ABSTRACT
Protein arginine methyltransferase (PRMT) is a family of nine proteins catalyzing the methylation of arginine residues. They were recently shown to be essential for proper regeneration of skeletal muscles. However, the mechanisms triggering the methylation event, as well as how the methylated substrates regulate muscle stem cell function and fate decision remain to be determined. This point-of-view will discuss the recent findings on the specific role of PRMT1, CARM1/PRMT4, PRMT5, and PRMT7 in muscle stem cell fate guidance, and shed light on the future challenges which could help defining the therapeutic potential of PRMT inhibitors against muscular disorders and aging.
Introduction
The process of tissue regeneration requires both the maintenance of the adult stem cell pool of the tissue and the preservation of the regenerative capacity of these cells throughout one individual's lifetime. The underlying mechanisms of the regeneration process are tightly regulated in a temporal and spatial manner. In response to injury, external signal(s) trigger the activation of the adult stem cell in their niche and direct their fate through differentiation and ultimately participate to tissue regeneration.
In skeletal muscle, the muscle stem cells (MSC) also called satellite cells are the main adult stem cells responsible for the muscle regeneration and are indispensable to fulfill this role.Citation1 Upon injury, quiescent MSC respond to cues and activate sequentially the expression of transcription factors called myogenic regulatory factors (MRF): Pax7, Myf5, MyoD, Myogenin, and Myf6, to guide the cell through self-renewal, proliferation, and differentiation. However, the exact molecular mechanisms governing the equilibrium of MSC fate decisions are not fully understood.
PRMTs, guardians of MSC fate
In the past few years, the protein arginine methyltransferase (PRMT) family, which catalyze the methylation of arginine residues, have been brought under the spotlight in the field of stem cell research.Citation2-5 The PRMT family comprises of nine members which are classed according to their catalytic activity: type I enzymes (ex: PRMT1) catalyze arginine asymmetrical dimethylation, type II enzymes (ex: PRMT5) catalyze arginine symmetrical dimethylation, and the unique type III enzyme PRMT7 catalyzes arginine monomethylation.Citation6 Several PRMT members have been shown to be essential for skeletal muscle regeneration in adults, although dispensable for myogenesis.
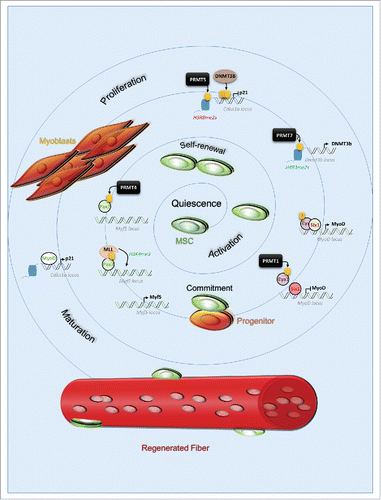
In response to injury, MSC exit quiescence to enter proliferation. The stem cells divide symmetrically to self-renew, or asymmetrically to give rise to progenitors. In adult mice, this cell fate decision is partially regulated by CARM1 (PRMT4), which promote expression of Myf5 via direct methylation of Pax7. Methylated Pax7 subsequently recruits the H3K4me3 methylation complex mixed lineage leukemia (MLL) at the Myf5 locus and activate its expression ().Citation4 Myf5 positive progenitors require the activation of MyoD to further differentiate into myoblasts. In progenitors, PRMT1 methylates Eya1, a tyrosine phosphatase, and co-factor of the transcription factor Six1. PRMT1 is required for the Eya1/Six1 complex to be recruited at the MyoD enhancer region for transcriptional activation ().Citation2 MyoD plays a pivotal role during MSC commitment, as it first allows progenitors expansion, but then represses the cell cycle permitting terminal differentiation. MyoD-mediated cell fate orchestration was shown to be dose-dependent, and regulated by a positive feedback loop. At low levels, MyoD enhances its activity by increasing p21 levels, as it suppresses the expression of cyclin-dependent kinases, CDKs, repressors of MyoD.Citation7 High levels of MyoD limit self-renewal and promote cell cycle arrest. Once the cell cycle is arrested, MyoD activates Myogenin expression, mandatory for differentiation progression and termination. The loss of PRMT1 in MSC causes an increase in the MSC/progenitor expansion and impairment of differentiation, suggesting PRMT1 might regulate the expression of other genes than MyoD.Citation2 Before its activation by MyoD, Cdkn1a expression is repressed epigenetically by PRMT5 and PRMT7. PRMT5 catalyzes the histone marks at the Cdkn1a locus, while PRMT7 promotes H4R3me2s at the Dnmt3b locus, DNMT3b expression, leading to DNMT3b-mediated methylation of CpG islands within Cdkn1a locus ().Citation3,5
Figure 1. Protein arginine methyltransferases govern muscle stem cell fate during regeneration. In response to injury, muscle stem cell activates to regenerate the injured muscle fibers. They undergo self-renewal to maintain the resident pool within their niche, and give rise to progenitors which will differentiate and proliferates as myoblasts to ultimately fuse and repair the damaged fiber. PRMTs are upstream regulators of muscle stem cells fate decisions during regeneration. They do so by catalyzing arginine methylation (yellow box) on substrates and consequently regulating the expression of essential factors for the progression of differentiation (Myf5), proliferation and maturation (MyoD, Cdkn1a). Some of these mechanisms are still not fully understood (indicated by (?)) such as: whether PRMT7 catalyzes H4R3me2s directly or indirectly, or if methylation of Eya1 is required for its recruitment at MyoD, and/or its association with Six1.
