ABSTRACT
The ETS family of transcription factors is a functionally heterogeneous group of gene regulators that share a structurally conserved, eponymous DNA-binding domain. DNA target specificity derives from combinatorial interactions with other proteins as well as intrinsic heterogeneity among ETS domains. Emerging evidence suggests molecular hydration as a fundamental feature that defines the intrinsic heterogeneity in DNA target selection and susceptibility to epigenetic DNA modification. This perspective invokes novel hypotheses in the regulation of ETS proteins in physiologic osmotic stress, their pioneering potential in heterochromatin, and the effects of passive and pharmacologic DNA demethylation on ETS regulation.
Introduction
The ETS family of transcription factors binds site-specific DNA via eponymous, structurally conserved DNA-binding domains that share low overall sequence homology ( and ). Although ETS members are not numerous (28 paralogs in humans), they are ubiquitously distributed in the metazoan,Citation1 and many are indispensable to life. Regardless of function, all ETS proteins show a highly conserved binding mode in which a recognition helix of their ∼80-residue ETS domain is inserted into the major groove of target DNA harboring the core consensus 5′-GGAA/T-3′, with additional interactions along the DNA backbone at flanking, sequence-variable minor groove positions (). The structural homology among ETS domains is remarkable in the context of the choreography that many ETS transcription factors execute in hematopoiesis, and the multi-step differentiation of blood cell lineages that is intricately controlled at the transcriptional level.Citation2,3 Differentiation of the hematopoietic stem cell and fate determination of downstream progenitors are driven by precise ebbs and flows of activity by ETS paralogs in conjunction with other transcription factors in a stage-specific and dosage-specific manner.Citation4
Figure 1. The DNA-binding domains of ETS transcription factors are sequence- and phylogenetically divergent, but strongly conserved in structure. (A) Sequence alignment for the 28 paralogous human ETS domains. Proteins were identified by UniProtKB identifier | residue numbering | protein name. Residues are colored by amino acid types. Asterisks denote positions with amino acid identity. (B) Phylogenetic tree constructed by the maximum-likelihood method, arranged with Ets-1 and PU.1 (Spi-1) at the ends. The horizontal distance (branch length) denotes phylogenetic distance defined as number of substitutions per position. (C) Structural alignment of the ETS domains of Ets-1 (silver; PDB: 1K79) and PU.1 (gold; PDB: 1PUE) from their co-crystal structures with DNA. The root-mean-square deviation is 1.4 Å, well below the experimental resolution of the models themselves. The target DNA from the Ets-1 co-crystal structure is shown to orient the viewer.
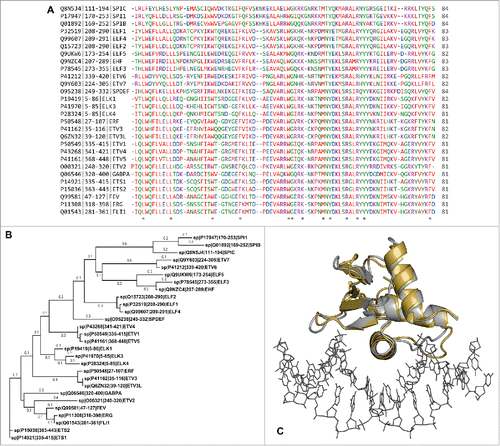
Of the hematopoietic ETS-family regulators, PU.1 (Spi-1) and Ets-1 draw one of the sharpest lines of contrasts (). Their ETS domains represent the most phylogenetically distant ETS sub-families, yet they are structurally superimposable when bound to DNA targets.Citation5 The two ETS paralogs drive cell fate specification coordinately, and often in opposing directions.Citation6-11 In T-cell differentiation, for example, an obligatory drop in PU.1 activity is concomitantly balanced by a surge in the Ets-1 activity,Citation11-13 and both are differentially required in the peripheral Th subsets.Citation14-16 Aberrant activity in either paralog is linked to a spectrum of diseases including rheumatism,Citation17 cancerCitation18-21 and Alzheimer disease.Citation22-24 Functional heterogeneity occurs even among very close ETS sequence homologs, as in the case of PU.1 and Spi-B, wherein Spi-B cannot compensate for the absence of PU.1-mediated B-cell signaling in PU.1-null mice.Citation25
Table 1. Biological and biochemical comparison of the ETS paralogs PU.1 and Ets-1.
Specificity determinants of ETS transcription factors
Given the significant overlap in expression of ETS proteins,Citation26 their DNA sequence preference,Citation27 and overall structure of their ETS domains on the one hand, and their general functional non-interchangeability on the other, the basis of their specificity has long been a subject of major interest. Currently, ETS proteins are grouped into classes (I–IV) according to their relative sequence preferences,Citation27 which correspond to their phylogenetic relatedness. The sharp and well-conserved delineation of major groove contacts at the 5′-GGAA/T-3′ core consensus, and minor groove backbone (sugar and phosphate) contacts at flanking bases where sequence variation occurs, has led to the notion of indirect readout. In contrast with direct readout of nucleobases at the core consensus, contacts with the DNA backbone are presumably parsed on the basis of some sequence-dependent DNA shape or propensity to adopt a preferred conformation (or ensemble of such). While indirect readout of bases flanking the core consensus has been unequivocally demonstrated,Citation28 its functional significance in target gene transactivation confirmedCitation29 and its thermodynamic basis understood in some detail,Citation30,31 the structural (and probably also dynamic) origin of indirect readout in ETS/DNA site recognition remains elusive. Most frustratingly, no predictive capability of how the structure of given ETS domain relates to its distinct spectrum of flanking sequence preferences has yet been achieved.
