Catechins-rich and -poor tea tree lines were selected through a colorimetric selection method from 160 tea tree lines used in previous studies. The selected tea tree lines, catechins-rich lines (HR-52, HR-29, HR-82, HR-123, and HR-55) and catechins-poor lines (HP-19, HP-108, HP-138, HP-150, and HP-18), were analyzed through morphological (leaf width, leaf length, leaf area, and shoot length) and genetic analysis. The cluster pattern of morphological characters showed a positive correlation between the catechins contents and the leaf morphology. In RAPD analysis, the amplified DNA bands did not appear with 10 primers, however, the various sizes of DNA bands were detected with 22 primers. The special DNA bands between the rich and poor lines were not found. These results suggest that the morphological and genetic character may be somewhat related to secondary metabolites of tea trees.
Introduction
Conventional tea tree breeding is well established, though time consuming and labor intensive due to its perennial nature and long gestation period (Mondal et al. Citation2004). Additionally, tea breeding has been slow due to a lack of reliable selection criteria (Islam et al. Citation2005). Particularly, studies concerning the selection and breeding of tea trees containing valuable plant secondary metabolites are scanty. Therefore, the development of methods for the rapid selection of catechins-rich tea trees is an important strategy for breeding new cultivars. In earlier research, the FBB colorimetric method was successfully used to select 160 tea trees on the basis of their contents as catechins-rich and -poor lines (Kim et al. Citation2010). Among the tea-tree lines selected by the colorimetric method, the content of catechins was high, containing 5-fold for EGC, 16-fold for EC, 47-fold for EGCG, and 4.4-fold for ECG content.
Biosynthesized plant secondary metabolites depend on both genetic and environmental factors, and large intraspecific variation in secondary chemistry has frequently been reported. The heritability of specific tree secondary metabolites is, however, mostly unknown (Laitinen et al. Citation2005).
Variation of catechins content may be a feature characteristic to the wild tea tree population and geographic regions or habitats of Hadong, Korea. A tea tree is an allogamous plant. Tea trees are propagated through seeding, which brings about high degrees of variability.
The characteristic analysis of leaf morphology and its genetic aspects is not well reported. Oh and Hong (Citation1999) reported a genetic relationship among Korean native tea tree (Camellia sinensis L.) using RAPD markers. Also, a few studies for the analysis on each characteristic of a tea leaf and on its genetic organization have been reported (Kaundun and Park Citation2002; Lee et al. Citation2002; Park et al. Citation2002). However, to our knowledge, the genetic and morphological basis for intraspecific variation in tea catechins biosynthesis has not yet been explored. In the study, the genetic and morphogenic characterization between catechins-rich and -poor tea tree lines is determined through morphological analysis and RAPD analysis.
Materials and methods
Plant materials
Among the selected 160 different tea tree lines, leaves from catechins-rich lines (HR-52, HR-29, HR-82 HR-123, and HR-55) and catechins-poor lines (HP-19, HP-108, HP-138, HP-150, and HP-18) were collected (). After sampling, the leaves and stems were stored at −70°C until further use.
Table 1. Catechins content of selected tea trees used in this study.
Morphological analysis of selected tea tree lines
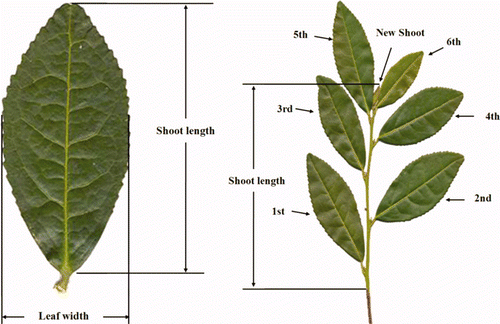
One-year-old leaves were used for the analysis. In order to survey the morphological characteristics of tea trees, the petiole length was measured with the Mitutoyo ruler without the maximum error. The name used for each part of a fresh 1-year-old shoot of a tea tree is shown in . The leaf area was measured by using the following formula:
Genetic relationship though RAPD analysis
100 mg of the leaf tissues were ground in liquid nitrogen and DNA extracted using DNeasy plant kit (QIAGEN, Germany). Extracted DNA was diluted with distilled water to 10 μg μl−1 and stored at − 20°C. Amplification reactions were performed in volumes of 20 μl including 20 μg of template DNA, 100 μM of arbitrary random primer (Bioneer Co, Korea), 200 μM of Dntp, 10 × reaction buffer, 1.0 U of Taq DNA polymerase (Bioneer Co, Korea), and ddH2O.
