynthesis of insect steroid hormone, ecdysteroid
Insect steroid hormones, ecdysteroids, control the timing of developmental progression, such as molting, metamorphosis, and reproductive maturation.Citation1,10-12 The last decade is a very fruitful period in the biology of ecdysteroids: several research groups including our group have successfully identified and characterized a number of genes essential for the ecdysteoid biosynthesis pathway.Citation10 Among the identified genes, neverland (nvd), spook (spo), spookier (spok), Cyp6t3, shroud (sro), phantom (phm), disembodied (dib), shadow (sad), and shade (shd) encode ecdysteroidogenic enzymes that catalyze the conversion steps from suitable sterol to 20-hydroxyecdysone and/or other ecdysteroids (). These enzyme genes, except for shd, are predominantly expressed in a specific endocrine organ called the prothoracic gland (PG). The expression levels of these genes fluctuate during developmental stages and well correlated to the ecdysteroid titer, indicating that their transcriptional activities are tightly regulated.Citation13-15 Therefore, the spatiotemporal regulation of ecdysteroidogenic genes is essential for ecdysteroid biosynthesis. This is also the case with steroidogenesis in vertebrates.
Figure 1. The ecdysteroid biosynthesis pathway and the role of Ouija board on spookier expression in the fruit fly Drosophila melanogaster. In the prothoracic gland (insect steroidogenic organ), cholesterol and other sterols are converted into ecdysteroids via several intermediate steps. The modifications on carbon residues and the responsible enzymes are indicated with red and green, respectively. In the nucleus, nvd and spok are located in the heterochromatin region. A zinc finger transcription factor named “Ouija board (Ouib)” specifically binds to the promoter region of “spookier” and induces expression of this gene. Both Ouib and Spok are essential for ecdysteroid biosynthesis during larval development.
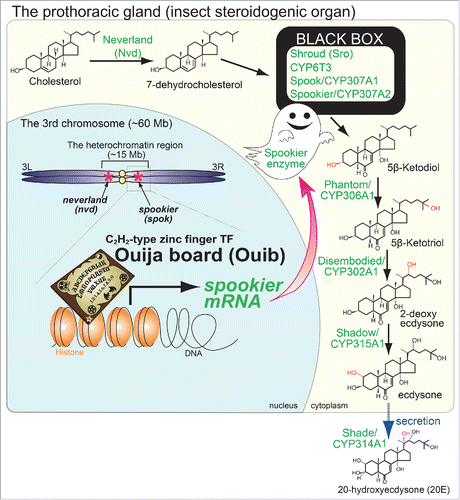
A number of ecdysteroidogenic TFs have also been reported. The first description of a steroidogenic TF was the discovery that βFTZ-F1 plays a role in expression of phm, dib, and sad.Citation16 βFTZ-F1 is an isoform of FTZ-F1, being transcribed from late-stage embryos to adults.Citation17,18 Since the discovery of βFTZ-F1, the list of ecdysteroidogenic transcription factors has grown subsequently until 2014, including Molting defective (Mld), DHR4, Broad, and Ventral vein lacking (Vvl).Citation1 Notably, all of these TFs are expressed not only in the PG, but also in other type of cells. Therefore, these TFs per se do not account for the spatial restriction of ecdysteroidogenic gene expression. It is likely that a combination of TFs can provide the PG-specific expression, along with epigenetic programming. On the other hand, it is also theoretically possible that there are unknown TFs whose expression and function are restricted to the PG.
Ouija board encodes the C2H2-type zinc finger type TF and is predominantly expressed in the ecdysteroid-producing organ, the prothoracic gland
In 2015, we reported a C2H2-type zinc finger protein gene CG11762, and named it ouija board (the reason is explained below).Citation3 ouib is predominantly expressed in the PG and Ouib protein is localized in the nuclei of the PG cells (Takemata, Kamiyama, YSN, and RN, unpublished observation). ouib null mutants arrest development in the 1st instar stage and the ecdysteroid titer is significantly reduced. Ecdysone signaling is also significantly reduced in these mutants, suggesting that Ouib is essential for development and involved in ecdysteroid biosynthesis. Considering the predicted primary structure of Ouib, we expected that Ouib might act as a TF that regulates the expressions of ecdysteroidogenic genes
To examine which step of the ecdysteroid biosynthesis pathway is regulated by Ouib, we measured the expression levels of ecdysteroidogenic genes in ouib null mutant larvae. Among six tested, the expression levels of spok, dib, and sad were significantly reduced. In particular, spok expression is drastically reduced as Spok protein level is almost abolished in the PG of ouib mutant larvae. Moreover, the qRT-PCR analysis revealed that spok expression temporally correlates well with ouib expression during the larval-to-pupal transition. These results indicate that ouib is required for spok expression in the PG.
