Abstract
L1 non-LTR retrotransposons are autonomously replicating genetic elements that profoundly affected their mammalian hosts having generated upwards of 40% or more of their genomes. Although deleterious, they remain active in most mammalian species, and thus the nature and consequences of the interaction between L1 and its host remain major issues for mammalian biology. We recently showed that L1 activity requires phosphorylation of one of its 2 encoded proteins, ORF1p, a nucleic acid chaperone and the major component of the L1RNP retrotransposition intermediate. Reversible protein phosphorylation, which is effected by interacting cascades of protein kinases, phosphatases, and ancillary proteins, is a mainstay in the regulation and coordination of many basic biological processes. Therefore, demonstrating phosphorylation-dependence of L1 activity substantially enlarged our knowledge of the scope of L1 / host interaction. However, developing a mechanistic understanding of what this means for L1 or its host is a formidable challenge, which we discuss here.
The autonomous L1 non-LTR retrotransposon encodes 2 proteins, ORF1p and ORF2p, essential for retrotransposition.Citation1 These proteins preferentially, though not exclusively, bind their encoding transcript (cis preference) to form the L1RNP retrotransposition intermediateCitation2–7 and [reviewed in 8, 9]. ORF2p contains highly conserved endonuclease and reverse transcriptase domains and is the L1 replicase.Citation1,10,11 The 3’ OH of endonuclease-nicked genomic DNA primes synthesis of a DNA copy of the L1 transcript, referred to as target site-primed reverse transcription (TPRT).Citation12 ORF1p (), the major component of the L1 RNP, is a trimerCitation13 that is mediated by a highly conserved coiled coil motif.Citation3,14–18 Its C-terminal half contains highly conserved domains that endow the protein with nucleic acid binding and chaperone activities, which are essential for retrotransposition.Citation9,19–26
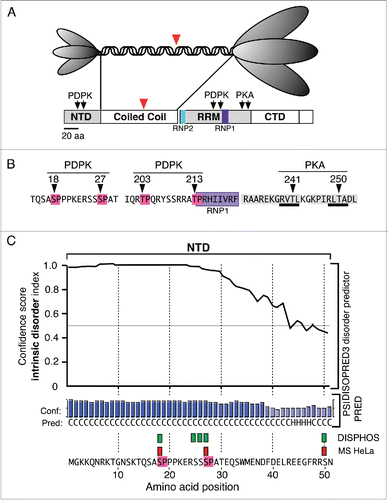
Figure 1. Structural Features of ORF1p (A). The top panel illustrates the trimeric structure of the protein revealed by atomic force microscopyCitation13 and the domains present in the monomer: N-terminal domain (NTD), coiled coil domain, RNA recognition domain (RRM), the C-terminal domain (CTD). The ovals that correspond to the NTD and the C-terminal half of the trimer cartoon are scaled to the their relative masses. The large red arrowheads indicate the amino-terminus of the protein that was expressed in E. coli for crystallization.Citation25 (B) The arrows indicate the locations of phosphokinase target sites and the amino acids corresponding to the PDPK sites are highlighted in red, those comprising the RNP1 motif in purple, those making up the PDPK docking sites in gray, and those corresponding to the PKA target sites are underlined in black. C. The results generated by the DISOPRED, PSIPRED and DISPHOS predictions programs (see text). The green rectangles correspond to the DISPHOS predictions for phosphorylated serines, the red rectangles correspond to those found by mass spectroscopy on ORF1p expressed in HeLa cells.