How does arginine methylation regulate muscle regeneration?
PRMTs were shown to be indispensable for proper muscle regeneration in mice using MSC-specific knockout model (Pax7-cre). It is now important to ask how each PRMT and their sequential methylation functionally intervene on specific substrates in a timely manner to govern MSC fate.
A good example is the case of PRMT5 and PRMT7, both epigenetically repressing Cdkn1a independently from p53. Although they regulate the same pathway, they act via different targets. Only PRMT5 binds to Cdkn1a locus directly. In the absence of the PRMT5, p21 levels increase, associated with decreased H3R8me2s at the Cdkn1a locus,Citation5 whereas in the absence of PRMT7, Cdkn1a-sustained expression was associated with reduction of H4R3me2s at Dnmt3b locus, repressing Dnmt3b expression and consequently hypomethylation of CpG islands at Cdkn1a locus.Citation3 Both epigenetically repress Cdkn1a, but the interplay between PRMT5- and PRMT7-mediated histone marks remains to be clarified. It is also of importance to assess whether PRMT7 catalyzes the mark H4R3me2s directly, since it is mainly described in the literature as a PRMT5-mediated mark. Because both enzymes are required for Cdkn1a silencing, PRMT5 and PRMT7 could act in synergy in this pathway. It becomes relevant to determine how these events are triggered and if they happen simultaneously or if PRMT7 is a priming enzyme for certain methyl-marks catalyzed by PRMT5.
Another interesting point arising is the therapeutic potential of PRMT1 inhibitors to expand MSC. Further investigation is required to understand how PRMT1-mediated methylation controls MSC fate. While Eya1 recruitment at MyoD promoter for its co-activation requires the presence of PRMT1,Citation3 the role of Eya1 methylation remains undefined. In the context of organogenesis, Eya1 was shown to activate Six1 through its phosphatase activity, as Six1 acts as a repressor until Eya1 is recruited.Citation8 In MSC, PRMT1-mediated methylation of Eya1 could be required for direct binding to Six1, as PRMT1 deletion results in the absence of Eya1 at MyoD promoter, whereas Six1 is still present but in the absence of Eya1 represses MyoD expression. The MyoD low levels cannot explain alone the observed phenotype, especially the increased self-renewal. Not mentioned in the original work, the depletion of PRMT1 in MSC also leads to an increase in Ezh2 expression (Blanc & Richard, unpublished data), which is known to repress differentiation genes, and maintain MSC identity and self-renewal capacity.Citation9 Mechanistically, PRMT1 directly binds the Ezh2 enhancer region, and PRMT1 null MSC shows a reduction of both H4R3me2a and H3K4me3 at the same locus, suggesting an epigenetic regulation. PRMT1 is responsible for nearly 85% of arginine methylation in the cell and consequently, has a high number of substrates. PRMT1 could act as an upstream epigenetic switch regulating several pathways and it may tune the balance between self-renewal, proliferation, and differentiation progression. If arginine methylation acts upstream of other epigenetic events, the identification of these downstream modifications is also crucial to understand muscle regeneration.
Challenges ahead
In MSC, PRMTs seem to be more pro-active after the initiation of the regeneration in response to injury. The existence of methylarginine erasers and the kinetic of methylarginine turnover being still under consideration, it is relevant to ask (1) how arginine methylation is regulated, (2) how does it affect other epigenetic events, and (3) what are the components recruited by the arginine methylation controlling MSC fate?
