Abstract
Phytoplasma has been newly detected in Elaeocarpus sylvestris, a tree species that grows on the island of Jeju, South Korea. Various symptoms of infection of E. sylvestris have appeared since 2013, including loss of leaf coloration, such as yellowing, darkening and branch dieback. We analyzed leaves and seeds of E. sylvestris sequences infected by phytoplasma, and found partial 16S rRNA sequences to be 99.5%, and secA genes 98.2% correlated with Malaysian periwinkle virescence group strain ELY-BN1, MaPV, MOP, MYD, and TtWB. Analyzing these sequences and through the virtual restriction fragment length polymorphism (virtual-RFLP), we report here the first occurrence of Elaeocarpus decline associated with a phytoplasma strain belonging to 16SXXXII groups, as well as the first detection of phytoplasma in the seeds of E. sylvestris in South Korea.
Introduction
Phytoplasmas are wall-less pleomorphic prokaryotes that were previously classified as mycoplasma-like organisms (MLOs) on account of their morphological similarity to mycoplasmas, first occurred in mulberry phloem with dwarf disease (Doi et al. Citation1967; McCoy et al. Citation1989; Bertaccini and Duduk Citation2009; Weintraub and Jones Citation2009; Pagliari and Musetti Citation2019). Phytoplasma chromosome size is about 600–880 kb, which is very small in different from other plant pathogens (Oshima et al. Citation2013).
Phytoplasma are pathogens that is the cause of over 1,000 diseases in different wild and cultivated plant species all over the world (Seemuüller et al. Citation2002; Bertaccini Citation2007; Hoshi et al. Citation2007; Kumari et al. Citation2019; Namba Citation2019). In South Korea, such diseases have included jujube witches’ broom (Kim Citation1968), paulownia witches’ broom (La Citation1968), mulberry dwarf (Chang and Kim Citation1971), and sumac witches’ broom (Kim Citation1980) causing significant economic losses from a lot of trees affected throughout the country (Jung et al. Citation2012). Currently, including crops, about 64 species of host plants infected with phytoplasma have been reported in Korea. (Chung et al. Citation2011; Jung et al. Citation2012; Lee et al. Citation2017). Plants infected with phytoplasma also show many symptoms, including witches' broom, yellowing, phyllody, virescence, shortened internodes, and little leaf, as well as declining and vascular discoloration (Bertaccini and Duduk Citation2009; Kumari et al. Citation2019; Dermastia Citation2019). Meanwhile, with the development of molecular biology, PCR technique and sequence analysis on 16S ribosomal DNA, is in commonly used molecular diversity and taxonomy classification on phytoplasma (Lee et al. Citation1998; Bertaccini and Duduk Citation2009).
Elaeocarpus sylvestris var. ellipticus (Thunb.) H. Hara is an evergreen tree belonging to Elaeocarpaceae, grown in southern China, south central Japan, Taiwan, Vietnam, and on Jeju Island in South Korea and it reproduces mostly by seed germination (Kim and Kim Citation2014). However since 2013, E. sylvestris trees on Jeju have shown decline, while some trees have died on streets, in parks or in their natural habitat (Lee et al. Citation2017). Since the 1980s, similar disease have also been reported in Japan named ELY (Elaeocarpus yellows), and E. zollingeri trees have been infected with phytoplasma and are in danger of decimation (Ikeda and Hashimoto Citation1996; Narazaki and Tsuda Citation2008; Kawabe Citation2010; Odawar City Cultural Property Protection Committee Citation2013; Kawabe et al. Citation2011; Satoh et al. Citation2014; Iwabuchi et al. Citation2018).
Until the present study, no research on the decline of E. sylvestris trees from phytoplasma has been conducted in Korea to date, even though the number of areas with damaged trees is increasing every year. The purpose of this study is to identify the cause of the decline in E. sylvestris trees using PCR assays, base sequences, and virtual-RFLP. In addition, we will determine whether the phytoplasma is found in the seeds of tree with decline symptoms.
