ABSTRACT
RBPJ is the central transcription factor that controls the Notch-dependent transcriptional response by coordinating repressing histone H3K27 deacetylation and activating histone H3K4 methylation. Here, we discuss the molecular mechanisms how RBPJ interacts with opposing NCoR/HDAC-corepressing or KMT2D/UTX-coactivating complexes and how this is controlled by phosphorylation of chromatin modifiers.
Signal transduction pathways and chromatin regulation determine gene expression programs during development and defects in chromatin regulation have been linked to disease occurrence, including carcinogenesis. The chromatin landscape of signal-dependent genes is controlled by activating and repressing chromatin modifiers that are recruited by transcription factors (TFs). How signaling to chromatin is conducted, remains largely unknown. RBPJ (recombination signal binding protein for immunoglobulin kappa J region) represents an excellent example to which extent a TF can act as a molecular switch, that either functions as an activator, in the presence of ligand, or as a repressor, in the absence of stimuli.
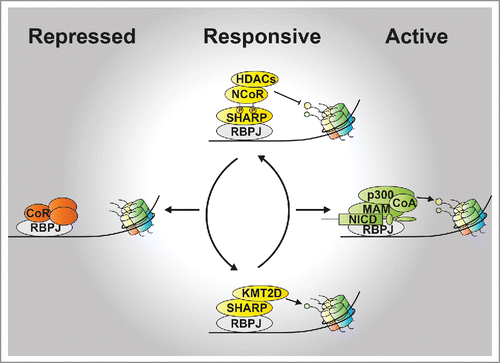
RBPJ, also known as CSL [CBF-1/RBPJ in vertebrates, Su(H) in Drosophila melanogaster, lag-1 in Caenorhabditis elegans], was originally identified by Honjo and colleagues as a protein able to bind the recombination signal sequence of the immunoglobulin Jκ segmentCitation1 and, subsequently shown to regulate peripheral nervous system development in Drosophila.Citation2 RBPJ is a signal-sensitive TF that binds both to proximal and distal enhancer elements characterized by the CGTGGGAA motif.Citation3 RBPJ is the main and key TF that governs the Notch signaling cascade. It actively represses transcription of target genes in absence of Notch signaling but, upon activation of the Notch pathway, it associates with the cleaved intracellular domain of Notch receptor (known as NICD or Notch-ICD; ). The RBPJ/NICD-containing coactivator complex displaces the RBPJ-associated repressor complex and mediates the transcriptional activation of Notch target genes ().Citation4 The signal is terminated by proteasomal degradation of the NICD that occurs via post-translational modifications (PTMs) of the NICD itselfCitation5,6 and, as consequence, the RBPJ-associated corepressor complex is reassembled at enhancer sites and repression of Notch target genes is reestablished ().
Figure 1. Model for the regulation of Notch target genes. Activation of the Notch pathway via binding of ligands to Notch receptor, leads to the release of the NICD which interacts with the transcription factor RBPJ. The RBPJ/NICD complex activates gene expression via recruiting MAM, the histone acetyltransferase (HAT) p300 and additional coactivators (CoA; active state, right). Upon termination of the signal, SHARP is recruited to RBPJ-bound enhancer sites where it keeps genes in a responsive state (responsive state, middle) via interaction with KMT2D or with phospho-NCoR. Furthermore, repression of Notch target genes can be enforced via interaction of RBPJ with corepressors (CoR; repressed state, left).