Combinatorial routes to ETS target specificity
A pervasive, though not universal, feature of ETS proteins is the presence of elements adjacent to their ETS domains that are structured in the absence of DNA, but unfold upon DNA binding. The energetic overhead to unfold these appending elements, which are most extensively characterized in Ets-1 (class I)Citation32-34 and ETV6 (class II),Citation35-37 result in reduced affinity to any given DNA site about an order of magnitude relative to truncated constructs. This auto-inhibitory mechanism serves as a handle for combinatorial control of ETS proteins through interactions with protein partners that displace the extra-ETS appendages. For example, Runx1/AML1/CBFα2/PEBP2 positively regulates Ets-1/DNA binding by displacing and destabilizing an extended inhibitory segment N-terminal to the ETS domain.Citation38-40 In ETS paralogs that are not auto-inhibited, such as PU.1, specific interactions with binding partners that positively regulate (e.g., IRF4/Pip)Citation41-43 or antagonize DNA binding (e.g., GATA-1)Citation44,45 are known. Another combinatorial strategy that modifies DNA site targeting is to couple binding to intrinsically low-affinity or nonspecific sites to specific interactions with binding partners. Such mechanisms are illustrated by the ability of Pax5 to recruit Ets-1 to a nonspecific sequence (5′-GGAG-3′).Citation46,47 Similar recruitment of PU.1 to intrinsically low-affinity sites has also been reported.Citation48 These and other interactions, all of which are functionally linked to cell fate specification or the regulation of lineage-specific target genes, have been well reviewed.Citation49
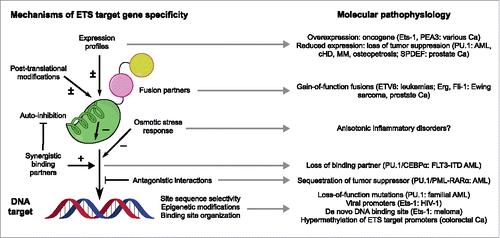
In recent years, high-throughput microarray and sequencing technologies have elevated investigations of ETS/DNA interactions to the genome-wide level. Detailed information on the localization, sequence characteristics of DNA targets, and associated binding partners is now available for ETS transcription factors in a range of cell types and developmental contexts.Citation27,50-55 Although various levels of redundancy and specificity are observed that correlate with the ontology of the genes involved, one recurring feature is the close correspondence between in vivo and in vitro DNA sequence preferences. Moreover, in the case of PU.1 and Ets-1, the information contents (a direct informatic measure of target specificity) of the in vivo sequences preferences shown by both proteins are over 15% higher (> 3 bits over a 10-bp sequence space) than their in vitro counterparts.Citation5 Given the vast number of sequence reads in the in vivo data, these two observations indicate that, integrated over the whole genome, combinatorial control refines, rather than usurps, the intrinsic selectivity of their ETS domains. Thus, while the target gene specificity of ETS proteins is functionally controlled by an inter-related web of interactions in vivo (), the intrinsic properties of ETS domains per se remain a keystone in understanding sequence usage of ETS transcription factors in the genome.
Figure 2. Selected mechanisms of ETS target gene specificity. ETS-dependent transcription is regulated at multiple levels, all of which can operate in a combinatorial fashion. The established molecular pathophysiology associated with some of these mechanisms are listed. The literature on ETS proteins is vast and this summary is only intended to be illustrative; readers interested in specific aspects or paralogs mentioned in this figure are referred to reviews and studies such as the following:Citation26,49,51,53,66,87-93 Note that auto-inhibition (indicated by the cartoon helix) is not a universal feature of ETS proteins; several paralogs, such as PU.1, are not auto-inhibited. Abbreviations: AML, acute myeloid leukemia; Ca, cancer; cHD, classical Hodgkin's disease; MM, multiple myeloma.
A deeper look into ETS/DNA recognition
To date, structures of ETS/DNA complexes, most of which involve high-affinity cognate sequences, have provided physical models of optimal interactions in DNA target recognition by representative ETS paralogs in each class. Do fundamental mechanisms exist that could explain DNA target selectivity across the spectrum of ETS paralogs? Such “molecular phenotypes” would reasonably reflect the selection pressures operating on functionally distinct ETS paralogs, and in turn provide insight into the biologic environment in which the proteins operate. To this end, we have identified two aspects that structural and biochemical data suggest unusual potential for insight: molecular hydration accompanying DNA recognition by ETS domains and their sensitivity to epigenetically modified DNA targets. We have been focusing our attention on Ets-1 and PU.1, which are attractive model systems for two reasons. First, they are archetypal representatives of the most phylogenetically most distant classes of ETS proteins,Citation49 so heterogeneity in their molecular phenotype should directly reflect the selection pressures in their evolutionary paths, even if the biologic basis of these pressures are not necessarily known. Second, these two ETS paralogs bind optimal DNA targets with indistinguishably high affinity,Citation56 so that heterogeneity between the two homologs that contribute to their DNA binding affinity and specificity would be biologically relevant.
Role of molecular hydration in DNA recognition by ETS proteins
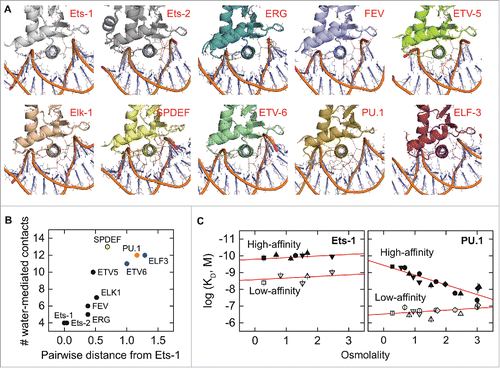
Transcription factors interact with and regulate their DNA targets in an aqueous milieu. Rather than a passive bathing medium, water molecules participate in protein/DNA interactions and can act as a major determinant of binding affinity and specificity, for example, by forming water-mediated contacts at the protein/DNA interface. One of the most intriguing differences between the co-crystal structures of PU.1 and Ets-1 is the abundance of interfacial water-mediated contacts in the PU.1 complex and the sparsity of such contacts in the Ets-1 complex.Citation47,57 The water-mediated contacts made by PU.1 are effectively replaced by direct protein-to-DNA contacts in Ets-1. To determine whether the crystallographic data indeed reflect a differential role for interfacial hydration in DNA recognition by the two proteins in solution, we interrogated DNA binding by the two proteins through osmotic stress.Citation56 Using physiologically compatible osmolytes to modulate the osmotic environment (water activity), the data indeed showed that high-affinity DNA binding by PU.1 was osmotic sensitive while binding by Ets-1 was not (). Moreover, the osmotic sensitivity of PU.1, wherein affinity was reduced by osmotic pressure, was dependent on the DNA sequence context, inferring a direct role of hydration in the specificity mechanism of PU.1.