Materials and methods
Diseased samples and extraction of DNA
Our investigation was conducted on E. sylvestris in Jeju island, we were collected 14 symptomatic samples which yellowing, darkening leaves and seeds near symptomatic leaves between April and September 2019 (). Samples were stored at 4 °C, while to identify phytoplasma infection, DNA was extracted from 0.1 g of leaf midribs, petioles, and separating the flesh and seeds of the drupe, using DNeasy Plant Mini Kit (Qiagen, Hilden, Germany).
Table 1. List of E. sylvestris samples used in this study.
Table 2. Classification of phytoplasma species reference based on 16S rDNA gene.
PCR
PCR with the phytoplasma universal primers P1 (5′-AAGAGTTTGATCCTGGCTCAGGATT-3′′) and P7 (5′-CGTCCTTCATCGGCTCTT-3′′) was used to amplify 16S rRNA, the 16S-23S intergenic spacer (ITS) region, tRNA-lle, and partial 23S rRNA (Deng and Hiruki Citation1991; Schneider et al. Citation1995). Nested PCR analysis was conducted using universal primer R16F2n (5′-GAAACGACTGCTAAGACTGG-3′′) and R16R2 (5′-TGACGGGCGGTGTGTACAAACCCCG-3′′) (Lee et al. Citation1993; Gundersen and Lee Citation1996). Reactions were carried out using a 25 µl volume reaction mixture containing 100 ng/µl DNA, Emerald- Amp GT PCR Master Mix [2 X Premix] (TaKaRa, Shiga, Japan) for 10 pmol of each primer, and the rest was filled with distilled water. Conditions for PCR amplification were as follows: one cycle for denaturation at 94 °C for 7 min, 35 cycles denaturation at 94 °C for 1 min, annealing at 55 °C (nested PCR 58 °C) for 2 min, and extension at 72 °C for 3 min, with a final extension step of 72 °C for 10 min (Lee et al. Citation1993; Gundersen and Lee Citation1996; Lee et al. Citation1998; Han Citation2005; Kamala-Kannan et al. Citation2011). The secA gene was amplified using the primers SecAfor1 (5′-GARATGAAAACTGGRGAAGG-3′) and SecArev3 (5′-GTTTTRGCAGTTCCTGTCATNCC-3′). PCR conditions with the primer pair SecAfor1/SecArev3 were 94 °C for 2 min followed by 35 cycles of 94 °C for 30 s, 53 °C for 60 s and 72 °C for 90 s, with a final extension step of 72 °C for 15 min (Hodgetts et al. Citation2008).
Comparison of sequencing analysis
To find taxonomic lineage of pathogen that caused the diseased E. sylvestris trees, we using the iPhyClassifier software from the 16SrRNA gene (Zhao et al. Citation2009). In addition, we using 17 key restriction enzymes: AluI, BamHI, BfaI, BstUI, DraI, EcoRI, HaeIII, HhaI, HinfI, HpaI, HpaII, KpnI, Sau3AI, MseI, RsaI, SspI and TaqI were used to compare the virtual-RFLP analysis results.
Amplified PCR products were purified using a PCR Purification Kit (Bioneer, Seoul, South Korea) and clone used to transform Escherichia coli JM109 cells according to manufacturer’s instructions (Qiagen, Valencia, CA, USA). The sequencing were performed by Macrogen Co. (Daejeon, korea). Sequence alignments were analyzed using GENETYX-WIN 4.0 (GENETYX, Tokoyo, Japan), and phylogenetic analysis was performed with MEGA ver. 10.0 software using the Neighbor-joining method, with the default values and 10,000 replications for bootstrap analysis.
Results
Symptomatology
The most of diseased E. sylvestris trees were observed in various locations including streets, parks, around the apartment and natural habitats on Jeju. In some street trees, symptomatic trees appeared to decline symptoms collectively, more than 70% trees showed that decline symptom from April to October. Early symptoms were as the previous year’s leaves began yellowing and darkening. A number of leaves fall sharply, the leafless branches becoming weak. After years of repetition of these symptoms the tree eventually dries up and dies over time ().