L1 has been active in the mammalian lineage since before its radiation 80–120 MYACitation27 and has had a defining effect on the genomes of most mammals, having generated upwards of 40% of their genomic mass, including humans.Citation28,29 Although deleterious,Citation16,30–32 they remain active in most species and are often the dominant, if not only, active autonomous transposable element, as is the case in humans where they can cause genetic diversity, rearrangements and defects, and can be activated in various somatic cells, including tumors.Citation8,33–42
L1 elements can be subject to various host repressive mechanisms.Citation31,43–47 However, the extent to which L1 appropriates or interacts with host factors and regulatory pathways is not clear. In this regard, we recently reported that ORF1p expressed in HeLa and insect cells is phosphorylated on multiple serines and threonines.Citation48 Some of these sites correspond to target and docking motifs for proline-directed protein kinases (PDPKs). This kinase family includes mitogen-activated protein kinases (MAPKs), cyclin-dependent kinases (CDKs), and glycogen synthase 3 (GSK3).Citation49
PDPKs phosphorylate serines or threonines with proline in the +1 position, known as S/T-P motifs, and ORF1p contains 4: S18P19, S27P28, T203P204, and T213P214 (). These sites are highly conserved as are 2 protein kinase A (PKA) threonine sites, T241 and T250, that are imbedded in 2 highly conserved PDPK docking sites (). Mass spectroscopy (MS) showed that S18, S27 and T203 were phosphorylated in HeLa cells, but we could not determine the state of T213 as the peptide containing this site was not recovered from ORF1p expressed in HeLa cells.Citation48
Mutational analysis showed that substitution at any one of the PDPK or PKA target sites by a non-phosphorylatable residue reduced retrotransposition to 0–40% of wild type. The double S18,27A mutant lacked all activity and was restored to 50% wild type activity by double substitution with the phosphomimetic aspartic acid (S18,27D). However, phosphomimetic substitution not only failed to restore activity at each threonine site but eliminated any residual activity exhibited by some of the threonine mutants including those at the PKA sites,Citation48 and Cook, P.R., unpublished observations]. These results suggest that a permanently acidic residue at any one of the threonine sites was not compatible with retrotransposition.
Equally important was our demonstration that peptidyl-prolyl cis / trans isomerase 1 (Pin1) can bind FLAG-tagged ORF1p present in HeLa cell extracts. This highly conserved 16-kDa protein is a crucial component of numerous PDPK-dependent pathways.Citation50,51 When bound to a phosphorylated PDPK site, it catalyzes conformational changes that enhance the efficacy of phosphorylation by generating distinct protein-specific effects. These include changes in the activity and stability of the target protein as well as promoting protein-protein interactions,Citation52,53 some of which can result in additional post translational modifications such as acetylation, ubiquitination and sumoylation.Citation50,51 Also, while Pin1 binding can protect a protein from de-phosphorylation, in other instances it enhances dephosphorylation. And finally, Pin1 is also subject to phosphorylation at several sites by various kinases. Notably serine 16, which resides in the amino-terminal WW domain, can be phosphorylated by 3 different kinases including PKA.Citation52,54 The WW domain mediates binding of Pin1 to the phospho-site of its target,Citation55 and binding is reduced when S16 is phosphorylated, an event that also alters the subcellular distribution of Pin1.Citation54 Thus, protein kinases could also affect ORF1p activity through their effect on Pin1.
Essentially all of the Pin1 binding apparently occurred at S18P19 and S27P28 under the conditions of our assay, as the doubly mutated S18,27A bound none and the doubly phosphomimetic S18,27D bound just trace amounts.Citation48 That S18,27D exhibited 50% of the wild type retrotransposition activity indicates that carboxylate moieties at these sites can partially substitute for phosphoryl groups. One interpretation of these results is that Pin1 functions, in part, to protect ORF1p from dephosphorylation. However, it seems that phosphorylation provides more than just an acidic group for if this were the case these sites would not have been conserved over the past ∼20 Myr of evolutionCitation48 as selection would likely have favored mutation to the far less energetically expensive option of amino acids with an acidic side chain.
Such considerations lead directly to the issue of the role of PDPK-mediated ORF1p phosphorylation in retrotransposition. In general, the physico-chemical effects of phosphorylation include altering protein activity as well as fostering protein-protein interactions including those between different regions of the same protein, either through direct structural changeCitation53 or mediated by factors such as Pin1 as mentioned above. Additionally, the effects of phosphorylation are often transient being reversible by protein phosphatases, which allows temporal (and spatial) coordination of multi-component and multi-step cellular processes. PDPKs are involved in pathways that regulate major biological processes such as development, differentiation, growth and cell division as well as cellular responses to stress, pathogens, inflammation and cancerCitation49,56,57 Therefore, ORF1p expressed during L1 replication in its normal and evolutionary relevant reproductive niche in the germline or during early embryonic development,Citation31,32,58,59 as well as ectopically when L1 is active in cancer and other somatic cellsCitation8,33–42 could usurp or compete for PDPKs, or their ancillary factors such as Pin1. Consequently, a comprehensive understanding of the consequences of ORF1p phosphorylation for both the activity of L1 and its interaction with its host presents a major challenge.
