ABSTRACT
Transcription factors TBP and TF(II)B assemble with RNA polymerase at the promoter DNA forming the initiation complex. Despite a high degree of conservation, the molecular binding mechanisms of archaeal and eukaryotic TBP and TF(II)B differ significantly. Based on recent biophysical data, we speculate how the mechanisms co-evolved with transcription regulation and TBP multiplicity.
Introduction
A fundamental difference between bacterial and eukaryotic/archaeal transcription initiation lies in the way the RNA polymerase (RNAP) is recruited to the promoter. In bacteria, promoter recognition is achieved by the RNAP holoenzyme, the complex formed by RNAP and one of the bacterial sigma factors. In contrast, archaeal and eukaryotic (nuclear) RNAP are recruited by basal transcription factors that are pre-assembled at the promoter DNA to form the pre-initiation complex (PIC). In both archaea and eukaryotes, PIC formation serves as an important point of transcriptional regulation. Hence, understanding the molecular mechanisms of basal transcription factors interacting with promoter DNA is a prerequisite to understand the functional basis of eukaryotic and archaeal transcriptional regulators. Here, we describe how recent single-molecule investigations provide new insights into transcription factor–DNA interactions, thereby revealing new levels for transcriptional regulation at the initiation phase of transcription. Based on these data, we discuss how the function of additional transcription factors co-evolved with the mode of the TBP–DNA interaction in the respective transcription system.
Conservation of archaeal and eukaryotic transcription machineries
In archaea and eukaryotes, transcription initiation starts with the recognition of the promoter DNA by the basal transcription factor TBP (TATA-binding protein) approximately 30 base pairs upstream of the transcription start site (TSS). TBP specifically recognizes a sequence motif rich in adenine and thymine, the TATA-box. TBP shows a high degree of conservation in sequence and on the structural level in the archaeal–eukaryotic lineage.Citation1 TBP binding to the promoter is essential for the recruitment of the second conserved basal transcription factor termed TFB (transcription factor B) in archaea and TFIIB in the eukaryotic RNAP II system. TFIIB paralogues are also present in the RNAP I and III transcription machineries, termed TAF1B and Brf1 in human, respectively.Citation2 TFB/TFIIB forms additional sequence-specific contacts with the DNA upstream or downstream of the TATA-box, a region termed B-recognition element (BRE), thereby orientating the PIC at the promoter. The molecular interactions and the domain organization of TFB/TFIIB are conserved in the archaeo–eukaryotic domain. Recruitment of the RNAP by TFB/TFIIB is promoted by intimate interactions of the B-linker, B-reader and B-ribbon domain of TFB/TFIIB with the RNAP. RNAP II is composed of 12 subunits and with the exception of subunits Rpb9 and Rpb8 that are absent in all or some archaeal RNAPs, respectively, the basic subunit composition is conserved between RNAP II and archaeal RNAPs.Citation1
The overall structural organization of TBP–DNA complexes and additional transcription factors like TFIIB and TFIIA was disclosed by X-ray crystallography.Citation3 These structures showed that the insertion of two sets of conserved phenylalanines of TBP into the minor groove leads to a pronounced bending of the DNA by approximately 90°. Stopped-flow measurements gave access to the kinetics of the eukaryotic TBP–DNA interaction suggesting that complex formation follows a three-step binding mechanism.Citation4 Whether these TBP–DNA intermediates differ in their DNA bending angle remained unclear until sophisticated bio-compatible single-molecule methods were developed. Förster resonance energy transfer (FRET) can be used to detect even minor conformational changes in biologic systems and has been applied successfully to gain insights into TBP-induced DNA bending on the single-molecule level (). This way, the dynamics of the DNA bending process, the heterogeneity of the bending angle and the influence of additional transcription factors like TFB/TFIIB could be monitored.Citation5-7 In our recent studies, we performed a direct comparison between archaeal and eukaryotic transcription initiation following the promoter bending pathways. Moreover, we were recently able to quantify the mechano-sensitivity of TBP-induced DNA bending. The question how initiation complex assembly is influenced by forces and torques that histone proteins or the DNA replication machinery exert when operating on DNACitation8 were a matter of discussion but could not be addressed mainly due to methodological limitations. Minor changes in DNA conformation such as introduced by TBP binding are beyond the sensitivity of typical force measurement approaches like atomic force microscopy, optical or magnetic tweezers. The advent of self-assembled programmable DNA devices allowed us to construct a DNA origami force clamp that exploits the entropic spring behavior of ssDNA to exert defined, tunable forces on the DNA of interest, in this case the TATA-box containing promoter DNA ( and ).Citation9 This nanoscopic force clamp requires no connection to the macroscopic world and enabled us to quantify the efficiency of TBP-induced DNA bending at different forces.