Detection of phytoplasma and molecular diversity
Polymerase chain reaction with phytoplasma universal primers (P1/P7) amplified the expected DNA fragment of 1.8 kb and targeted 16S rRNA, The 16S–23S ITS region, and partial 23S rRNA. Nested PCR with R16F2n/R16R2 primers resulted in the predicted 1.2 kb amplicon in leaves, both flesh and seeds detected with phytoplasma (). The phytoplasma universal primers (SecAfor1/SecArev3) also amplified the expected size of 0.84 kb and targeted the partial secA gene (). The nucleotide sequences of the 16S rRNA and secA genes of infected leaves and seeds on Jeju showed a 100.0% identity starins ESDP-JJ1, JJ2, JJ3, JS1, and JS2. The pairwise alignment of the 16S rRNA gene sequence with available sequences in the NCBI database showed that the phytoplasma was more than 99.5% correlated with Malaysian periwinkle virescence group strain ELY-BN1, MaPV, MOP, MYD, and TtWB (, ). The 16S rRNA and 16/23S ITS sequences of E. sylvestris decline phytoplasma (ESDP) were deposited in GenBank accession numbers: OL689203, OL689204, OL689205, OL689206 and OL689207. The 16SrRNA phytoplasma sequences were insufficient for identification of phytoplasmas belonging to the same group. Pairwise alignment of the secA gene sequence with sequences obtained from the NCBI database revealed that the phytoplasma was more than 98.2% associated with Malaysian periwinkle virescence group (). The secA gene sequence of E. sylvestris decline phytoplasma (ESDP) were deposited in GenBank accession numbers: OL689198, OL689199, OL689200, OL689201 and OL689202.
Figure 2. Agarose gel electrophoresis patterns of PCR products 16S rRNA gene and secA gene amplified by nested PCR using primer pair R16F2n/R16R2 and Direct SecAfor1/SecArev3 from E. sylvestris with decline symptoms in Jeju island, [Lane: M. molecular weight marker (100 bp), 1 ∼ 14: the symptomatic leaves and seeds, 15: jujube witches’ broom phytoplasma, 16: sumac witches’ broom phytoplasma].
![Figure 2. Agarose gel electrophoresis patterns of PCR products 16S rRNA gene and secA gene amplified by nested PCR using primer pair R16F2n/R16R2 and Direct SecAfor1/SecArev3 from E. sylvestris with decline symptoms in Jeju island, [Lane: M. molecular weight marker (100 bp), 1 ∼ 14: the symptomatic leaves and seeds, 15: jujube witches’ broom phytoplasma, 16: sumac witches’ broom phytoplasma].](/cms/asset/3d090da2-4c79-427f-bd33-2dc1ad154e0b/tfst_a_2029774_f0002_c.jpg)
Figure 3. Virtual RFLP profiles of phytoplasma associated with E. sylvestris tree decline (Accession No: OL689203, OL689204, OL689205, OL689206 and OL689207) in-silico digestions. Sequences from strains ESDP-JJ1, JJ2, JJ3, JS1 and JS2 were used key restriction enzymes: AluI, BamHI, BfaI, BstUI, DraI, EcoRI, HaeIII, HhaI, HinfI, HpaI, HpaII, KpnI, Sau3AI, MseI, RsaI, SspI and TaqI were used to compare the RFLP pattern results. MW: ΦX174DNA digested with HaeIII.
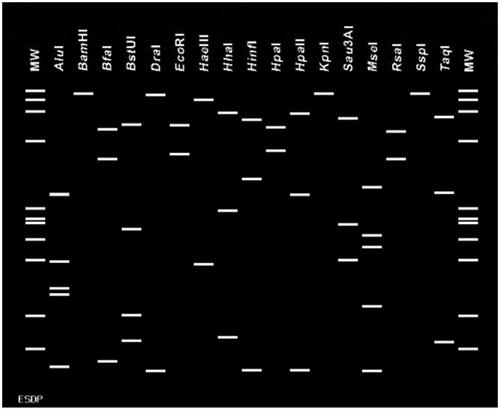
Figure 4. Phylogenetic relationship of E. sylvestris from nucleotide sequences of 16S rRNA gene, performed with MEGA ver. 10.0 software using the neighbor – joining method with the default values and 10,000 replications for bootstrap analysis.