In the rest of this commentary we will focus on the first issue; i.e., the possible direct effects of phosphorylation on the structure and function of the protein. To do so we determined the relationship of the 6 conserved phosphorylation sites shown in to structural features of ORF1p. In particular, we sought to determine the location of these sites relative to intrinsically disordered regions of the protein. This aspect of protein structure has generated increasing interest over the last 15 or so years as such regions characterize many proteins involved in regulatory processes including cell signaling, membrane trafficking, cell cycle control, and transcriptional and translational control.Citation53,60–66 Underlying their role in these processes is the fact that intrinsically disordered regions often mediate protein-protein interactions. Furthermore, because such regions are also preferred sites for post translational modification including phosphorylation, the nature and extent of these interactions can be subject to regulation.Citation63,64
The crystal structure of ORF1p does not include the NTD (or the first 8 heptads of the coiled coil), as it was determined on a protein that had been deleted of its amino-terminal third (red arrowheads, ).Citation25 Therefore, we used the DISOPRED3 and PSIPRED v3.3 programs from the PSIPRED protein sequence analysis workbench to assess the extent of intrinsically disordered sequence in the NTD.Citation67 Because we know the phosphorylation status of the NTD, we could corroborate the PSIPRED prediction by whether the NTD was highly ranked as a possible phosphorylation site by DISPHOS (DISorder-enhanced PHOSphorylation). This is because this program scores the likelihood of phosphorylation by including estimates of intrinsic disorder in its prediction algorithm.Citation63
The upper part of shows that DISOPRED predicts with high confidence that the structure of most of the NTD, particularly the region that contains the PDPK SP sites, is intrinsically disordered. Furthermore, the secondary structure predictor, PSIPRED, estimates that most of the sequence lacks defined secondary structure. In agreement, DISPHOS predicts 5 sites with ≥0 .8 confidence (green rectangles) to be candidate for phosphorylation. Three of these sites (red rectangles) correspond to those that MS showed were phosphorylated in HeLa and insect cells.Citation48
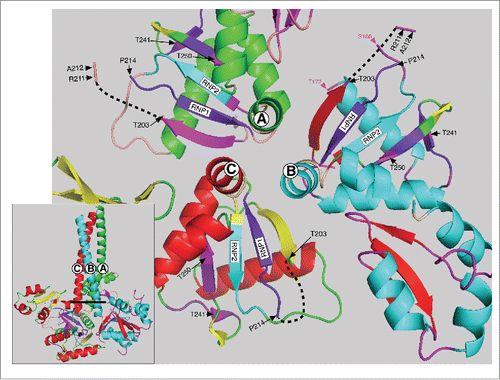
The two PDPK T/P sites at positions 203 and 213 also reside in an intrinsically disordered region of the protein. NMR analysis showed that amino acids P204 through T213 comprise the disordered loop L(β2-β3).Citation25 And shows that amino acids that comprise this region are missing from the crystal structure of all 3 monomers (upper left insert ). This region is exposed to the solvent in the 3 monomers () and is thus likely accessible to PDPK kinases. Furthermore, requisite PDPK docking sites with their imbedded highly conserved PKA target sites at T241 and T250 are near the T203 and T213 PDPK targets. These threonines also flank 3 highly conserved arginines (R 206, 210 and 211, upper left insert Fig, 2), each critical for RNA binding.Citation24,25 Therefore, a phosphate introduced at either T203 or T213 could interact with the guanidino group of one or more of these arginines, via hydrogen bonded “salt bridges”,Citation68 and potentially affect their roles in RNA binding. The possibility of such an interaction is supported by the strong inhibitory effect of the phosphomimetic aspartic acid at either site.