The strikingly different responses of the two homologs to their osmotic environment, which is fully compensated to yield indistinguishable binding affinities under normo-osmotic conditions, are a provocative observation. Higher-order organisms maintain a homeostatic environment in which intracellular parameters are controlled.Citation58 Osmotic pressure is one such parameter. Hyperosmotic stress and its attendant perturbation on cell volume trigger signaling pathways mediated by the transcription factor NFAT5/TonEBP that restore isotonicity through the accumulation of compatible osmolytes.Citation59,60 As a result, the compensated (isotonic) but now hyperosmolar conditions would necessarily perturb biomoleular interactions that involve sufficiently large hydration changes. The affinity of PU.1 for its optimal cognate sequence is reduced ∼10-fold across 0.5 osmolal,Citation61 a physiologically relevant level of osmotic stress in lympocytes.Citation62 We have found through analysis of microarray data that PU.1 target genes are disproportionately represented in osmotically sensitive (NFAT5-dependent) genes in primary murine macrophages.Citation61 Significantly, other transcription factors that are co-expressed with PU.1, such as the interferon regulatory factors, NF-κB2, and Stat proteins, show no such overlap. While PU.1 may interact with NFAT5 by direct contact or via post-translational modification, no such interactions are currently known. A more intriguing scenario is that PU.1 and NFAT5 targets overlap by virtue of the osmotic sensitivity of their regulators. It would therefore be possible for genetic networks to interact through “fields” and in a manner that requires no direct contact between or post-translational modification of the macromolecular components involved.
Significance of molecular hydration in ETS activity under normo-osmotic conditions
In addition to perturbing affinity under hyperosmotic stress, the osmotic sensitivity of DNA target recognition provides insight into differences in binding behavior under normo-osmotic conditions. Specifically, we have observed that the different disposition of hydration water directly impacts on the mechanisms of DNA target recognition as manifest in their kinetics of association and dissociation ().Citation61 Under normo-osmotic conditions, PU.1 engages sequence-specific target sites about ∼100 more slowly than Ets-1, but once formed, the complex is correspondingly more persistent than Ets-1. The starkly different kinetic profiles establish that interfacial hydration defines different mechanisms of target recognition by the two ETS homologs. In addition, the persistence of the PU.1/DNA complex against dissociation is consistent with PU.1 as a strong pioneer transcription factor,Citation63 by anchoring target genes in chromatin during recruitment of other transcription factors and remodeling proteins (such as histone acetyltransferases).Citation64 Currently, the pioneer status of Ets-1 is controversial: Although it appears to co-localize with nucleosomes in enhancer regions in developing thymocytes,Citation52 it does not exhibit functional pioneer activity in a defined reporter assay.Citation65 An intrinsic mechanism for resisting nucleosomal dynamics by the ETS domain to secure accessible proximal binding sites for other proteins represents an intriguing component of pioneering activity of “master” transcription factors such as PU.1.
Molecular hydration as a unifying feature in ETS evolution
Is heterogeneity in molecular hydration a general feature in the broader ETS family? A survey of binary and ternary co-crystal structures of ETS domains shows a range in both the density and pattern of interfacial hydration, as may be expected from the amino acid diversity in their DNA-binding surfaces. To examine this heterogeneity systematically, we considered the correlation between the number of water-mediated contacts in DNA co-crystal complexes of ETS paralogs as a function of their evolutionary relatedness. Taking Ets-1 as reference, we found that the density of water-mediated contacts for ETS paralogs is positively correlated with its pairwise phylogenetic distance from Ets-1 (). This is a remarkable correlation. The physicochemical diversity of the crystals (e.g., symmetry, packing, co-solvents, and overall hydration) strongly discount against the observed correlation as a crystallographic artefact. There is also no systematic differences in the resolution of the structures that would account for a bias in discernable hydration. Beyond several water-mediated contacts involving the sidechains and backbone of absolutely or highly conserved residues that are observed in all the structures, a significant diversity in bridging pattern is observed at all levels of hydration, suggesting that interfacial hydration is highly adaptive. While the evolution of ETS paralogs is undoubtedly subject to different selection pressures, which are not universally shared, it appears that as a general feature interfacial hydration is incrementally incorporated in the evolution of the ETS family. The biophysical and biological implications of this relationship are currently unknown and ripe for hypothesis.