It is thus imperative to determine first the status of arginine methylation on histones in quiescent and differentiated MSC on a genome-wide scale to identify the transcriptional targets and other relevant histone marks. It can be achieved by studying the temporal pattern of arginine methylation catalyzed by each PRMT family member during MSC differentiation. Second, identifying the methylarginine readers—as well as putative erasers—and how they subsequently regulate the myogenic differentiation program is crucial to understand how it might affect human muscle biology. It is especially relevant to understand how these arginine methylation changes during aging (sarcopenia), or in MSC-related diseases (ex: Duchenne Muscular Dystrophy; DMD), as PRMT1 inhibition enhances MSC expansion,Citation2 while the loss of PRMT5 mimics DMD,Citation5 and absence of PRMT7 causes sarcopenia and premature MSC aging.Citation3
The expansion of MSC ex vivo and the maintenance of their identity and capacity to regenerate muscles afterward is currently a major challenge in the field of regenerative medicine. This reason is why a better understanding of the temporal deposition of arginine methylation in MSC during regeneration is crucial to define how PRMT activity can be manipulated, and ultimately taken advantage for future therapies. It is particularly relevant now that newly developed PRMT selective inhibitors are becoming available, and how they can be used for the development of new stem cell-based therapies. It is particularly the case for PRMT1, whose depletion in MSC leads to an increase of their expansion in vivo and ex vivo. In the case of DMD, patients are born with normal motor functions, as well as functional MSC. However, the muscles undergo constant regeneration and will eventually result in the exhaustion of the MSC pool, leading to an increase of unrepaired muscles, muscle loss, impaired motors functions, and ultimately death of the patient. A promising approach in the field is to prevent the exhaustion of the MSC by combining genome editing and stem cell-based therapies. So far, it is impossible to efficiently expand enough corrected MSC ex vivo which maintain a long-term regenerative capacity once transplanted in vivo. Because PRMT1 has a substantial number of substrates and seems to be a key factor in the balance between proliferation and differentiation, it implies that a transient inhibition using a selective inhibitor would lead to sufficient expansion, preserving the self-renewing capacity. The challenge would then be to determine whether these cells once removed from the inhibitor would regain their ability to differentiate and thus participate in regeneration in vivo.
To conclude the exact role of arginine methylation, and how it modifies MSC fate, remains to be unraveled. A better understanding of arginine methylation and other post-translational modifications, coupled with development of PRMT inhibitors should give promising opportunities for stem cell-based therapy against diseases associated with exhaustion of the MSC pool and regenerative function, including sarcopenia and DMD.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was funded by CIHR MOP-67070 and MOP-93811.
References
- Lepper C, Partridge TA, Fan CM. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 2011; 138:3639-46; PMID:21828092; https://doi.org/10.1242/dev.067595
- Blanc RS, Vogel G, Li X, Yu Z, Li S, Richard S. Arginine methylation by PRMT1 regulates muscle stem cell fate. Mol Cell Biol 2017; 37:e00457-16; https://doi.org/10.1128/MCB.00457-16
- Blanc RS, Vogel G, Chen T, Crist C, Richard S. PRMT7 preserves satellite cell regenerative capacity. Cell Rep 2016; 14:1528-39; PMID:26854227; https://doi.org/10.1016/j.celrep.2016.01.022
- Kawabe Y, Wang YX, McKinnell IW, Bedford MT, Rudnicki MA. Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell 2012; 11:333-45; PMID:22863532; https://doi.org/10.1016/j.stem.2012.07.001
- Zhang T, Gunther S, Looso M, Kunne M, Kruger M, Kim J, Zhou Y, Braun T. Prmt5 is a regulator of muscle stem cell expansion in adult mice. Nat Commun 2015; 6:7140; PMID:26028225; https://doi.org/10.1038/ncomms8140
- Blanc RS, Richard S. Arginine methylation: The coming of age. Mol Cell 2017; 65:8-24; PMID:28061334; https://doi.org/10.1016/j.molcel.2016.11.003
- Guo K, Wang J, Andres V, Smith RC, Walsh K. MyoD-induced expression of p21 inhibits cyclin-dependent kinase activity upon myocyte terminal differentiation. Mol Cell Biol 1995; 15:3823-9; PMID:7791789; https://doi.org/10.1128/MCB.15.7.3823
- Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature 2003; 426:247-54; PMID:14628042; https://doi.org/10.1038/nature02083
- Juan AH et al. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev 2011; 25:789-94; PMID:21498568; https://doi.org/10.1101/gad.2027911