RBPJ consists of three main domains: the N-terminal domain (NTD) and the C-terminal domain (CTD) are similar to the Rel domain of Rel TFs; these domains, are separated by a β-trefoil domain (BTD) which is the characteristic of RBPJ and absent in Rel TFs.Citation7,8 While the NTD and BTD are both required for DNA binding, only the BTD interacts with the RAM domain of NICD. In addition, the CTD of RBPJ binds to the ankyrin repeats of NICD as well as to the coactivator Mastermind (MAM) the last of which, also interacts with the NTD of RBPJ.Citation7-10
The DNA-binding affinity of RBPJ is surprisingly low, indicating that there is a rapid exchange between ON and OFF-rates of RBPJ-containing complexes. Therefore, the equilibrium between RBPJ-repressor and RBPJ/NICD-activator complexes is decisive to determine the transcriptional output. There is no biophysical evidence that binding of either the corepressor SHARP (SMRT- and HDAC-associated repressor protein) or NICD to RBPJ influences its DNA-binding affinity. It remains to be elucidated how the versatile DNA binding of RBPJ is regulated, possibly the underlying mechanism includes also the action of cofactors able to read the chromatin signature. A second scenario envisages that RBPJ cooperates with other DNA-binding proteins, which as consequence reduce its diffusion rate and prolong its chromatin association. RBPJ can also compete with other TFs for binding to the same enhancer site. One good example of this mechanism is the tumor suppressor and TF Ikaros. Given that Ikaros expression strongly increases during lymphopoiesis, Ikaros abundance leads to displacement of RBPJ.Citation11,12 A third scenario is given by the observation that RBPJ/NICD-coactivator complexes dimerize at enhancer sites characterized by RBPJ-binding motifs which are oriented head-to-head.Citation13-15 The dimerization thus stabilizes the binding of each RBPJ monomer to its cognate motif on the DNA.
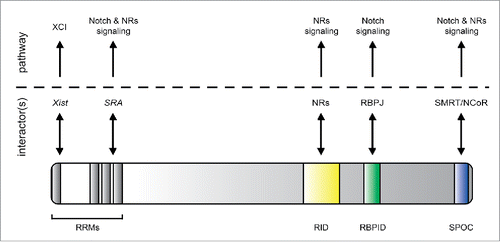
RBPJ either interacts with the SHARP-containing corepressor complex or the NICD-coactivator complex. The NICD-coactivator complex displaces the RBPJ-associated SHARP corepressor complex recruiting additional coactivators, among them MAM and the histone acetyltransferase (HAT) p300 play a key role for Notch target gene activation.Citation16 Several groups including our own have characterized the components of the RBPJ corepressor and coactivator complexes in recent years. Among the identified interactors, SHARP represents the key component of the corepressor complex in higher eukaryotes. SHARP directly interacts with RBPJCitation17 and, via its highly conserved C-terminal SPOC domain (Spen paralog and ortholog C-terminal domain), it recruits the additional corepressors CtIP/CtBP18 and ETOCitation19 as well as the NCoR/SMRT (nuclear receptor corepressor/silencing mediator for retinoid or thyroid-hormone receptors) complexCitation20,21 which link RBPJ to histone deacetylases (HDACs) (). Recently, the role of SHARP in regulating RBPJ-dependent transcription has been better defined and surprisingly, more than a simple corepressor, SHARP represents a hub for signaling events that direct gene expression both in a positive and negative fashion.Citation22 Focusing on the SPOC domain of SHARP, we have defined its interactome by mass-spectrometry (defined as SPOCome). Surprisingly, we identified not only the NCoR complex but also the KMT2D/UTX-containing coactivator complex (hereafter referred to as KMT2D complex).Citation22 KMT2D is known to methylate lysine 4 of histone 3 (H3K4me1, me2 and me3), which is associated with active chromatin.Citation23 On the other hand, SHARP is a strong transcriptional repressorCitation17 and an RBPJ-SPOC fusion protein acts as a super-repressor of Notch target genes both in vitro and in vivo.Citation22 Thus, identifying the activating KMT2D complex within the SPOCome was unexpected. Importantly, NCoR and KMT2D compete for the binding to SHARP and the phosphorylation-state of NCoR plays a key role in regulating this competition. NCoR is phosphorylated on two highly conserved C-terminal serine residues located within a highly conserved LSDSD motif at its very C-terminus.Citation24 Phospho-NCoR outcompetes KMT2D for binding to SPOCCitation22 explaining how dynamic complex composition is achieved. Thus, a phospho-dependent mechanism regulates the occupancy of opposing chromatin regulators (). These data suggest SHARP as a hub where signaling events converge and become integrated to determine time- and cell-specific transcriptional outputs. It remains to be established which kinase and phosphatase dynamically regulate the NCoR phosphorylation. So far, casein kinase-2β (CK2β) has been described to phosphorylate one serine residue within the LSDSD motif of NCoRCitation25 and we observed that casein kinase inhibition positively influences the expression of Notch target genes.Citation22 Since phosphorylation of both serine residues is required for tight binding of phospho-NCoR to SPOC, the identification of additional kinase(s) and unknown phosphatases represents an exciting challenge.