Figure 2. Structure of the ORF1p RRM The structure the C monomer RRM was displayed using MacPymol 1.7.6.3 based on the PDB file, 2yko.Citation25 The upper left insert shows the amino acids missing from the structures of the A, B, and C monomers (black dashed line, red box c, ) in the vicinity of T203 – T213. The lower right insert show the amino acid sequence encompassing the highly conserved PKA sites in the PDPK docking sites shown in
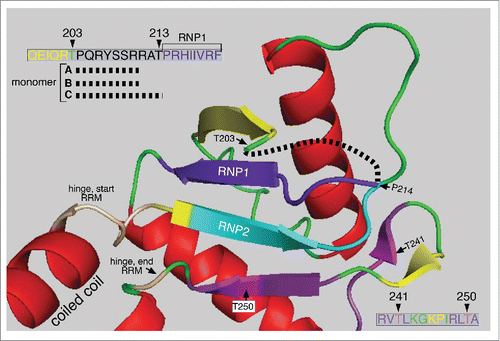
Figure 3. Top view of the C-terminal half of the ORF1p trimer. This structure (also based on the PDP file 2yko) shows a top view of the 3 trimers starting with the last heptad (LQWIEDY) of the coiled coil - as if the trimer was resected at the heavy line in the insert and then rotated toward the viewer. The hinge between the carboxy terminus of the coiled coil and the N-terminus of the RRM is in tan (see ) and the RNP2 and RNP1 motifs are respectively in teal and deep purple for each monomer as also indicated on . The PKA sites and surrounding amino acids are also colored the same for each monomer as indicated in the lower right insert of . The black dashed lines show the location of the missing amino acids (starting at P204) in each monomer (also see upper left insert, ) and red box c in . Other than these features the major secondary structures of monomers A, B and C are colored differently. For each of the respective monomers these are: α helices, green, teal, and red; β sheets, magenta, red, and yellow; loops, tan, magenta and green. The amino acid positions indicated in magenta in the monomer B structure flank the amino acids (167–172, corresponding to mauve box a, ) that are missing from the X-ray structure of the RRM just 3’ of RNP2.
Although the strength of the hydrogen bonded “salt bridge” between a carboxylate and arginine is somewhat less than with a -2 phosphate it is about the same as with a protonated -1 phosphate, and both ionic species co-exist at physiological pH given the pKa of ∼6.0 for the phosphate.Citation68 Thus, while formation and strength of such bonds will ultimately be a function of the protein micro-environment and include factors such as steric hindrance, solvation, etc., differences between the relative strengths of carboxylate vs. phosphate mediated salt bridges per se would not seem to be determinative of the effect of a phosphomimetic substitution at a phosphorylatable site.
Rather, it seems that it is the permanently acidic nature of the phosphomimetic carboxylate at T203 or T213 that is strongly inhibitory.Citation48 If so, and if phosphorylation does accompany retrotransposition, then L1 survival would require phosphate turnover. This would rationalize the presence of the predicted, highly conserved protein phosphatase 1 docking site at positions 282–290.Citation48 Demonstrating that cyclic phosphorylation plays a role in retrotransposition would represent a major advance in a mechanistic understanding of the role of ORF1p.
Intrinsically disordered N- and C-termini and inter-domain regions are often preferred sites of protein-protein interaction, particularly for a subset of proteins that includes kinases, regulatory proteins and DNA/RNA binding proteins.Citation53,66 To reiterate here, such interactions can occur even without the intercession of post-translational modification, although the latter, particularly phosphorylation / dephosphorylation, is an important means of regulating and coordinating multistep and multicomponent reactions. With respect to L1 activity, this could be the case for the assembly of the L1RNP, its interaction with host chromatin wherein retrotransposition is initiated, its ultimate disassembly during the reverse transcription of the L1 transcript, and the ORF1p mediated chaperone reactions during the various steps of generating a new L1 insert.