Figure 1. Modern single-molecule approaches to deduce biophysical parameters of TBP-induced promoter DNA bending. TBP associates with the DNA at the TATA-box. FRET between a donor and acceptor fluorophore placed on the DNA allows the quantification of TBP-induced DNA bending. TBP-free DNA exhibits a low FRET efficiency, while association of TBP results in a bent state with high FRET efficiency due to a closer spatial arrangement of the dyes. (A) FRET can be measured on the single-molecule level using for example total internal reflection (TIRF) microscopy. Here, the labeled DNA is immobilized on a biocompatible surface. Shown is a camera image of the immobilized DNA in the sample chamber with donor-labeled molecules in green, acceptor-labeled molecules in red and DNA that carry both dyes in yellow. (B) The FRET signal of hundreds of individual DNA molecules can be monitored simultaneously over time. This way, dynamic events like the association and dissociation of TBP can be detected by a rapid change in FRET efficiency. From these measurements, the distribution between low FRET (free DNA) and high FRET (TBP*DNA complex) states can be calculated (C) and the lifetime (τ) of the TBP–DNA complex can be derived (D). (E) The arrival of new nanotechnological tools allows force-dependent measurements of TBP-induced DNA bending. A nanosized force clamp built from DNA harbors single-stranded DNA sections that act as entropic springs, thereby exerting controlled tension on the double-stranded promoter DNA segment. TBP-induced conformational changes in the DNA can be monitored by single-molecule FRET. (F) The exerted force depends on the length of the spring and can be adjusted to forces in the low piconewton range. The high FRET population gradually disappears with increasing forces and bending is almost completely suppressed at 11.4 pN.
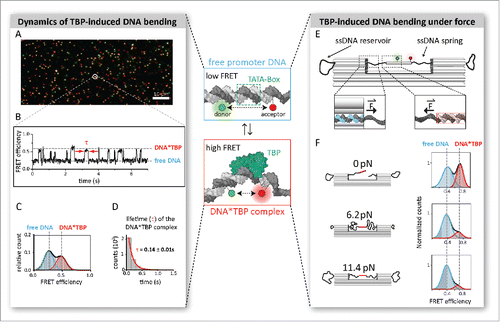
Figure 2. Molecular mechanism of promoter DNA bending by transcription initiation factors TBP and TFB. (A) Bending of the promoter DNA in the euryarchaeal transcription system (M. jannaschii) only requires TBP. The TBP–DNA interaction is highly dynamic with short complex life times (0.18 seconds). (B) In contrast, the crenarchaeal transcription system of S. acidocaldarius relies on the co-action of TBP and TFB to yield a bent TBP–TFB–DNA complex with increased stability (lifetime of 2.1 seconds) as compared with M. jannaschii. (C) While the archaeal systems show a one-step bending mechanism, eukaryotic TBP induces two interconverting states of DNA bending. Eukaryotic TBP–DNA complexes are highly stable for minutes. TFIIB binding stabilizes the fully bent state thereby converting the TBP–DNA complex into the transcriptional active TBP–TFIIB–DNA complex.