PCR was carried out in 20 μl reaction mixtures containing 0.3 μg each of chromosomal DNA, 10 mM Tris-HCl (pH 8.3), 5 mM KCl, 1.5 mM MgCl2, 100mM each of dNTP, 25 pmol primer, and 0.5 U of Taq DNA polymerase (Takara Co., Okayama, Japan). The mixture was overlaid with 20 μl of mineral oil (Sigma), and PCR was performed in a Takara PCR thermal as follows: initial denaturation at 94°C for 5 min followed by 45 cycles of 94°C for 1 min, 45°C for 1 min, and 72°C for 1 min, and a final 5 min extension at 72°C. Amplification was analysis by gel electrophoresis in 1% agarose gel (0.5 × TBE buffer, pH 8.0) and detected by UV transilluminator, staining in ethidium bromide. As a molecular weight marker, 100 bp and 1500 bp ladder (Bioneer Co., Korea) was used. The primers used were randomly constructed 10 mer primers, OPB, OPC, and OPD. They were synthesized by Takara Co., Ltd (Okayama, Japan). In all, 36 primers that gave clear and reproducible fragment patterns were selected for our final analysis.
For each accession, polymorphism was scored as 1 for the presence and 0 for absence of a band. RAPD data generated with 30 primers was used to compile a binary matrix for cluster analysis using the NTSYS-pc (Numerical Taxonomy and Multivariate Analysis System, Biostatistics, New York, USA, software version 2.02j package). Similarity measures were using UPGMA (unweighted pair group method with arithmetic averages). Genetic similarity among accessions was calculated according to the Dice similarity coefficient (Dice Citation1945). The similarity coefficients were then used to construct a dendrogram using the UPGMA (Unweighted Pairwise Group Method with Arithmetical averages) through NTSYS-pc. The screening of the entire set of samples was done three times to assess repeatability of the RAPD profiles and identical RAPD patterns were obtained.
Statistical analysis
Data are expressed as an average of at least three experiments. Each numeral value represents the mean and the standard deviation (SD).
Results and discussion
Morphological characters in selected tea tree lines
Morphological characters such as shoot length, leaf width, leaf length, and leaf area were investigated from the fresh 1-year-old shoot (). The leaf width of the catechins-rich lines is 18.7 mm, and that of the selected catechins-poor lines is 19.5 mm. It was shown that as a whole, there is no big difference in the leaf width. It was checked that even though the leaf width of the catechins-rich tea tree is a bit wide as a whole, the width of the 6th leaf of the catechins-poor line is rather wide.
Table 2. Morphology analysis from selected tea tree lines.
As a whole, the leaf length of the selected catechins-rich line was longer than that of the catechins-poor lines. The 2nd leaf is the largest one and is 54.5 and 48.3 mm wide. The leaf length of the selected catechins-rich line was 47.9 mm and that of the selected catechins poor line was 42.2 mm. According to the statistics, the three groups were generated. It was checked that 4th, 5th, 3rd, and 2nd were grouped similarly. However, they were not grouped as the catechins-rich and -poor tea tree lines.
According to the observation, the average area of 1st, 2nd, 3rd, and 6th leaves is 7.23 cm2 and is wider than that of the average area (4.75 cm2) of the 4th and 5th. Moreover, the area gets small abruptly in the 5th leaf. The tendency appeared in the whole two groups of selected catechins-rich and -poor tea trees. It was particularly noticeable in the selected catechins-poor tea trees. As the statistical data on the basis of leaf length, the four of them such as 3rd, 5th, 1st, and 2nd are grouped similarly.