Figure 3. Crystallographic interfacial hydration correlates positively with phylogenetic relatedness among the ETS domains of paralogs. (A) Co-crystal structures of the ETS domains of nine ETS paralogs, oriented identically with the recognition helix perpendicular to the plane of the page. Water-mediated contacts are shown as cyan spheres, defined operationally as crystallographic water within hydrogen-bonding distance (red dashes, ≤ 3.4 Å) of a protein and DNA contact, or another interfacial water that meets this criterion. To avoid ambiguity, water-mediated contacts involving only three or more consecutive bridging water are not counted. Interfacial water density is weakly correlated with overall hydration of the asymmetric unit, and there is no significant difference in interfacial hydration density ( ± 1) between different biological units. (B) Relationship between crystallographic interfacial hydration and pairwise phylogenetic distance from Ets-1, chosen as reference. The primary sequences of the 28 human ETS paralogs were analyzed by ClustalW using the neighbor-joining method. The results were expressed as a distance matrix from whose elements are pairwise distances (number of substitutions per position). ETS paralogs are formally categorized into classes I–IVCitation27 by color in order from black, blue, orange, to yellow. (C) Differential sensitivity to osmotic pressure in site-specific binding by the ETS domains of PU.1 and Ets-1 as reported by Wang et al.Citation56 The measured in vitro affinity is expressed as the logarithm of the dissociation constant (KD). High- (solid symbols) and low-affinity DNA (open) refer to defined cognate (not nonspecific) sequences harboring the 5′-GGAA-3′ consensus. The different symbols refer to the set of physiologically compatible osmolytes used to exert osmotic stress. The osmotic insensitivity of Ets-1 is not modified by the presence of auto-inhibition.Citation56
Differential tolerance to CpG methylation
While the importance of epigenetic regulation of ETS-dependent transcription is well established in hematopoiesis and cancer,Citation55,66-68 the mechanisms by which ETS activity are modulated at epigenetically modified DNA, with or without nucleosome, are not well understood. Genomic surveys have found that hematopoietic ETS transcription factors are over-represented in hypermethylated regions.Citation53 While several close class I ETS paralogs (such as Ets-1 and GABPα) have been reported to be inhibited by CpG methylation at their cognate sites, whether inhibition is a universal property of ETS proteins remains unknown. We have directly studied the binding properties of PU.1 and Ets-1 to hemi-methylated and fully methylated cognate DNA harboring a site-specific CpG dinucleotide (5′-CGGAA-3′) that frequently occurs in cognate ETS binding sites.Citation69 While any CpG methylation affected binding, PU.1 and Ets-1 responded qualitatively differently to hemi-methylated sites. Hemi-methylation of the sense (5′-GGAA-3′) strand was strongly inhibitory to Ets-1 binding, but hemi-methylation of the anti-sense strand (5′-GGAA-3′) was not and vice versa for PU.1. In addition, auto-inhibition was operative in Ets-1 with respect to binding to CpG-methylated sites. Finally, PU.1 was significantly more robust than Ets-1 in binding fully methylated DNA. Overall, our targeted studies showed that significant heterogeneity exists in their intrinsic sensitivity to CpG methylation among ETS transcription factors. They confirmed the strong inhibition of Ets-1 (and by extension, other class I ETS members) by full CpG methylation and provided a basis for the genomic-wide observation PU.1 to autonomously engage methylated DNA in vivo.Citation48
Mechanistically, we found by molecular simulations that the asymmetric effect of hemi-methylation on the DNA-binding affinity of ETS paralogs may be explained by structural perturbations on DNA backbone geometry. While hemi-methylation of either strand significantly perturbs backbone geometry out of the unmethylated configuration, full methylation produces a compensatory effect that brings backbone geometry back closer to unmethylated DNA. In light of the plasticity in interfacial hydration, we speculate that hydration waters serve as adapters that moderate the perturbative effects of DNA methylation on binding for hydration-rich ETS paralogs such as PU.1. In addition, the compensatory relationship between hemi-methylation and full methylation on DNA backbone structure suggests new biological implications in view of the semi-conservative nature of DNA replication. Immediately following DNA synthesis with unmethylated nucleotides, the DNA daughter strands are hemi-methylated until re-methylated by DNA methyltransferase I (DNMT1). The exact same site in passively de-methylated genome, therefore, presents a heterogeneous substrate for ETS paralogs (and probably other DNA-binding proteins) depending on the stage in the cell cycle or exogenous treatment with DNMT1 inhibitors (“hypomethylating agents” such as azacitidine and decitabine). Interest in this area is heightened by the advent of hypomethylating agents as clinical drugs in hematologic cancers, such as azacitidine in myelodysplastic syndromeCitation70 and a growing list of other malignancies.
Chemical biology of ETS proteins
Target-specific control of transcriptional pathways has long been a goal in experimental research and therapy. Despite the success and ubiquity of gene-based approaches to knock-in, known-down, and knock-out specific genes in vitro and in vivo, as well as the intense efforts to deliver genetic and other macromolecular payloads efficiently and without toxicity into cells and tissues, low-molecular weight molecules (i.e., chemical control) remain the preferred modality of intervention. With few exceptions, the clinically successful pharmacology of nuclear receptors has not been reprised for most of other transcription factors, particularly wildtype forms which lack endogenous ligands as templates for drug development.Citation71-74 A fruitful avenue in the case of inhibition is to target the cognate DNA site to which the transcription factor binds. Thanks to the considerable advance over the past two decades in sequence-specific targeting of DNA-binding ligands, particularly the hairpin polyamidesCitation75 and heterocyclic diamidines,Citation76 proofs-of-concept have been achieved for several ETS proteins. They include the inhibition of Ets-1 with a designed polyamideCitation77 and ERG using designed heterocyclic diamidines.Citation78 We have demonstrated the inhibition of PU.1 using diamidines of a different class.Citation79,80 Viable chemical biology of ETS proteins is a challenge in need of actionable targets, and identification of the molecular heterogeneity among ETS domains could clarify such targets and strategies for control.
Concluding remarks
Although all ETS paralogs display highly homologous backbone structures and engage target DNA in an essentially identical conformation, they harbor a spectrum of distinct physical chemistry that is likely reflected in their functional phenotypes. Targeted studies of interactions by ETS domains in new areas such as molecular hydration and epigenetically modified DNA are sparking novel perspectives and opportunities for new insights into the diversity of this important family of transcriptional regulators.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
We are grateful to Drs. W. David Wilson (GSU), David W. Boykin (GSU), and James K. Bashkin (University of Missouri, St. Louis), and their research personnel whose collaboration make our work possible.
Funding
Our work on ETS transcription factors is supported by NSF grant MCB 15451600 and NIH grant R21 HL129063. H.M.K. is supported by a GSU Molecular Basis of Disease Fellowship.