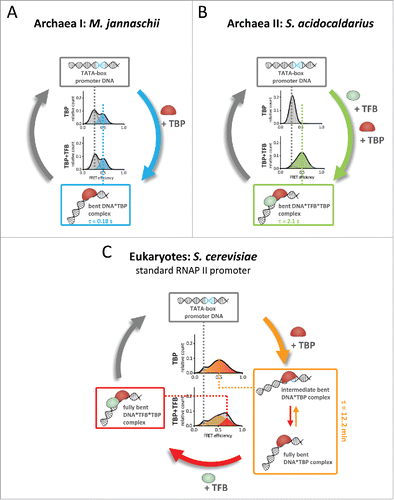
Mechanistic differences between eukaryotic and archaeal TBP action
We and others showed that eukaryotic TBP forms long-lived complexes with TATA-containing promoter DNAs that are stable for minutes to hours.Citation5-7 Archaeal TBP, however, associates with and bends the DNA only transiently. In contrast to the eukaryotic RNAP II system, no intermediate states were observed rendering TBP-induced bending a single step process ( and ). This observation was made for TBPs isolated from two evolutionary distant branches of the archaea, both hyperthermophilic organisms: Methanocaldococcus jannaschii (belonging to the euryarchaeota) and Sulfolobus acidocaldarius (belonging to the crenarchaeota). This suggests that the one-step bending mechanism and short complex lifetime in the range of milliseconds is a general hallmark of the archaeal TBP-promoter DNA interaction. Using archaeal TBP from M. jannaschii, we could demonstrate that DNA bending is force-dependent and almost completely suppressed if forces reach 10 pN. Here, a stabilization of the short-lived archaeal TBP–DNA interaction by additional factors—especially in the context of histone-bound DNA—seems to be important to ensure efficient initiation complex formation.
Intriguingly, and unlike all other TBP–DNA interactions described so far, DNA bending by S. acidocaldarius TBP in vitro strictly required the presence of TFB ().Citation7 Efficient bending of the DNA requires an intact TFB:TBP interface as well as TFB binding to the BRE.
Similar to S. acidocaldarius TFB, yeast TFIIB plays a role in TBP promoter DNA bending. In our measurements the yeast TBP–DNA complex is characterized by two interconverting TBP–DNA complexes and TF(II)B is able to shift the equilibrium toward the most bent state (). While TF(II)B does not control the initial bending, it still has regulatory potential on fast time scale as it influences the millisecond equilibrium between different bending states. Consequently, it can increase the number of transcriptional competent early initiation complexes as RNAP II is most likely recruited to the fully bent TBP/TF(II)B complex. The same is true for TFB in the Sulfolobus system albeit the archaeal system appears to be a more “simplified” version of this regulatory mechanism as it is lacking the intermediate bent state and functions in an on/off mode.
The TBP–TATA box interaction co-evolved with transcription regulation in eukaryotes and archaea
One major feature distinguishing the archaeal and the eukaryotic RNAP II transcription machinery is the incorporation of eukaryotic TBP into larger complexes such as TFIID or SAGA-like complexes that encompass several TBP-associated factors (TAFs). At least two mechanisms to regulate TFIID recruitment appear to have evolved. First, the N-terminal domain of TAF1 mimics the TATA-box DNA, thereby denying access of promoter DNA to the concave DNA-binding surface of TBP.Citation10 This blockage is relieved by TFIIA.Citation11 Second, TFIIA appears to change the equilibrium between different conformations of human TFIID towards the DNA binding-competent form.Citation12 These mechanisms both involve conformational changes of TFIID. On the other hand, genomic distribution of TFIID is actively controlled by the remodeler Mot1 and NC2,Citation13 which might have evolved as an answer to the need of fast removal of TBP from the promoter. Mot1 is able to displace TBP from TATA-containing promoters bound by the SAGA-complex but not from TATA-less promoters that are recognized by TFIID.Citation14 In summary, transcription initiation by RNAP II involves tight control of promoter-access and removal of TBP-containing complexes.
The notion that the behavior of TFIIB is representing a regulatory checkpoint was furthermore supported by another recent single-molecule study that monitored the binding of basal transcription factors of the human RNAP II system to promoter DNA.Citation15 Zhang et al. observed the rapid association and dissociation of human TFIIB on promoter DNA in the presence of TFIID and TFIIA. Stable association of TFIIB is only observed when TFIIF/RNAP II is added, suggesting that TFIIB can act as checkpoint for PIC formation responding to the availability of preformed TFIIF–RNAP II complexes.