Ouija board is specifically required for the expression of spookier in the ecdysteroid biosynthesis pathway
We have also conducted in vitro reporter assays for Ouib transcriptional activity on spok, and found that a critical sequence of 15 bp within 300 bp of the spok promoter region is sufficient for Ouib-dependent gene expression in cultured cells. Furthermore, our EMSA experiment has confirmed the direct binding of Ouib Zinc finger domain to the 15 bp region.
To address how specific ouib is for regulating spok expression, we performed ecdysteroid-feeding rescue experiments. We put ouib mutant larvae on food supplemented with 20-hydroxyecdysone or several upstream precursors. Indeed, the larval arrest phenotype of ouib mutant larvae is rescued by administration of 5β-ketodiol and 20E, but not by cholesterol or 7-dehydrocholesterol. This is consistent with the hypothesized catalytic step mediated by the enzyme Spok, as this is known to act in the conversion step from 7-dehydrocholesterol to 5β-ketodiol (). Furthermore, we have performed the rescue experiments by transgenic expression. Surprisingly, overexpression of the equivalent isoform of spok almost fully restores the developmental arrest phenotype of ouib mutants and they grew up to the adult stage. These results suggest that the function of Ouib mainly dedicates to spok expression.
In general, a transcription factor has multiple target genes. In contrast, our data suggest that this is not always the case. All of our data strongly support the idea that Ouib is essential for development via mediating a transcriptional activation of only one ecdysteroidogenic enzyme Spok. Because this zinc finger TF can induce the expression of the gene “spookier,” we named its TF after “Ouija board,” which was an instrument believed to be used in Western culture to attempt communication with the dead ().
The “catalytic step-specific” transcriptional regulation for steroid hormone biosynthesis
Ouib is the first invertebrate TF specialized for steroid hormone biosynthesis. Moreover, it provides the first example of the “catalytic-step specific” transcriptional regulation for steroid hormone biosynthesis. While all of the previously known steroidogenic TFs regulate multiple enzyme genes in insects as well as mammals, our study sheds light on a specialized TF that has been evolved only for one target gene in a specific catalytic step. Although we cannot completely rule out the possibility that Ouib is also involved in direct transcriptional regulation of genes other than spok, particularly dib and sad, further studies are required to clarify the extent to which Ouib regulates other genes, and the functional importance of these genes, through a transcriptome analysis/ChIP-seq analysis together with eventual mutational analysis of any identified targets.
What is the biological significance of such catalytic-step specific regulations at the transcription level? It should be noted that Spo/Spok act in the catalytic step referred to as “BLACK BOX,” where the intermediates from 7-dC to 5β-ketodiol are presumably unstable so that they have not fully been identified.Citation19 Considering that the “BLACK BOX” has been thought to act as the rate-limiting step in the ecdysteroid biosynthesis pathway,Citation20 it raises an interesting possibility that this bottleneck for ecdysteroid biosynthesis depends on catalytic-step specific transcriptional regulation. Identification of the substrates of Spo/Spok would lead to characterization of the reaction steps in the “BLACK BOX.”
A transcriptional regulation of ecdysteroidogenic genes located in the heterochromatin region
Previous studies reported that the expression of spok is regulated by another zinc finger protein Mld.Citation19,21,22 Therefore, the transcription of spok is dependent on the interaction between at least two zinc-finger proteins, Ouib and Mld. In contrast to Ouib, Mld appears not to be specific for the regulation of spok expression. First, Mld, unlike Ouib, is expressed in several other tissues during development besides the PG. Citation21 Second, the mld loss-of-function phenotype is not rescued by overexpressing either spo or spok.Citation19 Third, consistent with results of the transgenic experiment, Mld is essential for regulating expression not only spok but also another ecdysteroidogenic genes such as nvd.Citation22 Therefore, Ouib and Mld overlap in their regulation of spok expression, but also have distinct functions during development. It would be intriguing to examine a functional relationship between Ouib and Mld for induction of spok expression in the PG.