The average leaf morphology of fresh shoots at 1 year old in catechins-rich and -poor tea tree lines is 74.02 mm and 52.84 mm, respectively. The shoot length of the HR-55 line was measured to be 89.4 mm long resulting in the largest value. The shoot length of the HP-150 line was measured to be 30.7 mm resulting in the smallest value. The leaf morphology of a fresh shoot at 1-year-old of the HP-150 line is longer by 2.91 times compared with the HR-55 line. The results of statistical analysis show that other characters were not correlated among the selected lines ().
Figure 2. Morphology analysis from selected tea tree lines. (a) The average shoot length from selected tea tree lines. (b) The cluster analysis based on morphology analysis from selected tea tree lines. 1: HR-52, 2: HR-29, 3: HR-82, 4: HR-123, 5: HR-55, 6: HP-19, 7: HP-108, 8: HP-138, 9: HP-150 and 10: HP-18. This cluster analysis based on each variable was done by using the SPSS 17.0 statistics program.
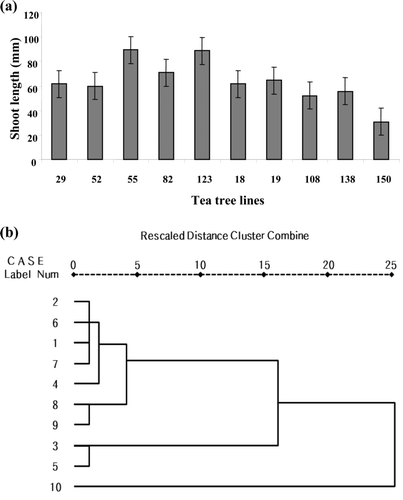
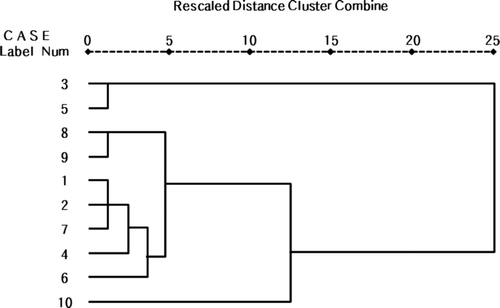
The cluster pattern was analyzed by utilizing four (leaf width, leaf length, leaf area, and shoot length) variables (; Table 3). As the result, they are generated into two large groups. Especially, 3rd (HR-82) and 5th (HR-55) line showed morphologically different groups compared with other selected trees. Furthermore, as it is a catechins-rich line, the internode distance is long, and the leaf area is narrow so that there is a negative correlation between the catechins content and the leaf morphology (). The other characters show no high correlation between the catechins content and the leaf morphology.
Figure 3. The cluster analysis based on four variables (leaf width, leaf length, leaf area, and shoot length) from selected tea tree lines. 1: HR-52, 2: HR-29, 3: HR-82, 4: HR-123, 5: HR-55, 6: HP-19, 7: HP-108, 8: HP-138, 9: HP-150, and 10: HP-18. This cluster analysis based on each variable was done by using the SPSS 17.0 statistics program.
Haughn et al. (Citation1991) also reported that glucosinolate content of Arabidopsis thaliana mutants does not result in any obvious changes in morphology or growth rate. According to Kim et al. (Citation2007), the family relation of a plant is mainly checked by analyzing the morphological characteristics, biochemical isoenzymes, and DNA of the plant. However, studies on the pattern tendency between highly functional selected trees have not been reported. In this study, fresh shoot at 1-year-old tree was observed to have a slight positive correlation with catechins. However, it was not clearly separated in parts of the content and pattern as a whole. It is thought that further research needs to be done.