In contrast, regulation of transcription initiation in archaea appears to follow entirely different pathways. The rapid association and dissociation of the archaeal basal transcription factor-promoter DNA complexes seems to be functionally linked with the bacteria-type mechanism of transcription repression where repressors block the access of basal transcription factors to promoter elements.Citation16 In addition, it enables transcription activation via stabilization of the ternary complex. In M. jannaschii, the transcriptional activator Ptr2 stabilizes the TBP–DNA interaction and is able to stimulate transcription by a factor of 40.Citation17 Similarly, in Pyrococcus furiosus, TFB-RF1 aids ternary complex formation by stabilizing weak TFB–BRE interactions.Citation18
Evolution of TBP and TFB/TFIIB multiplicity in eukaryotes and archaea
Eukaryotes partition their transcriptional space by using different sets of basal transcription factors and RNAPs. The most conserved and evolutionary oldest of these partitions is the devolvement of rRNA, mRNA and tRNA/5S rRNA transcription to the RNAP I, II and III systems, respectively. Recognition of their cognate promoters involves additional basal transcription factors interacting with specific promoter motifs. Several TAFs in TFIID mediate contacts to promoter motifs: The TAF1/2 heterodimer interacts with the initiator (Inr), while the winged helix of TAF1 appears to contact also the downstream promoter element (DPE).Citation19 These additional promoter elements allow the RNAP II machinery to act independent of a TATA-box being present in the promoter. In fact, only 10–15% of the mammalian promoters contain a TATA-box,Citation20,21 while “TATA-like elements” with up to two mismatches appear to be generally present in all RNAP II promoters in yeast based on ChIP-exo data.Citation22 The RNAP III machinery uses independent basal transcription factors such as TFIIIC, TFIIIA and SNAPc acting on different classes of RNAP III promoters, thereby taking over the function of initial promoter recognition.
It is remarkable that these additional interactions between basal transcription factors and promoter elements allowed TBP to serve as single basal transcription factor for all three eukaryotic RNAPs. However, in several eukaryotic lineages, paralogues of TBP evolved that lead to further partitioning of the transcriptional space.Citation23 A good example is TRF2 driving transcription of ribosomal protein genes in Drosophila.
In archaea, TBP functions generally as a monomeric protein, and TAF homologues are absent. Many archaeal species possess multiple TBP and TFB paralogues, and their initial discovery prompted the idea that they function analogous to bacterial sigma factors. Pioneering studies have provided insight on TBP and TFB multiplicity in several archaea species.Citation24-27 Because TBP and particularly TFB multiplicity seems to have originated many times independently throughout all archaeal lineages, the pioneering studies may not have given us the complete picture. It is noteworthy, however, that thus far none of these studies achieved to establish consensus promoter motifs for the different paralogues. The original concept of multiple TBPs and TFBs acting analogous to bacterial sigma factors might thus be somewhat misleading. The TATA-box consensus shows strong conservation from archaea to eukaryotes, reflecting the restrictions on the physicochemical properties of the DNA to allow bending. In contrast, the BRE consensus identified for Sulfolobus TFBCitation28 and human TFIIBCitation29 differ strongly, highlighting the greater potential of the BRE compared with the TATA-box for discriminatory binding between different TFB paralogues. Indeed, the seven TFB paralogues present in the euryarchaeon Halobacterium preferentially bind to slightly different BRE sequences in vivo.Citation30 It is tempting to speculate that the greater role of TFB and its interaction with the BRE in DNA bending by TBP that we observed for S. acidocaldarius (an organism with a single TBP, but multiple TFB paralogues) might improve the partitioning of transcription by multiple TFBs.Citation7 Halobacterium is the archaeon for which TBP and TFB multiplicity has been studied most intensely, and it highlights the important role of protein–protein interactions in the functional diversification of TBP and TFB paralogues. Next to seven TFB paralogues, Halobacterium also encodes six TBP paralogues. Seven specific pairs of TFB and TBP (out of the 42 possible combinations) were identified by co-immunoprecipitation and ChIP-seq experiments.Citation24,30 In a few archaeal species such as Methanosarcina, multiple TBP paralogues exist alongside a single TFB paralogue.Citation27 Without much variation in the TATA-box being possible and no specific TBP–TFB pairs, how might the multiple TBPs in Methanosarcina achieve their role in transcription regulation? Part of the answer might be transcriptional activators acting by recruitment of TBP similar to Ptr2. The number of biochemically characterized transcription activators in archaea, however, is thus far very limited.