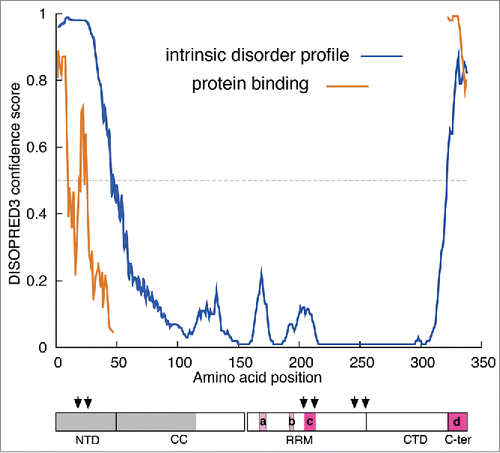
shows that similar to the NTD the C-terminal region (C-ter) is also disordered and that the intrinsically disordered regions predicted by DISOPRED correspond reasonably well with those regions missing from one or more monomers of the X-ray structure indicated by mauve and red boxes (see legend to and cf. ). As intrinsically disordered regions are correlated with protein surfaces that can mediate protein-protein interactions, DISOPRED also evaluates this possibility, and indicated that regions in both the N- and C- terminal regions of ORF1p score highly as candidates for such interaction (). Mutational and functional analysis coupled with protein pull-downs could reveal whether the N- or C-terminal regions of ORF1p do in fact mediate interaction with other cellular proteins that participate in retrotransposition. Therefore, it would be informative to determine whether these regions were involved in the recently described co-precipitation of ORF1p and various cellular proteins.Citation69–71
Figure 4. Intrinsic disorder profile of ORF1p The profiles of intrinsically disordered sequence and potential protein binding surface were generated by DISOPRED3. The cartoon showing the monomer domains is the same as that in , annotated as follows: The gray box indicates the region of the protein that had been deleted prior to expression in E. coli.Citation25 The mauve and red boxes indicate the amino acids missing from the X-ray structure in the 2yko PDB file – (a), positions 167–172, monomer B; (b), 191–193, and 190–194, monomers A and B respectively; (c), 204–210, 204–210, 203–212 monomers A, B, and C respectively; (d), 323–338, monomers A, B, and C.
In summary, the presence highly conserved phosphorylatable residues in intrinsically disordered regions of ORF1p means that a battery of well-developed paradigms and mechanismsCitation53 may be relevant to advancing and refining our understanding of how this protein functions in retrotransposition and how L1 activity affects and responds to the host. Although the vast landscape of phosphorylation mediated effects would not seem to provide an easily negotiated path to understanding these processes, one would hope that the extensive knowledge base on the physico-chemical effects of protein phosphorylation could provide a template for devising useful approaches for determining a mechanistic understanding of how ORF1p functions in retrotransposition.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Charles E. Jones for his comments.
Funding
This research was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases (grant ID: HNK6H7–107053).