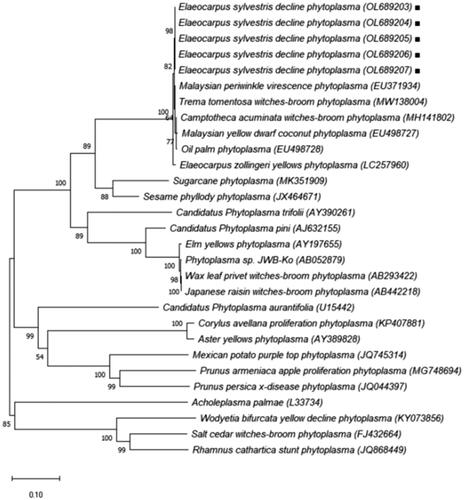
Figure 5. Phylogenetic relationship of E. sylvestris from nucleotide sequences of secA gene, performed with MEGA ver. 10.0 software using the neighbor – joining method with the default values and 10,000 replications for bootstrap analysis.
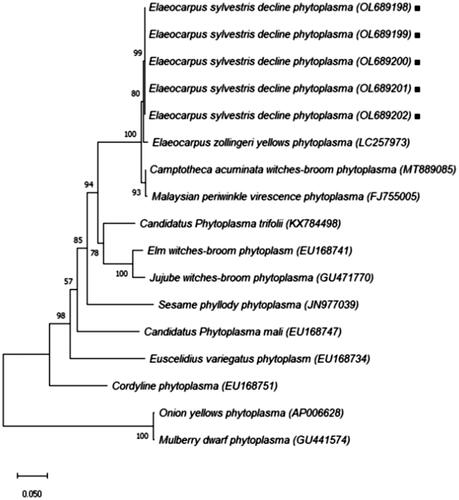
The virtual-RFLP patterns derived from the query 16S rDNA gene (F2nR2 fragment) of strain Elaeocarpus sylvestris decline phytoplasma (ESDP) is clearly different RFLP band pattern reported Malaysian periwinkle virescence (MaPV), Malayan Oil Palm (MOP), Malayan Yellow Dwarf (MYD) ().
Discussion
The results of the pairwise alignment of the 16SrRNA and the secA gene sequence revealed that the phytoplasma in each was 99.5%, 98.2% linked to others with Candidatus phytoplasma malaysianum (16SrXXXII). In the virtual-RFLP results, it was different from the previously reported 16SrXXXII-A (MaPV), 16SrXXXII-B (MYD), 16SrXXXII-C (MOP) group (Nejat et al. Citation2013). Among the 17 restriction enzymes, in BstUI enzyme case, 16S rDNA gene of strain ESDP were distinguished from 16SrXXXII-A (MaPV), 16SrXXXII-B (MYD), 16SrXXXII-C (MOP), and restriction enzyme BfaI can be distinguish 16SrXXXII-C (MYD). In addition, HhaI enzyme also distinguish 16SrXXXII-A (MaPV), 16SrXXXII-B (MOP), the other enzymes showed the same patterns. As a result of this, we propose that EDSP strain a new subgroup within the 16SrXXII group. Associated with Malaysian periwinkle virescence group (16SrXXXII) Strains including Malaysian periwinkle virescence (MaPV), Malaysian Oil Palm (MOP), Malayan Yellow Dwarf (MYD), Camptotheca acuminate witches'-broom (CAWB), Trema tomentosa witches'-broom (TtWB), and Elaeocarpus zollingeri yellows (ELY) (Nejat et al. Citation2009, Citation2013; Iwabuchi et al. Citation2018; Yu et al. Citation2020), among these, E. zollingeri yellows (ELY) closely hosts related and symptoms are similar to E. sylvestris decline in Korea. PCR-RFLP and virtual RFLP analysis also showed that the korean and Japanese diseased Elaeocarpus displayed a band pattern that was similar to the analysis results with the restriction enzymes RsaI, HpaII, and MseI (Satoh et al. Citation2014). However, using the AluI enzyme, it was showed that different band pattern from E. sylvestris decline in Korea. Japanese studies of E. zollingeri yellows (ELY) reported phytoplasma infection by types other than Ca. phytoplasma malaysianum, including Ca. phytoplasma asteris (Kawabe et al. Citation2001, Citation2010). In our studies, however, only Ca. phytoplasma malaysianum was detected. Further research is needed on this.
