ABSTRACT
Transcription regulation is an important mechanism that controls pluripotency and differentiation. Transcription factors dictate cell fate decisions by functioning cooperatively with chromatin regulators. We have recently demonstrated that BEND3 (BANP, E5R and Nac1 domain) protein regulates the expression of differentiation-associated genes by modulating the chromatin architecture at promoters. We highlight the collaboration of BEND3 with the polycomb repressive complex in coordinating transcription repression and propose a model highlighting the relevance of the BEND3-PRC2 axis in gene regulation and chromatin organization.
Abbreviations: BEND3, BANP, E5R and Nac1 domain; rDNA, ribosomal DNA; PRC2, Polycomb Repressive Complex 2; H3K27me3, Histone H3 Lysine 27 methylation; PcG, Polycomb group.
Introduction
Transcription factors regulate gene expression by either activating or repressing promoter activity and thereby control initiation of transcription. Repression of transcription is often associated with altering the chromatin architecture through the recruitment of chromatin-modifying complexes. BEN domain-containing factors are relatively newly identified as DNA binding modules that are transcriptional repressors [Citation1]. The BEN domain is predicted to function as an adaptor for higher-order chromatin organization and recruitment of chromatin complexes to regulate gene expression. In this article, we present how the quadruple BEN domain-containing repressor, BEND3, associates with the polycomb repressive complex to mediate transcriptional repression at differentiation-associated genes.
We have reported previously that BEND3 binds to heterochromatic structures and causes transcriptional repression by chromatin compaction at those sites [Citation2]. Furthermore, we found that BEND3 associates with rDNA, mediates rRNA gene repression and mediates rDNA silencing by stabilizing the nucleolar-remodeling complex (NoRC) [Citation3]. Finally, we demonstrated that the epigenetic changes in the rDNA regions were important for this repression. We and others recently reported that BEND3 is highly upregulated in stem cells, induced pluripotent stem cells, and pluripotent embryonal carcinoma cells [Citation4–6]. BEND3 is associated with genes involved in differentiation, and the expression of BEND3 resulted in enhanced accumulation of H3K27me3 marks at select promoters. Similarly, the Zhu laboratory proposed that BEND3 causes polycomb stabilization to safeguard the differentiation process [Citation5]. BEND3 has also been implicated in generating a repressive chromatin by facilitating polycomb recruitment and H3K27me3 deposition to specific sites that are devoid of H3K9me3 or DNA methylation [Citation7]. All the above supports the model that epigenetic changes are important means by which BEND3 mediates repression.
Polycomb proteins are epigenetic modifiers that regulate the expression of developmental genes in a cell and tissue context-dependent manner. These groups of proteins are required for embryonic development and play pivotal roles in stem cells [Citation8]. In mice as well as in humans, PRC2 is required for embryonal stem cell (ESC) fate specification and also shown to regulate the expression of developmental genes [Citation9–11]. A plethora of transcription factors have been reported to be master regulators in stem cells, including pluripotent and multipotent stem cells. PcGs exist in the form of two complexes, polycomb repressive complexes 1 and 2 [Citation12]. The PRC1 complex has an E3 ligase activity, and one of the main substrates is the monoubiquitinated H2A at lysine 119, while PRC2 contains methyltransferase activity catalyzed by the methyltransferase EZH1/2 that is responsible for H3K27 di- and trimethylation [Citation13]. It is generally believed that PRC2 is involved in recruiting PRC1 to promoters of common target genes. Both are crucial for gene repression, although newer data have suggested nonoverlapping functions in embryonic and adult stem cells [Citation14].
EZH2 is highly expressed in embryonic tissues and in actively proliferating cells [Citation15]. Many PRC2-associated factors have been shown to regulate the PRC2 enzymatic activity and its recruitment to specific gene loci. For example, the association of Pcl2 bound to PRC2 has also been reported to bind to pluripotent genes to negatively regulate its expression in embryonic stem cells. It is believed that the PRC2-containing PCL2 proteins at the promoter of pluripotent genes can rapidly repress these genes upon receiving a signal for differentiation [Citation16]. Similarly, the transcription repressor Jarid2 is a PRC2 interactor in ESCs and the model is that Jarid2 stabilizes PRC2ʹs occupancy at chromatin [Citation17,Citation18]. Interestingly, genome wide data of PRC2 components (Ezh2, Suz12) and those of H3K27me3 in pluripotent cells show the association of these with a subset of CpG islands, specifically the ones that lack transcriptional activators [Citation19]. There are data supporting that association with various factors dictates the functionality of PRC2. In this point of view, we discuss the functional relevance of PRC2 association with a transcription repressor and how this might influence the maintenance of pluripotency.