Figure 2. Schematic representation of SHARP and its functional network. SHARP is characterized by RNA recognition motifs (RRMs), the nuclear receptor interaction domain (RID) the RBPJ interaction domain (RBPID) and the SPOC domain. The RRMs of SHARP interact with the long non-coding RNAs Xist and SRA (steroid receptor coactivator). Xist links SHARP to X chromosome inactivation (XCI), whereas SRA involves SHARP in the regulation of the nuclear receptors (NRs)- and Notch signaling. The RID involves SHARP in NRs signaling, whereas the RBPID links SHARP to Notch signaling. The SPOC domain interacts with SMRT/NCoR linking SHARP to both NRs- and Notch-signaling. Additionally, the SPOC domain can interact with KMT2D, CtIP/CtBP and ETO (not shown).
The KMT2D/NCoR competition has a significant impact on the chromatin environment at the enhancers of Notch target genes. While the NCoR complex contains HDAC activities that remove acetylation from the histone tails [for example acetylation of lysine 27 of the histone 3 (H3K27ac)] having a negative impact on transcription, the KMT2D complex, promoting H3K4 methylation states, has a positive function in transcription (). The bivalent function of the RBPJ/SHARP complex gives a significant contribution in defining the dynamics of the transcriptional RBPJ-dependent switch at Notch target genes. While the RBPJ/SHARP complex keeps Notch target genes responsive via KMT2D-dependent histone methylation, activation of Notch signaling leads to NICD-dependent displacement of SHARP with concomitant recruitment of MAM and, consequently, of the HAT p30016,26 leading to full gene activation via histone acetylation (). Proteasomal-dependent NICD degradation terminates the signal allowing subsequently the recruitment of SHARP that drives histone deacetylation via the HDAC-containing phospho-NCoR complex (). Subsequently, two different scenarios can arise: Notch target genes can be maintained in a responsive state via SHARP-dependent recruitment of the KMT2D complex which function is to maintain H3K4 methylation; a recruitment which is dependent on the phospho-status of NCoR and eventually on PTMs of KMT2D (). Alternatively, responsiveness may be reduced via recruitment of histone demethylases (KDMs) leading to strong repression of Notch target genes. For example, KDM5A, the mammalian homolog of Drosophila Lid, is a direct interactor of RBPJ that decreases H3K4me3 at Notch-dependent enhancers.Citation27 Furthermore, repression of Notch target genes can be enforced by KDM1A, the homolog of Drosophila Lsd1, which is a negative regulator of Notch signaling.Citation28 Finally, additional mechanisms may be involved in modulating distinct Notch-dependent gene expression programs including, for example, Polycomb (PcG) complexes, histone variants and/or DNA methylation. Notably, the histone variant H2A.Z has been proposed to contribute to gene poising and activation but if and how it regulates Notch-dependent transcription has never been investigated.
The NCoR/KMT2D phospho-dependent switch is one of the first examples how changes of environmental signaling are integrated at the chromatin level. So far, mostly phosphorylation of transcription factors like NF-AT or FoxO have been described. Another example of a similar mechanism has been observed for the circadian gene expression program, which is regulated via acetylation of the histone methyltransferase KMT2A.Citation29 The existence of different intermediate RBPJ-associated complexes, namely the RBPJ/SHARP/KMT2D and RBPJ/SHARP/NCoR, suggests an additional layer of regulation of the Notch response that not only depends uniquely on the proteolytic release of the NICD but also on the specific time window when the release occurs. As a consequence, transcriptional responses may occur faster or slower based on the dynamic availability of RBPJ/SHARP-associated complex(es). Such regulation would be relevant during differentiation when not only gene activation is important but also gene repression as well as the timeframe required for turning ON and OFF a specific gene. Recycling RBPJ/SHARP-associated complexes might be relevant during somitogenesis when cycling expression of Notch target genes regulates the budding off of somites. Similar mechanisms might be used by other signaling pathways involved in somitogenesis like Wnt or FGF signaling pathways.