It is of interest to note that the gene loci of spok, as well as nvd, are located in the pericentric heterochromatin region on the 3rd chromosome of D. melanogaster (). It is therefore feasible to consider that some chromatin remodeling would be necessary to regulate the expression of spok and nvd. Consistent with this notion, it has been reported that dATAC histone acetylase complex and the insulator protein CTCF regulates ecdysteroidogenic gene expression.Citation23-25 However, to our knowledge, it is largely unknown how the heterochromatin region is decondensed to allow TFs to bind to the promoter region for inducing gene expression. We assume that there is a regulatory mechanism by which Mld and Ouib temporally decondense the heterochromatin status of nvd locus and/or spok locus. Such a heterochromatin remodeling system may contribute to the spatiotemporal dynamics of ecdysteroid biosynthsesis by restricting the expression of nvd and spok. In any case, the interactions between chromatin status and transcriptional activity should be tightly regulated in the earlier steps of ecdysteroid biosynthesis.
Evolutionarily divergence of transcriptional regulation in steroid hormone biosynthesis
Intriguingly, spok has evolved only in Diptera as a palalog of spook (spo)/CYP307A family. In D. melanogaster, while spo is expressed during early embryonic development and adult ovaries, spok is expressed in the PG throughout the larval stages.Citation19,26 Therefore, duplicated genes are retained but each has acquired a different temporal and tissue-specific expression profile. In accordance with the target, orthologs of ouib and mld are found only in Diptera so far.Citation3,19 In contrast, Lepidoptera has only one spo gene and has no orthologs of ouib or mld in the genome. These studies suggest an evolutionary split between Diptera and Lepidoptera in how the ecdysone biosynthesis pathway is regulated during development. Although recently evolved in Diptera lineage, ouib, mld, and their target spok are essential for D. melanogaster development. This idea is supported by a report that the newly-evolved, non-conserved genes are indeed essential.Citation27 Our study reveals a species-specific aspect of ecdysteroid biosynthesis and further studies required to comprehensively understand the molecular mechanisms of steroid hormone biosynthesis in animals.
Are there any other TFs specialized for ecdysteroidogenic enzymes other than Spok? Quite recently, a genome-wide transcriptome analysis and a genome-wide in vivo RNAi interference screen were reported and have revealed a number of genes with potential roles in steroidogenesis and developmental timing.Citation28,29 Since some of the newly identified genes indeed encode TFs, it would be intriguing to examine if these TFs regulate ecdysteroidogenic enzyme genes in the PG cells. In addition, we are curious to examine if the Drosophilidae genome has any paralogous zinc finger genes to ouib. Because the ZAD-Zinc finger genes are strikingly duplicated and diversified in the insect lineage,Citation30 we hypothesize that any of these zinc finger genes could also be specifically involved in ecdysteroid biosynthesis in the PG.
Abbreviations
| Mld | = | Molting defective |
| Ouib | = | Ouija board |
| PG | = | prothoracic gland |
| Spo | = | Spook |
| Spok | = | Spookier |
| TF | = | transcription factor |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Outa Uryu for critical comments on the manuscript.