Conclusions
With the arrival of the “single-molecule biochemistry” era the dynamics, assembly pathways, three-dimensional architecture and spatio-temporal distribution of transcriptional complexes can be explored with unseen precision in vitro and in vivo. In addition, the powerful marriage of DNA nanotechnology and smFRET provides the opportunity to study interaction of basal transcription factors (and histones) with DNA at biologic relevant forces. We think that these new developments might ultimately facilitate a more complete characterization of transcriptional regulators affecting basal transcription factor recruitment.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Finn Werner, Thomas Fouqueau (both University College London) and Robert Reichelt (University of Regensburg) for critical reading of the manuscript and Philip Nickels (LMU, Munich) for help with the production of figures.
Funding
DG acknowledges funding by DFG (SFB960 TP A7). This work was further supported by a DFG Research Fellowship DFG (BL 1189/1-1) to FB.
References
- Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol 2011; 9(2):85-98; PMID: 21233849; https://doi.org/10.1038/nrmicro2507
- Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell 2012; 45(4):439-446; PMID: 22365827; https://doi.org/10.1016/j.molcel.2012.01.023
- Cramer P. A tale of chromatin and transcription in 100 structures. Cell 2014; 159(5):985-994; PMID: 25416940; https://doi.org/10.1016/j.cell.2014.10.047
- Delgadillo RF, Whittington JE, Parkhurst LK, Parkhurst LJ. The TATA-binding protein core domain in solution variably bends TATA sequences via a three-step binding mechanism. Biochemistry 2009; 48(8):1801-1809; PMID: 19199812; https://doi.org/10.1021/bi8018724
- Schluesche P, Stelzer G, Piaia E, Lamb DC, Meisterernst M. NC2 mobilizes TBP on core promoter TATA boxes. Nat Struct Mol Biol 2007; 14(12):1196-1201; https://doi.org/10.1038/nsmb1328
- Blair RH, Goodrich JA, Kugel JF. Single-molecule fluorescence resonance energy transfer shows uniformity in TATA binding protein-induced DNA bending and heterogeneity in bending kinetics. Biochemistry 2012; 977:203-215; PMID: 22934924
- Gietl A, Holzmeister P, Blombach F, Schulz S, von Voithenberg LV, Lamb DC, Werner F, Tinnefeld P, Grohmann D. Eukaryotic and archaeal TBP and TFB/TF(II)B follow different promoter DNA bending pathways. Nucleic Acids Res 2014; 42(10):6219-6231; PMID: 24744242; https://doi.org/10.1093/nar/gku273
- Becker NB, Everaers R. DNA nanomechanics in the nucleosome. Structure 2009; 17(4):579-589; PMID: 19368891; https://doi.org/10.1016/j.str.2009.01.013
- Nickels PC, Wünsch B, Holzmeister P, Bae W, Kneer LM, Grohmann D, Tinnefeld P, Liedl T. Molecular force spectroscopy with a DNA origami-based nanoscopic force clamp. Science 2016; 354(6310):305-307; PMID: 27846560; https://doi.org/10.1126/science.aah5974
- Liu D, Ishima R, Tong KI, Bagby S, Kokubo T, Muhandiram DR, Kay LE, Nakatani Y, Ikura M. Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell 1998; 94(5):573-583; PMID: 9741622; https://doi.org/10.1016/S0092-8674(00)81599-8
- Bagby S, Mal TK, Liu D, Raddatz E, Nakatani Y, Ikura M. TFIIA-TAF regulatory interplay: NMR evidence for overlapping binding sites on TBP. FEBS Lett 2000; 468(2–3):149-154; PMID: 10692576; https://doi.org/10.1016/S0014-5793(00)01213-8
- Cianfrocco MA, Kassavetis GA, Grob P, Fang J, Juven-Gershon T, Kadonaga JT, Nogales E. Human TFIID binds to core promoter DNA in a reorganized structural state. Cell 2013; 152(1–2):120-131; PMID: 23332750; https://doi.org/10.1016/j.cell.2012.12.005
- Auble DT. The dynamic personality of TATA-binding protein. Trends Biochem Sci 2009; 34(2):49-52; PMID: 19038550; https://doi.org/10.1016/j.tibs.2008.10.008