RBPJ can be seen as the prototype of such a group of TFs that are able to switch their function from repression to activation of transcription. Among those, TCF/LEF (T-cell factor/lymphoid enhancer factor), which is involved in the Wnt signaling pathway, switches from a negative to a positive transcriptional regulator upon interaction with β-catenin. As in the case of RBPJ, also the TCF/LEF switch involves a dynamic exchange of HDAC-containing corepressors with HAT-containing coactivators. Such dynamic exchange may be regulated by PTMs of chromatin modifiers. Differently to RBPJ, the switch of Gli TFs is regulated via proteolytic events. Gli proteins are converted into repressors via proteasome-dependent degradation but, upon activation of the Hedgehog signaling pathway, Gli proteins are stabilized and act as transcriptional activators. The switch of Gli proteins illustrates once more how PTMs, in this case ubiquitination, of transcriptional regulators convert a repressor into an activator. Together, in our view, RBPJ-associated factors actively shape the chromatin landscape at Notch target genes even in the absence of a Notch signal. This process is able to incorporate signaling inputs thereby modifying the outcome of the Notch response. So far we know that at the heart of this mechanism is the phosphorylation of the NCoR-corepressor.
Abbreviations
| BTD | = | β-trefoil domain |
| CoA | = | coactivator |
| CoR | = | corepressor |
| CK2β | = | casein kinase 2β |
| CSLCBF-1/RBPJSu(H) | = | CBF-1/RBPJ/Su(H)/lag-1 |
| CTD | = | C-terminal domain |
| CtIP | = | CtBP-interacting protein |
| CtBP | = | C-terminal binding protein |
| ETO | = | 8–21 |
| HAT | = | histone acetyltransferase |
| HDACs | = | histone deacetylases |
| KDM | = | lysine demethylase |
| KMT | = | lysine (K) methyltransferase |
| MAM | = | Mastermind |
| NCoR/SMRT | = | nuclear receptor corepressor/silencing mediator for retinoid or thyroid-hormone receptors |
| NICD | = | Notch intracellular domain |
| NR | = | nuclear receptor |
| NTD | = | N-terminal domain |
| PcG | = | Polycomb |
| PTM | = | post-translational modification |
| RBPID | = | RBPJ interaction domain |
| RBPJ | = | recombination signal binding protein for immunoglobulin kappa J region |
| RID | = | nuclear receptor interaction domain |
| RRM | = | RNA recognition motif |
| SHARP | = | SMRT- and HDAC-associated repressor protein |
| SPOC | = | Spen paralog and ortholog C-terminal domain |
| SPOCome | = | SPOC interactome |
| SRA | = | steroid receptor coactivator |
| Su(H) | = | suppressor of hairless |
| TCF/LEF | = | T-cell factor/lymphoid enhancer factor |
| TF | = | transcription factor |
| XCI | = | X-chromosome inactivation |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors are grateful to Dr Bernd Baumann for his helpful comments and suggestions.
Funding
Funding was provided by the collaborative research grant TRR81 and the Heisenberg program (BO 1,639/5–1), by the Deutsche Forschungsgemeinschaft (DFG), the Excellence Cluster for Cardio Pulmonary System (ECCPS) and the Max-Planck society to T.B. This work was also supported by the DFG through a collaborative research grant (SFB 1074/A3) and by the BMBF (Federal Ministry of Education and Research, research nucleus SyStAR) to FO. Funding for open access charge was provided by the DFG collaborative research TRR81.