Genetic relationship in selected tea tree lines
DNA was extracted from leaves of the 10 catechins-rich tea tree lines and then analyzed by RAPD analysis using 30 random primers. The amplified DNA bands did not appear in the 8 primers. The various sizes of DNA bands were produced in the 22 primers ().
Table 3. The community similarity among the four major stands within selected tea tree lines.
Table 4. RAPD primers and their sequences used to detect genetic diversity.
Of the primer contents the GC (%) contents ranged from 64.1% and did not show any effect on amplification. The size range of amplified products was 100 to 1500 bp from RAPD primers. The band pattern is located mainly between 300 and 1000 bp. The selected tea tree line used in the present study, produced almost identical banding pattern within each species in certain primer. And the number of polymorphic fragments for each primer also varied ().
Figure 4. Polymorphisms of selected tea tree lines by RAPD analysis using various primers. OPB18 (a); OPC4 (b); OPC7 (c); and OPC10 (d).
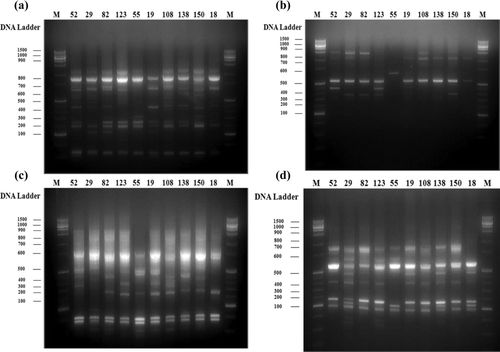
When using 22 primers, banding pattern showed reliability and repeatability. The amplification of the genomic DNA generated a total of 722 bands with an average of 32 bands per primer and obtained 97 (13.43%) polymorphic bands patterns with an average of 4.4 bands per primer. Maximum number of polymorphic bands was scored with primer OPC 13 (6 bands, 26.09%), however, minimum polymorphic bands produced by primer is OPB 10 (3 band, 7.32%). However, OPC 8 primer did not show any polymorphism.
The genetic diversity between catechins-rich and -poor tea tree lines was determined through RAPD-PCR analysis. As shown in , the special DNA band between the catechins-rich and -poor tea tree lines was not found. On the basis of the results of the RAPD analysis, the genetic similarity among selected trees was further analyzed. It was not a complete separation as a result of systems analysis, but the genetic similarity among 10 selected lines showed a range between 0.28 and 0.93. The lowest similarity showed 0.28 in the No. 5 (HR-55), No. 6 (HP-19) and No. 9 (HP-150) lines among selected trees. It was known that two genetic groups are classified through the experiment. The first group was classified as No. 5 (HR-55), No. 8 (HP-138), No. 7 (HP-108), No. 4 (HR-123), and No. 3 (HR-82) line. The second group is classified into No. 2 (HR-29), No. 1 (HR-52), No. 10 (HP-18), No. 9 (HP-150), and No. 6 (HP-19) lines, and the third group is classified into No. 3(HR-19) and No. 4 (HR-131) lines (). Variation within an individual tree and between branches at the same level of Betula tree canopy was low for most of the studied compounds and compound groups (Laitinen et al. Citation2005). Reichardt et al. (1981) found 25 times greater amounts of papyriferic acid in juvenile stems than in mature twigs. The results of this study agree with previous studies that catechins vary due to significant genotype × environment (different fertilization, defoliation, and moisture, etc.) interactions.
Figure 5. A UPGMA phenogram of catechins-rich and -poor tea tree lines based on RAPD analysis. 1: HR-52, 2: HR-29, 3: HR-82, 4: HR-123, 5: HR-55, 6: HP-19, 7: HP-108, 8: HP-138, 9: HP-150, and 10: HP-18.