References
- Degnan BM, Degnan SM, Naganuma T, Morse DE. The ETS multigene family is conserved throughout the Metazoa. Nucleic Acids Res 1993; 21:3479-3484; PMID:8346026; https://doi.org/10.1093/nar/21.15.3479
- Zhu J, Emerson SG. Hematopoietic cytokines, transcription factors and lineage commitment. Oncogene 2002; 21:3295-3313; PMID:12032771; https://doi.org/10.1038/sj.onc.1205318
- Orkin SH. Transcription factors and hematopoietic development. J Biol Chem 1995; 270:4955-4958; PMID:7890597; https://doi.org/10.1074/jbc.270.10.4955
- Ciau-Uitz A, Wang L, Patient R, Liu F. ETS transcription factors in hematopoietic stem cell development. Blood Cells Mol Dis 2013; 51:248-255; PMID:23927967; https://doi.org/10.1016/j.bcmd.2013.07.010
- He G, Tolic A, Bashkin JK, Poon GM. Heterogeneous dynamics in DNA site discrimination by the structurally homologous DNA-binding domains of ETS-family transcription factors. Nucleic Acids Res 2015; 43:4322-4331; PMID:25824951; https://doi.org/10.1093/nar/gkv267
- Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 1994; 265:1573-1577; PMID:8079170; https://doi.org/10.1126/science.8079170
- Nutt SL, Metcalf D, D'Amico A, Polli M, Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp Med 2005; 201:221-231; PMID:15657291; https://doi.org/10.1084/jem.20041535
- Dahl R, Simon MC. The importance of PU.1 concentration in hematopoietic lineage commitment and maturation. Blood Cells Mol Dis 2003; 31:229-233; PMID:12972030; https://doi.org/10.1016/S1079-9796(03)00152-9
- DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science 2000; 288:1439-1441; PMID:10827957; https://doi.org/10.1126/science.288.5470.1439
- Mak KS, Funnell AP, Pearson RC, Crossley M. PU.1 and haematopoietic cell fate: dosage matters. Int J Cell Biol 2011; 2011:808524; PMID:21845190; https://doi.org/10.1155/2011/808524
- Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV. Precise developmental regulation of ETS family transcription factors during specification and commitment to the T cell lineage. Development 1999; 126:3131-3148; PMID:10375504
- Mouly E, Chemin K, Nguyen HV, Chopin M, Mesnard L, Leite-de-Moraes M, Burlen-defranoux O, Bandeira A, Bories JC. The Ets-1 transcription factor controls the development and function of natural regulatory T cells. J Exp Med 2010; 207:2113-2125; PMID:20855499; https://doi.org/10.1084/jem.20092153
- Anderson MK, Weiss AH, Hernandez-Hoyos G, Dionne CJ, Rothenberg EV. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity 2002; 16:285-296; PMID:11869688; https://doi.org/10.1016/S1074-7613(02)00277-7
- Chang HC, Sehra S, Goswami R, Yao W, Yu Q, Stritesky GL, Jabeen R, McKinley C, Ahyi AN, Han L et al. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat Immunol 2010; 11:527-534; PMID:20431622; https://doi.org/10.1038/ni.1867
- Chang H-C, Zhang S, Thieu VT, Slee RB, Bruns HA, Laribee RN, Klemsz MJ, Kaplan MH. PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity 2005; 22:693-703; PMID:15963784; https://doi.org/10.1016/j.immuni.2005.03.016
- Grenningloh R, Kang BY, Ho I-C. Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J Exp Med 2005; 201:615-626; PMID:15728239; https://doi.org/10.1084/jem.20041330
- Dozmorov MG, Wren JD, Alarcon-Riquelme ME. Epigenomic elements enriched in the promoters of autoimmunity susceptibility genes. Epigenetics 2014; 9:276-285; PMID:24213554; https://doi.org/10.4161/epi.27021
- Gilliland DG. The diverse role of the ETS family of transcription factors in cancer. Clin Cancer Res 2001; 7:451-453; PMID:11297232
- Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, Akashi K, Fiering S, Tenen DG. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet 2004; 36:624-630; PMID:15146183; https://doi.org/10.1038/ng1361
- Tatetsu H, Ueno S, Hata H, Yamada Y, Takeya M, Mitsuya H, Tenen DG, Okuno Y. Down-regulation of PU.1 by methylation of distal regulatory elements and the promoter is required for myeloma cell growth. Cancer Res 2007; 67:5328-5336; PMID:17545613; https://doi.org/10.1158/0008-5472.CAN-06-4265
- Yuki H, Ueno S, Tatetsu H, Niiro H, Iino T, Endo S, Kawano Y, Komohara Y, Takeya M, Hata H et al. PU.1 is a potent tumor suppressor in classical Hodgkin lymphoma cells. Blood 2013; 121:962-970; PMID:23212521; https://doi.org/10.1182/blood-2012-05-431429
- Gjoneska E, Pfenning AR, Mathys H, Quon G, Kundaje A, Tsai L-H, Kellis M. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer's disease. Nature 2015; 518:365-369; PMID:25693568; https://doi.org/10.1038/nature14252
- Gomez-Nicola D, Fransen NL, Suzzi S, Perry VH. Regulation of microglial proliferation during chronic neurodegeneration. J Neurosci 2013; 33:2481-2493; PMID:23392676; https://doi.org/10.1523/JNEUROSCI.4440-12.2013
- Jantaratnotai N, Ling A, Cheng J, Schwab C, McGeer PL, McLarnon JG. Upregulation and expression patterns of the angiogenic transcription factor Ets-1 in Alzheimer's disease brain. J Alzheimers Dis 2013; 37:367-377; PMID:23948889; https://doi.org/10.3233/JAD-122191