References
- Niwa YS, Niwa R. Transcriptional regulation of insect steroid hormone biosynthesis and its role in controlling timing of molting and metamorphosis. Dev Growth Differ 2015; 58:94-105; PMID:26667894; http://dx.doi.org/10.1111/dgd.12248
- Gehring WJ. The animal body plan, the prototypic body segment, and eye evolution. Evol Dev 2012; 14:34-46; PMID:23016973; http://dx.doi.org/10.1111/j.1525-142X.2011.00528.x
- Komura-Kawa T, Hirota K, Shimada-Niwa Y, Yamauchi R, Shimell M, Shinoda T, Fukamizu A, O'Connor MB, Niwa R. The Drosophila zinc finger transcription factor Ouija board controls ecdysteroid biosynthesis through specific regulation of spookier. PLOS Genet 2015; 11:e1005712.; PMID:26658797; http://dx.doi.org/10.1371/journal.pgen.1005712
- Omura T, Morohashi K. Gene regulation of steroidogenesis. J Steroid Biochem Mol Biol 1995; 53:19-25; PMID:7626452; http://dx.doi.org/10.1016/0960-0760(95)00036-Y
- Luo X, Ikeda Y, Parker KL. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell 1994; 77:481-490; PMID:8187173; http://dx.doi.org/10.1016/0092-8674(94)90211-9
- Sadovsky Y, Crawford PA, Woodson KG, Polish JA, Clements MA, Tourtellotte LM, Simburger K, Milbrandt J. Mice deficient in the orphan receptor steroidogenic factor 1 lack adrenal glands and gonads but express P450 side-chain-cleavage enzyme in the placenta and have normal embryonic serum levels of corticosteroids. Proc Natl Acad Sci U S A 1995; 92:10939-10943; PMID:7479914; http://dx.doi.org/10.1073/pnas.92.24.10939
- Yazawa T, Mizutani T, Yamada K, Kawata H, Sekiguchi T, Yoshino M, Kajitani T, Shou Z, Umezawa A, Miyamoto K. Differentiation of adult stem cells derived from bone marrow stroma into Leydig or adrenocortical cells. Endocrinology 2006; 147:4104-4111; PMID:16728492; http://dx.doi.org/10.1210/en.2006-0162
- Yamada M, Murata T, Hirose S, Lavorgna G, Suzuki E, Ueda H. Temporally restricted expression of transcription factor βFTZ-F1: significance for embryogenesis, molting and metamorphosis in Drosophila melanogaster. Development 2000; 127:5083-5092; PMID:11060234
- Markov GV, Tavares R, Dauphin-Villemant C, Demeneix BA, Baker ME, Laudet V. Independent elaboration of steroid hormone signaling pathways in metazoans. Proc Natl Acad Sci U S A 2009; 106:11913-11918; PMID:19571007; http://dx.doi.org/10.1073/pnas.0812138106
- Niwa R, Niwa YS. Enzymes for ecdysteroid biosynthesis: their biological functions in insects and beyond. Biosci Biotechnol Biochem 2014; 78:1283-1292; PMID:25130728; http://dx.doi.org/10.1080/09168451.2014.942250
- Uryu O, Ameku T, Niwa R. Recent progress in understanding the role of ecdysteroids in adult insects: Germline development and circadian clock in the fruit fly Drosophila melanogaster. Zool Lett 2015; 1:32; http://dx.doi.org/10.1186/s40851-015-0031-2
- Faunes F, Larraín J. Conservation in the involvement of heterochronic genes and hormones during developmental transitions. Dev Biol 2016; in press; PMID:27297887; http://dx.doi.org/10.1016/j.ydbio.2016.06.013
- Parvy J-P, Wang P, Garrido D, Maria A, Blais C, Poidevin M, Montagne J. Forward and feedback regulation of cyclic steroid production in Drosophila melanogaster. Development 2014; 141:3955-3965; PMID:25252945; http://dx.doi.org/http://dx.doi.org/10.1242/dev.102020
- Moeller ME, Danielsen ET, Herder R, O'Connor MB, Rewitz KF. Dynamic feedback circuits function as a switch for shaping a maturation-inducing steroid pulse in Drosophila. Development 2013; 140:4730-4739; PMID:24173800; http://dx.doi.org/http://dx.doi.org/10.1242/dev.099739
- Ou Q, Magico A, King-Jones K. Nuclear receptor DHR4 controls the timing of steroid hormone pulses during Drosophila development. PLOS Biol 2011; 9:e1001160; PMID:21980261; http://dx.doi.org/http://dx.doi.org/10.1371/journal.pbio.1001160