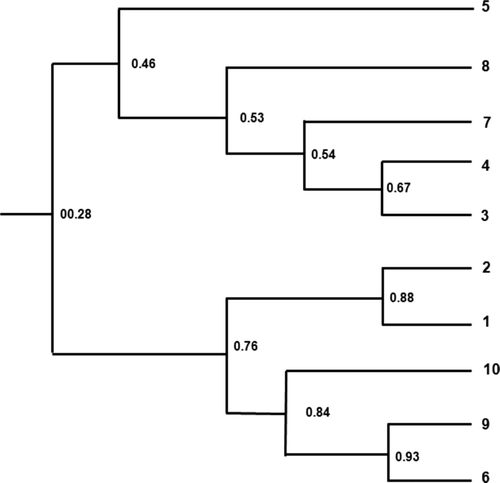
The genetic diversity among selected tea tree lines was analyzed by RAPD analysis. However, it was difficult to determine the genetic difference among selected lines. However, it is expected to get better results when utilizing more primers and other genetic analysis methods. Although these lines showed positive correlation between the catechins contents and the leaf morphology, comparison of the external morphological characteristics with RAPD analysis is needed for more reasonable conclusions. It is expected that these results can be used as the basic data to determine the genetic characteristics among bio-active compounds contained in tea trees. Moreover, this result can be also utilized in the breeding of highly valuable tea trees.
Acknowledgement
This research was supported by the Ministry for Food, Agriculture, Forestry and Fisheries.
References
- Dice , L R . 1945 . Measures of the amount of ecologic association between species . Ecology , 26 : 297 – 302 .
- Haughn , G W , Davin , L , Giblin , M and Underhill , E W . 1991 . Biochemical genetics of plant secondary metabolites in Arabidopsis thaliana . Plant Physiol , 97 : 217 – 226 .
- Islam , G MR , Iqbal , M , Quddus , K G and Ali , M Y . 2005 . Present status and future needs of tea industry in Bangladesh . Proc Pak Acad Sci , 42 : 305 – 314 .
- Kaundun , S S and Park , Y G . 2002 . Genetic structure of six Korean tea populations as revealed by RAPD-PCR markers . Crop Sci , 42 ( 2 ) : 594 – 601 .
- Kim , H J and Lee , S Y . 2007 . Genetic relationships based on morphological characteristics in Korean native Sedum sarmentosum . Kor J Hort Sci Technol , 25 ( 2 ) : 103 – 109 .
- Kim , Y D , Min , J Y , Jeong , M J , Song , H J , Hwang , J G , Karigar , C S , Cheong , G W and Choi , M S . 2010 . Rapid selection of catechins-rich tea-trees (Camellia sinensis L.) by a colorimetric method . J Wood Sci , 56 ( 5 ) : 411 – 417 .
- Lee , Y H , Song , K W , Chung , Y M and Choi , J H . 2002 . Analysis of genetic relationships among leaf characteristics in collected varieties of native tea by RAPD . J Kor Tea Soc , 18 ( 3 ) : 99 – 109 .
- Laitinen , M L , Julkunen-Tiitto , R , Tahvanainen , J , Heinonen , J and Rousi , M . 2005 . Variation in birch (Betula pendula) shoot secondary chemistry due to genotype, environment, and ontogeny . J Chem Ecol , 31 ( 4 ) : 697 – 717 .
- Mondal , T K , Bhattacharya , A , Laxmikumaran , M and Ahuja , P S . 2004 . Recent advances of tea (Camellia sinensis) biotechnology . Plant Cell Tiss Organ Cult , 76 : 195 – 254 .
- Oh , M J and Hong , B H . 1999 . Genetic relationship among Korean Native tea tree (Camellia sinensis L.) using RAPD markers . Kor J Breed , 27 ( 2 ) : 140 – 147 .
- Park , Y G , Kaundun , S S and Zhyvoloup , A . 2002 . Use of the bulked genomic DNA-based RAPD methodology to assess the genetic diversity among abandoned Korea tea plantations. Genet Res . Crop Evolut , 49 : 159 – 165 .
- Reichardt , P B . 1981 . Papyriferic acid: a triterpenoid from Alaskan paper birch . J Org Chem , 46 : 4576 – 4578 .