In Japan, insect vectors of ELY has not yet to be determined (Satoh et al. Citation2014; Iwabuchi et al. Citation2018). Likewise, it is still unknown whether Leafhoppers carry phytoplasma disease in Korea. Preventing the spread of phytoplasma disease requires identifying the insect hosts associated with it.
As for the reports about phytoplasma transmission in seeds, including apricot, peach, corn, and carrot (Necas et al. Citation2008; Çağlar et al. Citation2019; Carminati et al. Citation2019). Theoretically speaking, Phytoplasma transmission via seeds has not yet been reported in Korea (Shin Citation1980). However, seeds of diseased E. sylvestris, phytoplasma was detected by the PCR analysis performed in this study. This trees mostly reproduces by seed germination, but the phytoplasma was detected in the seeds. There are a risk of the spread of phytoplasma in the end.
Phytoplasma were present in trees displaying decline symptoms. The present study is the first confirmed report of Ca. phytoplasma malaysianum (16SrXXXII) in E. sylvestris leaves and seeds on Jeju Island in South Korea. In response to this, considering that various symptoms and signs of decline are evident, we provisionally propose to name it “Elaeocarpus sylvestris decline phytoplasma (ESDP).”
Conclusions
This study was conducted to discover phytoplasma in Elaeocarpus sylvestris growing in Jeju Island, various decline symptoms observed incluing yellowing, darkening and branch dieback. Trees infected with phytoplasma die progressing rapidly and a number of trees are being cut down. As a result of PCR using phytoplasma universial primer P1/P7, R16F2n/R2, and SecAfor1/SecArev3 phytoplasma detected in leaves and seeds. We analyzed that E. sylvestris decline sequences, and found partial 16S rRNA sequences to be 99.5%, and secA genes 98.2% similarities with Ca. phytoplasma malaysianum (16SrXXXII). As a result of virtual-RFLF analysis with 17 key restriction enzymes with 16S rRNA gene, ESDP strain was presumed a new subgroup within the 16SrXXXII group.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Bertaccini A, Duduk B. 2009. Phytoplasma and phytoplasma diseases: a review of recent research. Phytopathol Mediterr. 48:355–378.
- Bertaccini A. 2007. Phytoplasmas: diversity, taxonomy, and epid emio- logy. Front Biosci. 12(1):673–689.
- Çağlar BK, Satar S, Bertaccini A, Elbeaino T. 2019. Detection and seed transmission of Bermudagrass phytoplasma in maize in Turkey. J Phytopathol. 167(4):248–255.
- Carminati G, Satta E, Paltrinieri S, Bertaccini A. 2019. Simultaneous evaluation of ‘Candidatus Phytoplasma’ and ‘Candidatus Liberibacter solanacearum’ seed transmission in carrot. Phyt Moll. 9(1):141–142.
- Chang BH, Kim CJ. 1971. Studies on the dwarf disease of mulberry tree. Seri J Korea. 13:17–21.
- Chung BN, Jeong MI, Choi GS. 2011. Characterization of phytoplasma disease occurred on floricultural crops in Korea. Res Plant Dis. 17(3):265–271.
- Deng S, Hiruki C. 1991. Amplification of 16S rRNA genes from culturable and non-culturable Mollicutes. J Microbiol Methods. 14(1):53–61.
- Dermastia D. 2019. Plant hormones in phytoplasma infected plants. Front Plant Sci. 10:477.
- Doi Y, Teranaka M, Yora K, Asuyama H. 1967. Mycoplasma or PLT-group like micro- organisms found in the phloem elements of plants infected with mulberry dwarf, potato witches’ broom, aster yellows and paulownia witches’ broom. Jpn J Phytopathol. 33(4):259–266.