References
- Hamaguchi Y, Matsunami N, Yamamoto Y, Honjo T. Purification and characterization of a protein that binds to the recombination signal sequence of the immunoglobulin J kappa segment. Nucleic Acids Res 1989; 17:9015-9026; PMID:2511556; http://dx.doi.org/10.1093/nar/17.22.9015
- Furukawa T, Maruyama S, Kawaichi M, Honjo T. The Drosophila homolog of the immunoglobulin recombination signal-binding protein regulates peripheral nervous system development. Cell 1992; 69:1191-1197; PMID:1617729; http://dx.doi.org/10.1016/0092-8674(92)90640-X
- Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M. Recognition sequence of a highly conserved DNA binding protein RBP-J kappa. Nucleic Acids Res 1994; 22:965-971; PMID:8152928; http://dx.doi.org/10.1093/nar/22.6.965
- Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci 2009; 66:1631-1646; PMID:19165418; http://dx.doi.org/10.1007/s00018-009-8668-7
- Hein K, Mittler G, Cizelsky W, Kühl M, Ferrante F, Liefke R, Berger IM, Just S, Sträng JE, Kestler HA et al. Site-specific methylation of Notch1 controls the amplitude and duration of the Notch1 response. Sci Signal 2015; 8:ra30; PMID:25805888; http://dx.doi.org/10.1126/scisignal.2005892
- Borggrefe T, Lauth M, Zwijsen A, Huylebroeck D, Oswald F, Giaimo BD. The Notch intracellular domain integrates signals from Wnt, Hedgehog, TGFbeta/BMP and hypoxia pathways. Biochim Biophys Acta 2016; 1863:303-313; PMID: 26592459; http://dx.doi.org/10.1016/j.bbamcr.2015.11.020
- Nam Y, Weng AP, Aster JC, Blacklow SC. Structural requirements for assembly of the CSL.intracellular Notch1.Mastermind-like 1 transcriptional activation complex. J Biol Chem 2003; 278:21232-21239; PMID:12644465; http://dx.doi.org/10.1074/jbc.M301567200
- Kovall RA, Hendrickson WA. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J 2004; 23:3441-3451; PMID:15297877; http://dx.doi.org/10.1038/sj.emboj.7600349
- Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 2006; 124:973-983; PMID:16530044; http://dx.doi.org/10.1016/j.cell.2005.12.037
- Wilson JJ, Kovall RA. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell 2006; 124:985-996; PMID:16530045; http://dx.doi.org/10.1016/j.cell.2006.01.035
- Geimer Le Lay AS, Oravecz A, Mastio J, Jung C, Marchal P, Ebel C, Dembélé D, Jost B, Le Gras S, Thibault C et al. The tumor suppressor Ikaros shapes the repertoire of notch target genes in T cells. Sci Signal 2014; 7:ra28; PMID:24643801; http://dx.doi.org/10.1126/scisignal.2004545
- Chari S, Winandy S. Ikaros regulates Notch target gene expression in developing thymocytes. J Immunol 2008; 181:6265-6274; PMID:18941217; http://dx.doi.org/10.4049/jimmunol.181.9.6265
- Nam Y, Sliz P, Pear WS, Aster JC, Blacklow SC. Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc Natl Acad Sci U S A 2007; 104:2103-2108; PMID:17284587; http://dx.doi.org/10.1073/pnas.0611092104
- Hass MR, Liow HH, Chen X, Sharma A, Inoue YU, Inoue T, Reeb A, Martens A, Fulbright M, Raju S et al. SpDamID: Marking DNA Bound by Protein Complexes Identifies Notch-Dimer Responsive Enhancers. Mol Cell 2015; 59:685-697; PMID:26257285; http://dx.doi.org/10.1016/j.molcel.2015.07.008
- Liu H, Chi AW, Arnett KL, Chiang MY, Xu L, Shestova O, Wang H, Li YM, Bhandoola A, Aster JC et al. Notch dimerization is required for leukemogenesis and T-cell development. Genes Dev 2010; 24:2395-2407; PMID:20935071; http://dx.doi.org/10.1101/gad.1975210