- Garrett-Sinha LA, Su GH, Rao S, Kabak S, Hao Z, Clark MR, Simon MC. PU.1 and Spi-B are required for normal B cell receptor-mediated signal transduction. Immunity 1999; 10:399-408; PMID:10229183; https://doi.org/10.1016/S1074-7613(00)80040-0
- Hollenhorst PC, Jones DA, Graves BJ. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res 2004; 32:5693-5702; PMID:15498926; https://doi.org/10.1093/nar/gkh906
- Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Simon MC. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J 2010; 29:2147-2160; PMID:20517297; https://doi.org/10.1038/emboj.2010.106
- Szymczyna BR, Arrowsmith CH. DNA binding specificity studies of four ETS proteins support an indirect read-out mechanism of protein-DNA recognition. J Biol Chem 2000; 275:28363-28370; PMID:10867009; https://doi.org/10.1074/jbc.M004294200
- Li SL, Schlegel W, Valente AJ, Clark RA. Critical flanking sequences of PU.1 binding sites in myeloid-specific promoters. J Biol Chem 1999; 274:32453-32460; PMID:10542290; https://doi.org/10.1074/jbc.274.45.32453
- Poon GM, Macgregor RB, Jr. Base coupling in sequence-specific site recognition by the ETS domain of murine PU.1. J Mol Biol 2003; 328:805-819; PMID:12729756; https://doi.org/10.1016/S0022-2836(03)00362-0
- Poon GM, Macgregor RB, Jr. A thermodynamic basis of DNA sequence selectivity by the ETS domain of murine PU.1. J Mol Biol 2004; 335:113-127; PMID:14659744; https://doi.org/10.1016/j.jmb.2003.09.046
- Desjardins G, Okon M, Graves BJ, McIntosh LP. Conformational dynamics and the binding of specific and nonspecific DNA by the autoinhibited transcription factor Ets-1. Biochemistry 2016; 55:4105-4118; PMID:27362745; https://doi.org/10.1021/acs.biochem.6b00460
- Lee GM, Donaldson LW, Pufall MA, Kang HS, Pot I, Graves BJ, McIntosh LP. The structural and dynamic basis of Ets-1 DNA binding autoinhibition. J Biol Chem 2005; 280:7088-7099; PMID:15591056; https://doi.org/10.1074/jbc.M410722200
- Pufall MA, Lee GM, Nelson ML, Kang H-S, Velyvis A, Kay LE, McIntosh LP, Graves BJ. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science 2005; 309:142-145; PMID:15994560; https://doi.org/10.1126/science.1111915
- Coyne HJ, 3rd, De S, Okon M, Green SM, Bhachech N, Graves BJ, McIntosh LP. Autoinhibition of ETV6 (TEL) DNA binding: appended helices sterically block the ETS domain. J Mol Biol 2012; 421:67-84; PMID:22584210; https://doi.org/10.1016/j.jmb.2012.05.010
- De S, Chan AC, Coyne HJ, 3rd, Bhachech N, Hermsdorf U, Okon M, Murphy ME, Graves BJ, McIntosh LP. Steric mechanism of auto-inhibitory regulation of specific and non-specific DNA binding by the ETS transcriptional repressor ETV6. J Mol Biol 2014; 426:1390-406.
- De S, Okon M, Graves BJ, McIntosh LP. Autoinhibition of ETV6 DNA binding is established by the stability of its inhibitory helix. J Mol Biol 2016; 428:1515-1530; PMID:26920109; https://doi.org/10.1016/j.jmb.2016.02.020
- Shrivastava T, Mino K, Babayeva ND, Baranovskaya OI, Rizzino A, Tahirov TH. Structural basis of Ets1 activation by Runx1. Leukemia 2014; 28:2040-2048; PMID:24646888; https://doi.org/10.1038/leu.2014.111
- Kim WY, Sieweke M, Ogawa E, Wee HJ, Englmeier U, Graf T, Ito Y. Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J 1999; 18:1609-1620; PMID:10075931; https://doi.org/10.1093/emboj/18.6.1609
- Goetz TL, Gu TL, Speck NA, Graves BJ. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor alpha2. Mol Cell Biol 2000; 20:81-90; PMID:10594011; https://doi.org/10.1128/MCB.20.1.81-90.2000
- Yee AA, Yin P, Siderovski DP, Mak TW, Litchfield DW, Arrowsmith CH. Cooperative interaction between the DNA-binding domains of PU.1 and IRF4. J Mol Biol 1998; 279:1075-1083; PMID:9642085; https://doi.org/10.1006/jmbi.1998.1838
- Groß P, Yee AA, Arrowsmith CH, Macgregor RB, Jr. Quantitative hydroxyl radical footprinting reveals cooperative interactions between DNA-binding subdomains of PU.1 and IRF4. Biochemistry 1998; 37:9802-9811; PMID:9657694; https://doi.org/10.1021/bi9731448
- Escalante CR, Brass AL, Pongubala JM, Shatova E, Shen L, Singh H, Aggarwal AK. Crystal structure of PU.1/IRF-4/DNA ternary complex. Mol Cell 2002; 10:1097-1105; PMID:12453417; https://doi.org/10.1016/S1097-2765(02)00703-7
- Liew CW, Rand KD, Simpson RJY, Yung WW, Mansfield RE, Crossley M, Proetorius-Ibba M, Nerlov C, Poulsen FM, Mackay JP. Molecular analysis of the interaction between the hematopoietic master transcription factors GATA-1 and PU.1. J Biol Chem 2006; 281:28296-28306; PMID:16861236; https://doi.org/10.1074/jbc.M602830200
- Zhang P, Zhang X, Iwama A, Yu C, Smith KA, Mueller BU, Narravula S, Torbett BE, Orkin SH, Tenen DG. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood 2000; 96:2641-2648; PMID:11023493
- Fitzsimmons D, Lukin K, Lutz R, Garvie CW, Wolberger C, Hagman J. Highly cooperative recruitment of Ets-1 and release of autoinhibition by Pax5. J Mol Biol 2009; 392:452-464; PMID:19616560; https://doi.org/10.1016/j.jmb.2009.07.028
- Garvie CW, Hagman J, Wolberger C. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol Cell 2001; 8:1267-1276; PMID:11779502; https://doi.org/10.1016/S1097-2765(01)00410-5