BEND3 associates with the polycomb repressive complex proteins
Work from the Dejardin laboratory has shown that BEND3 helps the recruitment of the H3K27me3 methyltransferase EZH2 to chromatin sites, especially in the absence of constitutive heterochromatic marks, like H3K9me3 [Citation7]. Their observations prompted the authors to propose a role for BEND3 in switching from constitutive to facultative heterochromatin. It is worth noting that BEND3 associates with pericentric heterochromatin and at telomeres, and the association with heterochromatic sites increases dramatically upon the loss of enzymes that catalyze H3K9me3. Furthermore, the Zhu laboratory showed that loss of BEND3 resulted in the loss of SUZ12 (a subunit of PRC2) in the BEND3-occupied regions [Citation5]. This was concomitant with the loss of H3K27me3 at these sites, especially at the bivalent targets. Based on these and our own recent results, we examined the functional interaction of BEND3 with the PRC2 complex.
Using an artificially generated heterochromatin locus in vivo, we tethered BEND3 to the locus using a triple fusion protein (CFP-LacI-BEND3). We then addressed the association of polycomb proteins (YFP-EZH2) with the BEND3-tethered locus. We find that BEND3 associates with PRC2 at the in vivo reporter locus () and at other prominent heterochromatic foci, including at rDNA regions.
Figure 1. A heterochromatic locus stably integrated in U2OS 2-6-3 cells was visualized by LacI. LacI fusion proteins are forcibly tethered to the LacO repeats, and CFP is fused to LacI for visualizing the loci. The coding region of EZH2 and BEND3 was PCR amplified and cloned into pEYFP-C1 (Clontech) and pECFP-LacI (modified from pEGFP-LacI; kindly provided by Miroslav Dundr) [Citation27]. U2OS 2-6-3 cells were transiently transfected with 100 ng of CFP-LacI or CFP-LacI-BEND3 and 500 ng of YFP-EZH2. Images of EZH2 recruitment to the heterochromatin loci in CFP-LacI or CFP-LacI-BEND3 expressing cells. Note the robust overlap of BEND3 and EZH2 at the CLTon locus and at heterochromatic sites. The scale bar represents 5 μm.
![Figure 1. A heterochromatic locus stably integrated in U2OS 2-6-3 cells was visualized by LacI. LacI fusion proteins are forcibly tethered to the LacO repeats, and CFP is fused to LacI for visualizing the loci. The coding region of EZH2 and BEND3 was PCR amplified and cloned into pEYFP-C1 (Clontech) and pECFP-LacI (modified from pEGFP-LacI; kindly provided by Miroslav Dundr) [Citation27]. U2OS 2-6-3 cells were transiently transfected with 100 ng of CFP-LacI or CFP-LacI-BEND3 and 500 ng of YFP-EZH2. Images of EZH2 recruitment to the heterochromatin loci in CFP-LacI or CFP-LacI-BEND3 expressing cells. Note the robust overlap of BEND3 and EZH2 at the CLTon locus and at heterochromatic sites. The scale bar represents 5 μm.](/cms/asset/a7120750-c918-4d74-adc8-c013a3bcc43f/ktrn_a_2105128_f0001_oc.jpg)
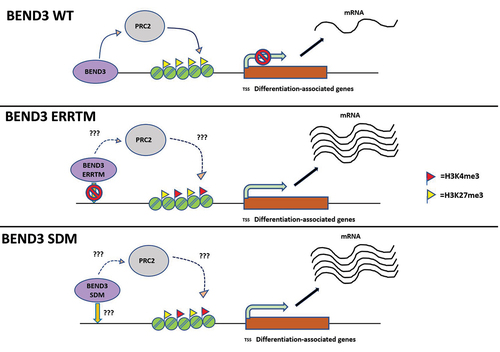
Furthermore, we found that BEND3-occupied gene promoters also show enrichment of PRC2 components at these promoters. PcG decorates H3K27me3 at bivalent gene promoters, promoters that have both activating H3K4me3 and repressing H3K27me3 marks [Citation19]. At these bivalent genes, activation and repression of genes provide a means for epigenetic regulation to induce differentiation. The Zhu laboratory proposed that the gene bivalency prevents gene activation during differentiation [Citation5]. We showed that the expression of BEND3 causes increased H3K27me3 at BEND3-occupied promoters of prodifferentiation genes [Citation4]. Based on these, we propose that BEND3 is required to fine-tune the expression of prodifferentiation genes by modulating the levels of H3K27me3. BEND3 enables polycomb recruitment and H3K27me3 deposition to BEND3-bound promoters to generate repressive chromatin. These data are consistent with the recent report that BEND3 function is largely dependent on PRC2 at the bivalent CpG islands that are enriched for BEND3 occupancy.