- Oswald F, Täuber B, Dobner T, Bourteele S, Kostezka U, Adler G, Liptay S, Schmid RM. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol Cell Biol 2001; 21:7761-7774; PMID:11604511; http://dx.doi.org/10.1128/MCB.21.22.7761-7774.2001
- Oswald F, Kostezka U, Astrahantseff K, Bourteele S, Dillinger K, Zechner U, Ludwig L, Wilda M, Hameister H, Knöchel W et al. SHARP is a novel component of the Notch/RBP-Jkappa signaling pathway. EMBO J 2002; 21:5417-5426; PMID:12374742; http://dx.doi.org/10.1093/emboj/cdf549
- Oswald F, Winkler M, Cao Y, Astrahantseff K, Bourteele S, Knöchel W, Borggrefe T. RBP-Jkappa/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol Cell Biol 2005; 25:10379-10390; PMID:16287852; http://dx.doi.org/10.1128/MCB.25.23.10379-10390.2005
- Salat D, Liefke R, Wiedenmann J, Borggrefe T, Oswald F. ETO, but not leukemogenic fusion protein AML1/ETO, augments RBP-Jkappa/SHARP-mediated repression of notch target genes. Mol Cell Biol 2008; 28:3502-3512; PMID:18332109; http://dx.doi.org/10.1128/MCB.01966-07
- Shi Y, Downes M, Xie W, Kao HY, Ordentlich P, Tsai CC, Hon M, Evans RM. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev 2001; 15:1140-1151; PMID:11331609; http://dx.doi.org/10.1101/gad.871201
- Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev 1998; 12:2269-2277; PMID:9694793; http://dx.doi.org/10.1101/gad.12.15.2269
- Oswald F, Rodriguez P, Giaimo BD, Antonello ZA, Mira L, Mittler G, Thiel VN, Collins KJ, Tabaja N, Cizelsky W et al. A phospho-dependent mechanism involving NCoR and KMT2D controls a permissive chromatin state at Notch target genes. Nucleic Acids Res 2016; 44:4703-4720; PMID:26912830; http://dx.doi.org/10.1093/nar/gkw105
- Mohan M, Herz HM, Shilatifard A. SnapShot: Histone lysine methylase complexes. Cell 2012; 149:498–498; ; http://dx.doi.org/10.1016/j.cell.2012.03.025
- Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A 2008; 105:10762-10767; PMID:18669648; http://dx.doi.org/10.1073/pnas.0805139105
- Yoo JY, Choi HK, Choi KC, Park SY, Ota I, Yook JI, Lee YH, Kim K, Yoon HG. Nuclear hormone receptor corepressor promotes esophageal cancer cell invasion by transcriptional repression of interferon-gamma-inducible protein 10 in a casein kinase 2-dependent manner. Mol Biol Cell 2012; 23:2943-2954; PMID:22675025; http://dx.doi.org/10.1091/mbc.E11-11-0947
- Jung C, Mittler G, Oswald F, Borggrefe T. RNA helicase Ddx5 and the noncoding RNA SRA act as coactivators in the Notch signaling pathway. Biochim Biophys Acta 2013; 1833:1180-1189; PMID: 23396200; http://dx.doi.org/10.1016/j.bbamcr.2013.01.032
- Liefke R, Oswald F, Alvarado C, Ferres-Marco D, Mittler G, Rodriguez P, Dominguez M, Borggrefe T. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev 2010; 24:590-601; PMID:20231316; http://dx.doi.org/10.1101/gad.563210
- Mulligan P, Yang F, Di Stefano L, Ji JY, Ouyang J, Nishikawa JL, Toiber D, Kulkarni M, Wang Q, Najafi-Shoushtari SH et al. A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol Cell 2011; 42:689-699; PMID:21596603; http://dx.doi.org/10.1016/j.molcel.2011.04.020
- Aguilar-Arnal L, Katada S, Orozco-Solis R, Sassone-Corsi, P. NAD(+)-SIRT1 control of H3K4 trimethylation through circadian deacetylation of MLL1. Nat Struct Mol Biol 2015; 22:312-318; PMID:25751424; http://dx.doi.org/10.1038/nsmb.2990