- Zentner GE, Henikoff S. Mot1 redistributes TBP from TATA-containing to TATA-less promoters. Mol Cell Biol 2013; 33(24):4996-5004; PMID: 24144978; https://doi.org/10.1128/MCB.01218-13
- Zhang Z, English BP, Grimm JB, Kazane SA, Hu W, Tsai A, Inouye C, You C, Piehler J, Schultz PG et al. Rapid dynamics of general transcription factor TFIIB binding during preinitiation complex assembly revealed by single-molecule analysis. Genes Dev 2016; 30(18):2106-2118; ; https://doi.org/10.1101/gad.285395.116
- Bell SD, Jackson SP. Transcription and translation in archaea: a mosaic of eukaryal and bacterial features. Trends Microbiol 1998; 6(6):222-228; PMID: 9675798; https://doi.org/10.1016/S0966-842X(98)01281-5
- Ouhammouch M, Dewhurst RE, Hausner W, Thomm M, Geiduschek EP. Activation of archaeal transcription by recruitment of the TATA-binding protein. Proc Natl Acad Sci USA 2003; 100(9):5097-5102; PMID: 12692306; https://doi.org/10.1073/pnas.0837150100
- Ochs SM, Thumann S, Richau R, Weirauch MT, Lowe TM, Thomm M, Hausner W. Activation of archaeal transcription mediated by recruitment of transcription factor B. J Biol Chem 2012; 287(22):18863-18871; PMID: 22496454; https://doi.org/10.1074/jbc.M112.365742
- Louder RK, He Y, López-Blanco JR, Fang J, Chacón P, Nogales E. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 2016; 531(7596):604-609; PMID: 27007846; https://doi.org/10.1038/nature17394
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engström PG, Frith MC et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 2006; 38(6):626-635; PMID: 16645617; https://doi.org/10.1038/ng1789
- Kadonaga JT. Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip Rev Dev Biol 2012; 1(1):40-51; PMID: 23801666; https://doi.org/10.1002/wdev.21
- Rhee HS, Pugh BF, Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 2012; 483(7389):295-301; PMID: 22258509; https://doi.org/10.1038/nature10799
- Duttke SH. Evolution and diversification of the basal transcription machinery. Trends Biochem Sci 2015; 40(3):127-129; PMID: 25661246; https://doi.org/10.1016/j.tibs.2015.01.005
- Facciotti MT, Reiss DJ, Pan M, Kaur A, Vuthoori M, Bonneau R, Shannon P, Srivastava A, Donohoe SM, Hood LE et al. General transcription factor specified global gene regulation in archaea. Proc Natl Acad Sci USA 2007; 104(11):4630-4635; PMID: 17360575; https://doi.org/10.1073/pnas.0611663104
- Santangelo TJ, Cubonová L, James CL, Reeve JN. TFB1 or TFB2 is sufficient for Thermococcus kodakaraensis viability and for basal transcription in vitro. J Mol Biol 2007; 367(2):344-357; PMID: 17275836; https://doi.org/10.1016/j.jmb.2006.12.069
- Micorescu M, Grünberg S, Franke A, Cramer P, Thomm M, Bartlett M. Archaeal transcription: function of an alternative transcription factor B from Pyrococcus furiosus. J Bacteriol 2008; 190(1):157-167; PMID: 17965161; https://doi.org/10.1128/JB.01498-07
- Reichlen MJ, Murakami KS, Ferry JG. Functional analysis of the three TATA binding protein homologs in Methanosarcina acetivorans. J Bacteriol 2010; 192(6):1511-1517; PMID: 20081030; https://doi.org/10.1128/JB.01165-09
- Bell SD, Kosa PL, Sigler PB, Jackson SP. Orientation of the transcription preinitiation complex in archaea. Proc Natl Acad Sci USA 1999; 96(24):13662-13667; PMID: 10570129; https://doi.org/10.1073/pnas.96.24.13662
- Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev 1998; 12(1):34-44; PMID: 9420329; https://doi.org/10.1101/gad.12.1.34
- Seitzer P, Wilbanks EG, Larsen DJ, Facciotti MT. A Monte Carlo-based framework enhances the discovery and interpretation of regulatory sequence motifs. BMC Bioinform 2012; 13:317; PMID: 23181585; https://doi.org/10.1186/1471-2105-13-317