- Pham TH, Minderjahn J, Schmidl C, Hoffmeister H, Schmidhofer S, Chen W, Längst G, Benner C, Rehli M. Mechanisms of in vivo binding site selection of the hematopoietic master transcription factor PU.1. Nucleic Acids Res 2013; 41:6391-6402; PMID:23658224; https://doi.org/10.1093/nar/gkt355
- Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem 2011; 80:437-471; PMID:21548782; https://doi.org/10.1146/annurev.biochem.79.081507.103945
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 2010; 7:532-544; PMID:20887958; https://doi.org/10.1016/j.stem.2010.07.016
- Hollenhorst PC, Chandler KJ, Poulsen RL, Johnson WE, Speck NA, Graves BJ. DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet 2009; 5:e1000778; PMID:20019798; https://doi.org/10.1371/journal.pgen.1000778
- Cauchy P, Maqbool MA, Zacarias-Cabeza J, Vanhille L, Koch F, Fenouil R, Gut M, Gut I, Santana MA, Griffon A et al. Dynamic recruitment of Ets1 to both nucleosome-occupied and -depleted enhancer regions mediates a transcriptional program switch during early T-cell differentiation. Nucleic Acids Res 2016; 44:3567-3585; PMID:26673693; https://doi.org/10.1093/nar/gkv1475
- Hogart A, Lichtenberg J, Ajay SS, Anderson S, Margulies EH, Bodine DM. Genome-wide DNA methylation profiles in hematopoietic stem and progenitor cells reveal overrepresentation of ETS transcription factor binding sites. Genome Res 2012; 22:1407-1418; PMID:22684279; https://doi.org/10.1101/gr.132878.111
- Boros J, Donaldson IJ, O'Donnell A, Odrowaz ZA, Zeef L, Lupien M, Meyer CA, Liu XS, Brown M, Sharrocks AD. Elucidation of the ELK1 target gene network reveals a role in the coordinate regulation of core components of the gene regulation machinery. Genome Res 2009; 19:1963-73; PMID:19687146; https://doi.org/10.1101/gr.093047.109
- Pham TH, Benner C, Lichtinger M, Schwarzfischer L, Hu Y, Andreesen R, Chen W, Rehli M. Dynamic epigenetic enhancer signatures reveal key transcription factors associated with monocytic differentiation states. Blood 2012; 119:e161-e171; PMID:22550342; https://doi.org/10.1182/blood-2012-01-402453
- Wang S, Linde MH, Munde M, Carvalho VD, Wilson WD, Poon GM. Mechanistic heterogeneity in site recognition by the structurally homologous DNA-binding domains of the ETS family transcription factors Ets-1 and PU.1. J Biol Chem 2014; 289:21605-21616; PMID:24952944; https://doi.org/10.1074/jbc.M114.575340
- Kodandapani R, Pio F, Ni CZ, Piccialli G, Klemsz M, McKercher S, Maki RA, Ely KR. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature 1996; 380:456-460; PMID:8602247; https://doi.org/10.1038/380456a0
- Burg MB, Ferraris JD, Dmitrieva NI. Cellular response to hyperosmotic stresses. Physiol Rev 2007; 87:1441-1474; PMID:17928589; https://doi.org/10.1152/physrev.00056.2006
- Burg MB, Ferraris JD. Intracellular organic osmolytes: function and regulation. J Biol Chem 2008; 283:7309-7313; PMID:18256030; https://doi.org/10.1074/jbc.R700042200
- Ferraris JD, Burg MB. Tonicity-regulated gene expression. In: Methods in enzymology, Dieter H, Helmut S, eds. Cambridge, MA: Academic Press; 2007; 279-296.
- Poon GM. Sequence discrimination by DNA-binding domain of ETS family transcription factor PU.1 is linked to specific hydration of protein-DNA interface. J Biol Chem 2012; 287:18297-18307; PMID:22474303; https://doi.org/10.1074/jbc.M112.342345
- Bortner CD, Scoltock AB, Sifre MI, Cidlowski JA. Osmotic stress resistance imparts acquired anti-apoptotic mechanisms in lymphocytes. J Biol Chem 2012; 287:6284-95; PMID:22228768
- Iwafuchi-Doi M, Zaret KS. Pioneer transcription factors in cell reprogramming. Genes Dev 2014; 28:2679-92; PMID:25512556; https://doi.org/10.1101/gad.253443.114
- Goswami R, Kaplan MH. Gcn5 is required for PU.1-dependent IL-9 induction in Th9 cells. J Immunol 2012; 189:3026-3033; PMID:22904310; https://doi.org/10.4049/jimmunol.1201496
- Sherwood RI, Hashimoto T, O'Donnell CW, Lewis S, Barkal AA, van Hoff JP, Karun V, Jaakkola T, Gifford DK. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol 2014; 32:171-178; PMID:24441470; https://doi.org/10.1038/nbt.2798
- van Riel B, Rosenbauer F. Epigenetic control of hematopoiesis: the PU.1 chromatin connection. Biol Chem 2014; 395:1265-1274; PMID:25205721; https://doi.org/10.1515/hsz-2014-0195
- Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 2012; 149:467-482; PMID:22500808; https://doi.org/10.1016/j.cell.2012.01.056
- Alvarez-Errico D, Vento-Tormo R, Sieweke M, Ballestar E. Epigenetic control of myeloid cell differentiation, identity and function. Nat Rev Immunol 2015; 15:7-17; PMID:25534619; https://doi.org/10.1038/nri3777
- Stephens DC, Poon GM. Differential sensitivity to methylated DNA by ETS-family transcription factors is intrinsically encoded in their DNA-binding domains. Nucleic Acids Res 2016; 44:8671-8681; PMID:27270080; https://doi.org/10.1093/nar/gkw528
- Gurion R, Vidal L, Gafter-Gvili A, Belnik Y, Yeshurun M, Raanani P, Shpilberg O. 5-azacitidine prolongs overall survival in patients with myelodysplastic syndrome – a systematic review and meta-analysis. Haematologica 2010; 95:303-310; PMID:19773261; https://doi.org/10.3324/haematol.2009.010611