References
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH Jr. High frequency retrotransposition in cultured mammalian cells. Cell 1996; 7(5):917-27; PMID:8945518; http://dx.doi.org/10.1016/S0092-8674(00)81998-4
- Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol Cell Biol 1991; 11(9):4804-7; PMID:1715025; http://dx.doi.org/10.1128/MCB.11.9.4804
- Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. Embo J 1996; 15(3):630-9; PMID:8599946
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol 2001; 21(4):1429-39; PMID:11158327; http://dx.doi.org/10.1128/MCB.21.4.1429-1439.2001
- Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum Mol Genet 2005; 14(21):3237-48; PMID:16183655; http://dx.doi.org/10.1093/hmg/ddi354
- Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol 2006; 13(7):655-60; PMID:16783376; http://dx.doi.org/10.1038/nsmb1107
- Doucet AlJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, Kopera HC, Athanikar JN, Hasnaoui M, Bucheton A, Moran JV, et al. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet 2010; 6(10):e1001150; PMID:20949108; http://dx.doi.org/10.1371/journal.pgen.1001150
- Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. LINE-1 retrotransposition activity in human genomes. Cell 2010; 141(7):1159-70; PMID:20602998; http://dx.doi.org/10.1016/j.cell.2010.05.021
- Martin SL. Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1. RNA Biol 2010; 7(6):67-72; PMID:20009507; http://dx.doi.org/10.4161/rna.7.6.13766
- Mathias SL, Scott AF, Kazazian HH Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science 1991; 254(5039):1808-10; PMID:1722352; http://dx.doi.org/10.1126/science.1722352
- Feng Q, Moran JV, Kazazian HH Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 1996; 87(5):905-16; PMID:8945517; http://dx.doi.org/10.1016/S0092-8674(00)81997-2
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 1993; 72(4):595-605; PMID:7679954; http://dx.doi.org/10.1016/0092-8674(93)90078-5
- Martin SL, Branciforte D, Keller D, Bain DL. Trimeric structure for an essential protein in L1 retrotransposition. Proc Natl Acad Sci USA 2003; 100(24):13815-20; PMID:14615577; http://dx.doi.org/10.1073/pnas.2336221100
- Demers GW, Matunis MJ, Hardison RC. The L1 family of long interspersed repetitive DNA in rabbits: sequence, copy number, conserved open reading frames, and similarity to keratin. J Mol Evol 1989; 29(1):3-19; PMID:2475641; http://dx.doi.org/10.1007/BF02106177
- Furano AV. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Progr Nucleic Acids Res Mol Biol 2000; 64:255-94; PMID:10697412; http://dx.doi.org/10.1016/S0079-6603(00)64007-2
- Boissinot S, Entezam A, Furano AV. Selection against deleterious LINE-1-containing loci in the human lineage. Mol Biol Evol 2001; 18(6):926-35; PMID:11371580; http://dx.doi.org/10.1093/oxfordjournals.molbev.a003893
- Boissinot S, Furano AV. The recent evolution of human L1 retrotransposons. Cytogenet Genome Res 2005; 110(1-4):402-6; PMID:16093692; http://dx.doi.org/10.1159/000084972
- Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res 2006; 16(1):78-87; PMID:16344559; http://dx.doi.org/10.1101/gr.4001406
- Kolosha VO, Martin SL. High-affinity, non-sequence-specific RNA binding by the open reading frame 1 (ORF1) protein from long interspersed nuclear element 1 (LINE-1). J Biol Chem 2003; 278(10):8112-7; PMID:12506113; http://dx.doi.org/10.1074/jbc.M210487200
- Basame S, Wai-lun Li P, Howard G, Branciforte D, Keller D, Martin SL. Spatial assembly and RNA binding stoichiometry of a LINE-1 protein essential for retrotransposition. J Mol Biol 2006; 357(2):351-7; PMID:16434051; http://dx.doi.org/10.1016/j.jmb.2005.12.063
- Martin SL, Cruceanu M, Branciforte D, Wai-Lun Li P, Kwok SC, Hodges RS, Williams MC. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J Mol Biol 2005; 348(3):549-61; PMID:15826653; http://dx.doi.org/10.1016/j.jmb.2005.03.003