BEND3 associates with multiple large complexes including NoRC, NuRD and PRC2 complexes, and we believe that this is highly context, tissue-, and developmental stage-specific interactions [Citation3,Citation7]. It is equally likely that these complexes coexist, resulting in significant crosstalk and collaboration at specific chromatin sites to mediate gene regulation. It is well established that PRC2 is critical for differentiation and development, [Citation20] and therefore, it was not entirely surprising to find a concerted role of BEND3 with PRC2 in mediating these processes. Significantly, BEND3 is autoregulatory and binds its own promoter (). It is important to note that the PRC2 complex is enriched at the BEND3 promoter (), further supporting our observations that the association of BEND3 with PRC2 is a mechanism for transcription repression at the BEND3 gene locus ()
DNA-binding domain of BEND3 is critical for transcription repression
We have previously shown that BEN-domain 4 (BD4) is critical for heterochromatin localization [Citation2]. Our recent collaborative work with the Lai laboratory further supported these observations [Citation6]. We found that the BD4 is a major determinant for in vivo association and repression of BEND3 target genes, including at the previously reported rDNA genomic sites and the calreticulin (CALR) gene [Citation3,Citation21]. BD4 was found to harbor specific DNA binding activity at the sequence YCCACGC [Citation6]. The Zhu laboratory also identified a sequence motif CCCACGCG (with two CpG sites) as the most enriched motif and found that BEND3 occupancy was most prominent at CpG island-associated genes in stem cells [Citation5]. Furthermore, they found that BD4 recognizes its target DNA in a DNA-methylation-sensitive manner. We also identified BEND3 binding to the TTAGGG repeat sites and at G-quadruplexes in the embryonal carcinoma cells [Citation4]. These results are consistent with BEND3 localization at the telomeric heterochromatin and to gene promoters [Citation4,Citation22].
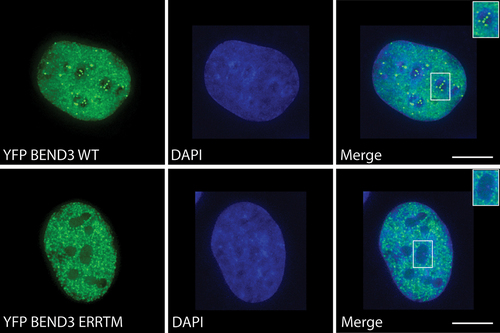
Three mutations within the BD4 residues (E807V, R810L and R814L) that are involved in sequence-specific base contacts have previously been demonstrated to abolish the repressive ability of BEND3 [Citation6]. We find that YFP-BEND3-ERR mutant fails to bind to rDNA, consistent with the data that these residues are critical for DNA binding and transcription repression (). As reported recently, DNA recognition by BEN domains is very similar among other BEN-domain containing factors, including in Drosophila Insv and Bsg25a [Citation23,Citation24] and these are distinguishable from the non-DNA-binding BEN domains, which likely facilitate protein-protein interactions.
Figure 3. YFP-BEND3- and YFP-BEND3-ERR-mutant distribution in the interphase cell. Note the lack of rDNA binding of the ERR mutant. The rDNA signal within the nucleolus (white box) is shown in the inset.
Future work would determine if the DNA binding ability of BEND3 is critical for PRC2 association with BEND3 and with specific gene promoters ().
SUMOylation of BEND3 is required for transcription repression
Our earlier work has demonstrated that SUMOylation of BEND3 at K20 and K512 is important for transcription repression [Citation2]. Furthermore, we showed that these post-translational modifications are required for its function as a repressor in a reporter assay. We also reported the relevance of SUMOylation in the stability of NoRC and its requirement for rDNA silencing. The mechanistic insights into how SUMOylation of BEND3 may impact its ability to associate with other interactors remains to be investigated. It has been suggested that transcription factor SUMOylation could facilitate repression by increased association with co-repressors or by altering the association with target sites on chromatin and with chromatin modifiers [Citation25].
To address how the SUMOylation of BEND3 regulates gene expression, it would be important to study the binding of BEND3-SUMO double mutant (SDM) to gene promoters where BEND3 has previously been shown to bind to and cause repression. Since the SUMO sites are not in the BD4 domain, our prediction would be that the SDM may continue to bind gene promoters. It would be important to pinpoint the domain within BEND3 that associates with the PRC2 complex. Functional analyses of the truncation mutants and the SDM mutant and their association with PRC2 would provide important insights into why SDM mutants fail to repress transcription ().