- Darnell JE, Jr. Transcription factors as targets for cancer therapy. Nat Rev Cancer 2002; 2:740-749; PMID:12360277; https://doi.org/10.1038/nrc906
- Koehler AN. A complex task? Direct modulation of transcription factors with small molecules. Curr Opin Chem Biol 2010; 14:331-340; PMID:20395165; https://doi.org/10.1016/j.cbpa.2010.03.022
- Ghosh D, Papavassiliou AG. Transcription factor therapeutics: long-shot or lodestone. Curr Med Chem 2005; 12:691-701; PMID:15790306; https://doi.org/10.2174/0929867053202197
- Hagenbuchner J, Ausserlechner MJ. Targeting transcription factors by small compounds – current strategies and future implications. Biochem Pharmacol 2016; 107:1-13; PMID:26686579; https://doi.org/10.1016/j.bcp.2015.12.006
- Melander C, Burnett R, Gottesfeld JM. Regulation of gene expression with pyrrole-imidazole polyamides. J Biotechnol 2004; 112:195-220; PMID:15288953; https://doi.org/10.1016/j.jbiotec.2004.03.018
- Reddy BS, Sharma SK, Lown JW. Recent developments in sequence selective minor groove DNA effectors. Curr Med Chem 2001; 8:475-508; PMID:11281837; https://doi.org/10.2174/0929867003373292
- Dickinson LA, Trauger JW, Baird EE, Dervan PB, Graves BJ, Gottesfeld JM. Inhibition of Ets-1 DNA binding and ternary complex formation between Ets-1, NF-kappaB, and DNA by a designed DNA-binding ligand. J Biol Chem 1999; 274:12765-12773; PMID:10212261; https://doi.org/10.1074/jbc.274.18.12765
- Nhili R, Peixoto P, Depauw S, Flajollet S, Dezitter X, Munde MM, Ismail MA, Kumar A, Farahat AA, Stephens CE et al. Targeting the DNA-binding activity of the human ERG transcription factor using new heterocyclic dithiophene diamidines. Nucleic Acids Res 2013; 41:125-138; PMID:23093599; https://doi.org/10.1093/nar/gks971
- Munde M, Wang S, Kumar A, Stephens CE, Farahat AA, Boykin DW, Wilson WD, Poon GM. Structure-dependent inhibition of the ETS-family transcription factor PU.1 by novel heterocyclic diamidines. Nucleic Acids Res 2014; 42:1379-1390; PMID:24157839; https://doi.org/10.1093/nar/gkt955
- Stephens DC, Kim HM, Kumar A, Farahat AA, Boykin DW, Poon GMK. Pharmacologic efficacy of PU.1 inhibition by heterocyclic dications: a mechanistic analysis. Nucleic Acids Res 2016; 44:4005-4013; PMID:27079976; https://doi.org/10.1093/nar/gkw229
- Testoni M, Chung EYL, Priebe V, Bertoni F. The transcription factor Ets1 in lymphomas: friend or foe? Leukemia Lymphoma 2015; 56:1975-1980; PMID: 25363344; https://doi.org/10.3109/10428194.2014.981670
- Cook WD, McCaw BJ, Herring C, John DL, Foote SJ, Nutt SL, Adams JM. PU.1 is a suppressor of myeloid leukemia, inactivated in mice by gene deletion and mutation of its DNA binding domain. Blood 2004; 104:3437-3444; PMID:15304397; https://doi.org/10.1182/blood-2004-06-2234
- Okuno Y, Yuki H. PU.1 is a tumor suppressor for B cell malignancies. Oncotarget 2012; 3:1495-1496; PMID:23455457; https://doi.org/10.18632/oncotarget.800
- Garvie CW, Pufall MA, Graves BJ, Wolberger C. Structural analysis of the autoinhibition of Ets-1 and its role in protein partnerships. J Biol Chem 2002; 277:45529-45536; PMID:12221090; https://doi.org/10.1074/jbc.M206327200
- Poon GM. DNA binding regulates the self-association of the ETS domain of PU.1 in a sequence-dependent manner. Biochemistry 2012; 51:4096-4107; PMID:22533913; https://doi.org/10.1021/bi300331v
- Jia X, Lee LK, Light J, Palmer AG, 3rd, Assa-Munt N. Backbone dynamics of a short PU.1 ETS domain. J Mol Biol 1999; 292:1083-1093; PMID:10512704; https://doi.org/10.1006/jmbi.1999.3123
- De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Basinko A, De Braekeleer M. ETV6 fusion genes in hematological malignancies: a review. Leuk Res 2012; 36:945-961; PMID:22578774; https://doi.org/10.1016/j.leukres.2012.04.010
- Oikawa T. ETS transcription factors: possible targets for cancer therapy. Cancer Sci 2004; 95:626-633; PMID:15298723; https://doi.org/10.1111/j.1349-7006.2004.tb03320.x
- Blair DG, Athanasiou M. ETS and retroviruses – transduction and activation of members of the ETS oncogene family in viral oncogenesis. Oncogene 2000; 19:6472-6481; PMID:11175363; https://doi.org/10.1038/sj.onc.1204046
- Camoes MJ, Paulo P, Ribeiro FR, Barros-Silva JD, Almeida M, Costa VL, Cerveira N, Skotheim RI, Lothe RA, Henrique R et al. Potential downstream target genes of aberrant ETS transcription factors are differentially affected in Ewing's sarcoma and prostate carcinoma. PLoS One 2012; 7:e49819; PMID:23185447; https://doi.org/10.1371/journal.pone.0049819
- Hollenhorst PC, Ferris MW, Hull MA, Chae H, Kim S, Graves BJ. Oncogenic ETS proteins mimic activated RAS/MAPK signaling in prostate cells. Genes Dev 2011; 25:2147-2157; PMID:22012618; https://doi.org/10.1101/gad.17546311
- Tondravi MM, McKercher SR, Anderson K, Erdmann JM, Quiroz M, Maki R, Teitelbaum SL. Osteopetrosis in mice lacking haematopoietic transcription factor PU.1. Nature 1997; 386:81-84; PMID:9052784; https://doi.org/10.1038/386081a0
- Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science 2013; 339:957-959; PMID:23348506; https://doi.org/10.1126/science.1229259