- Parvy JP, Blais C, Bernard F, Warren JT, Petryk A, Gilbert LI, O'Connor MB, Dauphin-Villemant C. A role for βFTZ-F1 in regulating ecdysteroid titers during post-embryonic development in Drosophila melanogaster. Dev Biol 2005; 282:84-94; PMID:15936331; http://dx.doi.org/http://dx.doi.org/10.1016/j.ydbio.2005.02.028
- Ueda H, Sonoda S, Brown JL, Scott MP, Wu C. A sequence-specific DNA-binding protein that activates fushi tarazu segmentation gene expression. Genes Dev 1990; 4:624-635; PMID:2113881; http://dx.doi.org/http://dx.doi.org/10.1101/gad.4.4.624
- Lavorgna G, Ueda H, Clos J, Wu C. FTZ-F1, a steroid hormone receptor-like protein implicated in the activation of fushi tarazu. Science 1991; 252:848-851; PMID:1709303; http://dx.doi.org/http://dx.doi.org/10.1126/science.1709303
- Ono H, Rewitz KF, Shinoda T, Itoyama K, Petryk A, Rybczynski R, Jarcho M, Warren JT, Marqués G, Shimell MJ et al. Spook and Spookier code for stage-specific components of the ecdysone biosynthetic pathway in Diptera. Dev Biol 2006; 298:555-570; PMID:16949568; http://dx.doi.org/http://dx.doi.org/10.1016/j.ydbio.2006.07.023
- Gilbert LI, Rybczynski R, Warren JT. Control and biochemical nature of the ecdysteroidogenic pathway. Annu Rev Entomol 2002; 47:883-916; PMID:11729094; http://dx.doi.org/http://dx.doi.org/10.1146/annurev.ento.47.091201.145302
- Neubueser D, Warren JT, Gilbert LI, Cohen SM. molting defective is required for ecdysone biosynthesis. Dev Biol 2005; 280:362-372; PMID:15882578; http://dx.doi.org/http://dx.doi.org/10.1016/j.ydbio.2005.01.023
- Danielsen ET, Moeller ME, Dorry E, Komura-Kawa T, Fujimoto Y, Troelsen JT, Herder R, O'Connor MB, Niwa R, Rewitz KF. Transcriptional control of steroid biosynthesis genes in the Drosophila prothoracic gland by Ventral veins lacking and Knirps. PLOS Genet 2014; 10:e1004343; PMID:24945799; http://dx.doi.org/http://dx.doi.org/10.1371/journal.pgen.1004343
- Pankotai T, Popescu C, Martín D, Grau B, Zsindely N, Bodai L, Tora L, Ferrús A, Boros I. Genes of the ecdysone biosynthesis pathway are regulated by the dATAC histone acetyltransferase complex in Drosophila. Mol Cell Biol 2010; 30:4254-4266; PMID:20584983; http://dx.doi.org/http://dx.doi.org/10.1128/MCB.00142-10
- Borsos BN, Pankotai T, Kovács D, Popescu C, Páhi Z, Boros IM. Acetylations of Ftz-F1 and histone H4K5 are required for the fine-tuning of ecdysone biosynthesis during Drosophila metamorphosis. Dev Biol 2015; 404:80-87; PMID:25959239; http://dx.doi.org/http://dx.doi.org/10.1016/j.ydbio.2015.04.020
- Fresan U, Cuartero S, O'Connor MB, Espinas ML. The insulator protein CTCF regulates Drosophila steroidogenesis. Biol Open 2015; 4:852-857; PMID:25979705; http://dx.doi.org/http://dx.doi.org/10.1242/bio.012344
- Namiki T, Niwa R, Sakudoh T, Shirai K-I, Takeuchi H, Kataoka H. Cytochrome P450 CYP307A1/Spook: a regulator for ecdysone synthesis in insects. Biochem Biophys Res Commun 2005; 337:367-374; PMID:16188237; http://dx.doi.org/http://dx.doi.org/10.1016/j.bbrc.2005.09.043
- Chen S, Zhang YE, Long M. New genes in Drosophila quickly become essential. Science 2010; 330:1682-1685; PMID:21164016; http://dx.doi.org/http://dx.doi.org/10.1126/science.1196380
- Ou Q, Zeng J, Yamanaka N, Brakken-Thal C, O'Connor MB, King-Jones K. The insect prothoracic gland as a model for steroid hormone biosynthesis and regulation. Cell Rep 2016; 16:247–62; PMID:27320926; http://dx.doi.org/10.1016/j.celrep.2016.05.053
- Danielsen ET, Moeller ME, Yamanaka N, Ou Q, Laursen JM, Soenderholm C, Zhuo R, Phelps B, Tang K, Zeng J et al. A Drosophila genome-wide screen identifies regulators of steroid hormone production and developmental timing. Dev Cell 2016; 37:558-570; PMID:27326933; http://dx.doi.org/http://dx.doi.org/10.1016/j.devcel.2016.05.015
- Chung H-R, Löhr U, Jäckle H. Lineage-specific expansion of the zinc finger associated domain ZAD. Mol Biol Evol 2007; 24:1934-1943; PMID:17569752; http://dx.doi.org/http://dx.doi.org/10.1093/molbev/msm121