- Gundersen DE, Lee IM. 1996. Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathol Mediterr. 35:144–151.
- Han SS. 2005. Specific primer for detection of Jujube witches’ broom phytoplasma group (16SrV) in Korea. Plant Pathol. 21(1):55–58.
- Hodgetts J, Boonham N, Mumford R, Harrison N, Dickinson M. 2008. Phytoplasma phylogenetics based on analysis of secA and 23S rRNA gene sequences for improved resolution of candidate species of 'Candidatus Phytoplasma'. Int J Syst Evol Microbiol. 58(Pt 8):1826–1837.
- Hoshi A, Ishii Y, Kakizawa S, Oshima K, Namba S. 2007. Hostparasite interaction of phytoplasmas from a molecular biological perspective. Bullet Insectol. 60:105–107.
- Ikeda T, Hashimoto H. 1996. On decline of street trees of Elaeocarpus sylvestris POIRET var. ellipticus HARA in relation to water relations in Japanese. J Jpn Soc. 78:478–480.
- Jung HY, Win NKK, Kim YH. 2012. Current status of phytoplasmas and their related diseases in Korea. Plant Pathol J. 28(3):239–247
- Kamala-Kannan S, Han SS, Lee KJ, Velmurugan P, Lee YH, Chae JC, Lee YS, Lee JY, Oh BT. 2011. Association of elm yellows subgroup 16SrV-B phytoplasma with a disease of Hovenia dulcis. J Phytopathol. 159(3):171–174.
- Kawabe Y. 2010. Phytoplasma and Elaeocarpus yellows in Japanese. J For. 1511:56–59.
- Kawabe Y, Kusunok M, Matsuura K, Ogawa S, Usami Y, Joe T. 2010. ホルトノキ萎黄病による街路樹の衰退枯死被害. 日本森林学会大会学術講演集. 第 121 回日本森林学会大会 p.148-148. CO1.
- Kawabe Y, Kusunoki M, Miyashita S, Kikuchi Y. 2001. Genetic diagnosis of phytoplasma diseases on trees. Tsukuba: Forestry and Forest Products Research Institute Kenkyu Seika Senshu 2000 (fiscal); p. 10–11.
- Kawabe Y, Tsuda J, Matsuura K, Ogawa S, Usami Y, Kusunoki M. 2011. Analysis of oxytetracycline and detection of phytoplasma after the trunk injection of oxytetracycline formulation in Elaeocarpus sylvestris var. ellipticus infected with Elaeocarpus yellows (in Japanese with English summary). J Tree Health. 15:97–101.
- Kim CJ. 1968. Witches’ broom of jujube tree (Zizyphus jujuba Mill. var. inermis Rehd.). Res Bull Choonchun Agri Coll. 2:47–53.
- Kim JS, Kim TY. 2014. Woody plants of Korean Peninsula. Dolbegae. p. 214.
- Kim YH. 1980. Studies on mycoplasma-like organism associated with witches’ broom of Rhus javanica. Kor J For. 47:1–15.
- Kumari S, Nagendran K, Rai AB, Singh B, Rao GP, Bertaccini A. 2019. Global status of phytoplasma diseases in vegetable crops. Front Microbiol. 10:1349–1349.
- La YJ. 1968. Insect transmission of paulownia witches’ broom disease in Korea. Kor Observer. 8:55–64.
- Lee IM, Hammond RW, Davis RE, Gundersen DE. 1993. Universal amplification and analysis of pathogen 16S rRNA for classification and identification of mycoplasma-like organisms. Mol Plant Pathol. 83(8):834–843.
- Lee IM, Gundersen DE, Davis RE, Bartoszyk IM. 1998. Revised classification scheme of phytoplasmas based on RFLP analysis of 16S rRNA and ribosomal protein gene sequences. Int J Syst Bacteriol. 48(4):1153–1169.
- Lee SK, Han SS, Seo ST, Lee DH, Lee SH. 2017. Witch’s broom of trees and shrubs in Korea. National Institute of Forest Science; p. 67–68.