Concluding remarks
BEND3 levels in cells are very precisely regulated, and this is corroborated by the fact that it is autoregulatory. Significantly, overexpression of BEND3 causes hyperchromatinization, with decoration of H3K27me3 genome-wide. We and others have demonstrated that BEND3 mediates transcription repression by virtue of association with the polycomb complex in a cell- and tissue-specific context. PcGs are known to modulate transcription by multiple mechanisms, including binding to a plethora of factors, altering chromatin architecture and imposing changes to the transcriptional machinery [Citation26]. BEND3 is highly upregulated in stem cells and shows excellent correlation with PRC2 association with bivalent gene promoters. It is known that PRC2-mediated repression is established during development, and this controls spatiotemporal gene activity. The co-occupancy of BEND3 and PRC2 complex proteins at bivalent gene promoters in pluripotent cells provides another dimension to how gene expression is intricately coordinated during development. Much work needs to be performed in order to understand how chromatin-dependent gene regulation impacts development.
Acknowledgments
We thank Drs K. Prasanth, E. Lai, M. Aladjem and M. Kamran for sharing reagents, discussions and suggestions. This work was supported by NSF (1243372 and 1818286) and NIH (GM125196) awards to SGP. The authors declare no competing financial interests.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Abhiman S, Iyer LM, Aravind L. BEN: a novel domain in chromatin factors and DNA viral proteins. Bioinformatics. 2008;24:458–461.
- Sathyan KM, Shen Z, Tripathi V, et al. A BEN-domain-containing protein associates with heterochromatin and represses transcription. J Cell Sci. 2011;124:3149–3163.
- Khan A, Giri S, Wang Y, et al. BEND3 represses rDNA transcription by stabilizing a NoRC component via USP21 deubiquitinase. Proc Natl Acad Sci U S A. 2015;112:8338–8343.
- Kurniawan F, Chetlangia N, Kamran M, et al. BEND3 safeguards pluripotency by repressing differentiation-associated genes. Proc Natl Acad Sci U S A. 2022;119. e2107406119
- Zhang J, Zhang Y, You Q, et al. Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation. Science. 2022;375:1053–1058.
- Zheng L, Liu J, Niu L, et al. Distinct structural bases for sequence-specific DNA binding by mammalian BEN domain proteins. Genes Dev. 2022;36:225–240.
- Saksouk N, Barth TK, Ziegler-Birling C, et al. Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol Cell. 2014;56:580–594.
- Aloia L, Di Stefano B, Di Croce L. Polycomb complexes in stem cells and embryonic development. Development. 2013;140:2525–2534.
- Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043.
- Kirmizis A, Bartley SM, Kuzmichev A, et al. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–1605.
- Wang H, Wang L, Erdjument-Bromage H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878.
- Blackledge NP, Rose NR, Klose RJ. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol. 2015;16:643–649.
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349.
- Luis NM, Morey L, Di Croce L, et al. Polycomb in stem cells: PRC1 branches out. Cell Stem Cell. 2012;11:16–21.
- Chou RH, Yu YL, Hung MC. The roles of EZH2 in cell lineage commitment. Am J Transl Res. 2011;3:243–250.
- Walker E, Chang WY, Hunkapiller J, et al. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2010;6:153–166.
- Kalb R, Latwiel S, Baymaz HI, et al. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol. 2014;21:569–571.
- Son J, Shen SS, Margueron R, et al. Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev. 2013;27:2663–2677.
- Ku M, Koche RP, Rheinbay E, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242.
- Shan Y, Liang Z, Xing Q, et al. PRC2 specifies ectoderm lineages and maintains pluripotency in primed but not naive ESCs. Nat Commun. 2017;8:672.
- Aghajanirefah A, Nguyen LN, Ohadi M. BEND3 is involved in the human-specific repression of calreticulin: implication for the evolution of higher brain functions in human. Gene. 2016;576:577–580.
- Khan A, Prasanth SG. BEND3 mediates transcriptional repression and heterochromatin organization. Transcription. 2015;6:102–105.
- Dai Q, Ren A, Westholm JO, et al. Common and distinct DNA-binding and regulatory activities of the BEN-solo transcription factor family. Genes Dev. 2015;29:48–62.
- Dai Q, Ren A, Westholm JO, et al. The BEN domain is a novel sequence-specific DNA-binding domain conserved in neural transcriptional repressors. Genes Dev. 2013;27:602–614.
- Rosonina E, Akhter A, Dou Y, et al. Regulation of transcription factors by sumoylation. Transcription. 2017;8:220–231.
- Aranda S, Mas G, Di Croce L. Regulation of gene transcription by Polycomb proteins. Sci Adv. 2015;1:e1500737.
- Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717.