- Iwabuchi N, Endo A, Kameyama N, Satoh M, Miyazaki A, Koinuma H, Kitazawa Y, Maejima K, Yamaji Y, Oshima K, et al. 2018. First report of “Candidatus Phytoplasma malaysianum” associated with Elaeocarpus yellows of Elaeocarpus zollingeri. J Gen Plant Pathol. 84(2):160–164.
- McCoy RE, Caudwell A, Chang CJ, 16 other authors. 1989. Plant diseases associated with mycoplasma-like organisms. Vol. 5. In: Whitcomb RF, Tully JG, editors. The mycoplasmas. San Diego, CA: Academic Press; p. 545–640.
- Namba S. 2019. Molecular and biological properties of phytoplasmas. Proc Jpn Acad Ser B Phys Biol Sci. 95(7):401–418.
- Narazaki K, Tsuda J. 2008. Studies on control technique of Elaeocarpus yellows (in Japanese). Bull Fukuoka Pref for Res Ctr. 9:21–24.
- Necas T, Maskova V, Krska B. 2008. The possibility of ESFY phytoplasma transmission: through flowers and seeds. Acta Horticulturae. 781(781):443–448.
- Nejat N, Sijam K, Abdullah SNA, Vadamalai G, Dickinson M. 2009. Phytoplasmas associated with disease of coconut in Malaysia: phylogenetic groups and host plant species. Plant Pathol. 58(6):1152–1160.
- Nejat N, Vadamalai G, Davis RE, Harrison NA, Sijam K, Dickinson M, Abdullah SN, Zhao Y. 2013. ‘Candidatus Phytoplasma malaysianum', a novel taxon associated with virescence and phyllody of Madagascar periwinkle (Catharanthus roseus). Int J Syst Evol Microbiol. 63(Pt 2):540–548.
- Odawar City Cultural Property Protection Committee. 2013. Cancellation of Odawara city designated historic site scenic natural monument (in Japanese). Vol 2. Odawara: Odawara City Cultural Property Protection Committee Summary 2012 (fiscal); p. 1–3.
- Oshima K, Maejima K, Namba S. 2013. Genomic and evolutionary aspects of phytoplasmas. Front Microbiol. 4:230.
- Pagliari L, Musetti R. 2019. Phytoplasmas: AN introduction. In: Musetti R, Pagliari L, editors. Phytoplasmas. Methods in molecular biology. New York, NY, USA: Humana Press; p. 1–6.
- Satoh M, Takahashi H, Chikamori M, Tani Y, Adachi N. 2014. Prevalence of Elaeocarpus yellows in Mt. Shiroyama in Tokushima City (in Japanese with English summary). Nat Sci Res Univ Tokushima. 28:21–25.
- Schneider B, Seemuller E, Smart CD, Kirkpatrick BC. 1995. Phylogenetic classification of plant pathogenic mycoplasma-like organisms or phytoplasma. Mol Diag Proc Mycoplasmol. 1:369–379.
- Seemuüller E, Garnier M, Schneider B. 2002. Mycoplasmas of plants and insects. In: Razin S, Herrmann R, editors. Molecular biology and pathology of mycoplasmas. London: Kluwer Academic/Plenum Publishers; p. 91–116.
- Shin HD. 1980. Impossibility of seed transmission in plant mycoplasmal diseases. Korea J Pl Prot. 19:141–143.
- Yu S, Tang Q, Wu Y, Lin M, Zhao R, Song W, Qin W. 2020. First report of phytoplasma belongs to 16SrXXXII group associated with witches’-broom symptoms in Trema tomentosa in China. Plant Disease, 105(4):1191–1191.
- Weintraub PG, Jones P. 2009. Phytoplasmas: Genomes, plant hosts and vectors.
- Zhao Y, Wei W, Lee IM, Shao J, Suo X, Davis RE. 2009. Construction of an interactive online phytoplasma classification tool, iPhyClassifier, and its application in analysis of the peach X-disease phytoplasma group (16SrIII). Int J Syst Evol Microbiol. 59(Pt 10):2582–2593.