- Januszyk K, Li PW, Villareal V, Branciforte D, Wu H, Xie Y, Feigon J, Loo JA, Martin SL, Clubb RT. Identification and solution structure of a highly conserved C-terminal domain within ORF1p required for retrotransposition of long interspersed nuclear element-1. J Biol Chem 2007; 282(34):24893-904; PMID:17569664; http://dx.doi.org/10.1074/jbc.M702023200
- Martin SL, Bushman D, Wang F, Li PW, Walker A, Cummiskey J, Branciforte D, Williams MC. A single amino acid substitution in ORF1 dramatically decreases L1 retrotransposition and provides insight into nucleic acid chaperone activity. Nucleic Acids Research 2008; 36(18):5845-54; PMID:18790804; http://dx.doi.org/10.1093/nar/gkn554
- Khazina E, Weichenrieder O. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc Natl Acad Sci USA 2009; 106(3):731-6; PMID:19139409; http://dx.doi.org/10.1073/pnas.0809964106
- Khazina E, Truffault V, Büttner R, Schmidt S, Coles M, Weichenrieder O. Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition. Nature Structural & Molecular Biology 2011; 18(9):1006-U64; PMID:21822284; http://dx.doi.org/10.1038/nsmb.2097
- Callahan KE, Hickman AB, Jones CE, Ghirlando R, Furano AV. Polymerization and nucleic acid-binding properties of human L1 ORF1 protein. Nucleic Acids Res 2012; 40(2):813-27; PMID:21937507; http://dx.doi.org/10.1093/nar/gkr728
- Burton FH, Loeb DD, Voliva CF, Martin SL, Edgell MH, Hutchison CA 3rd. Conservation throughout mammalia and extensive protein-encoding capacity of the highly repeated DNA long interspersed sequence one. J Mol Biol 1986; 187(2):291-304; PMID:3009828; http://dx.doi.org/10.1016/0022-2836(86)90235-4
- IHGS-Consortium. Initial sequencing and analysis of the human genome. Nature 2001; 409(6822):860-921; PMID:11237011; http://dx.doi.org/10.1038/35057062
- Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet 2012; 13(1):36-46; PMID:22124482
- Boissinot S, Davis J, Entezam A, Petrov D, Furano AV. Fitness cost of LINE-1 (L1) activity in humans. Proc Natl Acad Sci USA 2006; 103(25):9590-4; PMID:16766655; http://dx.doi.org/10.1073/pnas.0603334103
- Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 2004; 431(7004):96-9; PMID:15318244; http://dx.doi.org/10.1038/nature02886
- Soper SFC, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell 2008; 15(2):285-97; PMID:18694567; http://dx.doi.org/10.1016/j.devcel.2008.05.015
- Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH Jr. Isolation of an active human transposable element. Science 1991; 254(5039):1805-8; PMID:1662412; http://dx.doi.org/10.1126/science.1662412
- Boissinot S, Entezam A, Young L, Munson PJ, Furano AV. The insertional history of an active family of L1 retrotransposons in humans. Genome Res 2004; 14:1221-31; PMID:15197167; http://dx.doi.org/10.1101/gr.2326704
- Boissinot S, Chevret , Furano AV. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol 2000; 17(6):915-28; PMID:10833198; http://dx.doi.org/10.1093/oxfordjournals.molbev.a026372
- Moran JV, DeBerardinis RJ, Kazazian, Jr. HH. Exon shuffling by L1 retrotransposition. Science 1999; 283(5407):1530-4; PMID:10066175; http://dx.doi.org/10.1126/science.283.5407.1530
- Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell 2002; 110(3):315-25; PMID:12176319; http://dx.doi.org/10.1016/S0092-8674(02)00828-0
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 2005; 435(7044):903-10; PMID:15959507; http://dx.doi.org/10.1038/nature03663
- Ewing AD, Kazazian HH. High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res 2010; 20(9):1262-72; PMID:20488934; http://dx.doi.org/10.1101/gr.106419.110
- Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 2010; 141(7):1253-61; PMID:20603005; http://dx.doi.org/10.1016/j.cell.2010.05.020
- Tubio JMC, Li Y, Ju YS, Martincorena I, Cooke SL, Tojo M, Gundem G, Pipinikas CP, Zamora J, Raine K, Menzies A, et al. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 2014; 345(6196):1251343; PMID:25082706; http://dx.doi.org/10.1126/science.1251343
- Upton K, Gerhardt DJ, Jesuadian JS, Richardson SR, Sánchez-Luque FJ, Bodea GO, Ewing AD, Salvador-Palomeque C, van der Knaap MS, Brennan PM, Vanderver A, et al. Ubiquitous l1 mosaicism in hippocampal neurons. Cell 2015; 161(2):228-39; PMID:25860606; http://dx.doi.org/10.1016/j.cell.2015.03.026
- Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci USA 2006; 103(23):8780-5; PMID:16728505; http://dx.doi.org/10.1073/pnas.0603313103
- Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem 2006; 281(25):16837-41; PMID:16648136; http://dx.doi.org/10.1074/jbc.M602367200
- Muckenfuss H, Hamdorf M, Held U, Perkovic M, Löwer J, Cichutek K, Flory E, Schumann GG, Münk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem 2006; 281(31):22161-72; PMID:16735504; http://dx.doi.org/10.1074/jbc.M601716200
- Koito A, Ikeda T. Intrinsic immunity against retrotransposons by APOBEC cytidine deaminases. Front Microbiol 2013; 4:28; PMID:23431045
- Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet 2013; 14(7):447-59; PMID:23732335; http://dx.doi.org/10.1038/nrg3462
- Cook PR, Jones CE, Furano AV. Phosphorylation of ORF1p is required for L1 retrotransposition. Proc Natl Acad Sci U S A 2015; 112(14):4298-303; PMID:25831499; http://dx.doi.org/10.1073/pnas.1416869112
- Ubersax JA, Ferrell, Jr. JE. Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol 2007; 8(7):530-41; PMID:17585314; http://dx.doi.org/10.1038/nrm2203
- Lu KP, Liou YC, Zhou XZ. Pinning down proline-directed phosphorylation signaling. Trends Cell Biol 2002; 12(4):164-72; PMID:11978535; http://dx.doi.org/10.1016/S0962-8924(02)02253-5
- Liou YC, Zhou XZ, Lu KP. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem Sci 2011; 36(10):501-14; PMID:21852138; http://dx.doi.org/10.1016/j.tibs.2011.07.001
- Litchfield DW, Shilton BH, Brandl CJ, Gyenis L. Pin1: Intimate involvement with the regulatory protein kinase networks in the global phosphorylation landscape. Biochim Biophys Acta 2015; 1850(10):2077-86; PMID:25766872; http://dx.doi.org/10.1016/j.bbagen.2015.02.018
- Nishi H, Shaytan A, Panchenko AR. Physicochemical mechanisms of protein regulation by phosphorylation. Frontiers in Genetics 2014; 5:270; PMID:25147561; http://dx.doi.org/10.3389/fgene.2014.00270
- Lu PJ, Zhou XZ, Liou YC, Noel JP, Lu KP. Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. J Biol Chem 2002; 277(4):2381-4; PMID:11723108; http://dx.doi.org/10.1074/jbc.C100228200
- Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 1999; 283(5406):1325-8; PMID:10037602; http://dx.doi.org/10.1126/science.283.5406.1325
- Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev 2011; 75(1):50-83; PMID:21372320; http://dx.doi.org/10.1128/MMBR.00031-10
- Koivomagi M, Valk E, Venta R, Iofik A, Lepiku M, Morgan DO, Loog M. Dynamics of Cdk1 substrate specificity during the cell cycle. Mol Cell 2011; 42(5):610-23; PMID:21658602; http://dx.doi.org/10.1016/j.molcel.2011.05.016
- Trelogan SA, Martin SL. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc Natl Acad Sci USA 1995; 92(5):1520-4; PMID:7878012; http://dx.doi.org/10.1073/pnas.92.5.1520
- Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, Kazazian HH Jr. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes & Development 2009; 23(11):1303-12; PMID:19487571; http://dx.doi.org/10.1101/gad.1803909
- Iakoucheva LM, Brown CJ, Lawson JD, Obradović Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol 2002; 323(3):573-84; PMID:12381310; http://dx.doi.org/10.1016/S0022-2836(02)00969-5
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradović Z. Intrinsic disorder and protein function. Biochemistry 2002; 41(21):6573-82; PMID:12022860; http://dx.doi.org/10.1021/bi012159+
- Radivojac P, Obradovic Z, Smith DK, Zhu G, Vucetic S, Brown CJ, Lawson JD, Dunker AK. Protein flexibility and intrinsic disorder. Protein Sci 2004; 13(1):71-80; PMID:14691223; http://dx.doi.org/10.1110/ps.03128904
- Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 2004; 32(3):1037-49; PMID:14960716; http://dx.doi.org/10.1093/nar/gkh253
- Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J 2007; 92(5):1439-56; PMID:17158572; http://dx.doi.org/10.1529/biophysj.106.094045
- Lobanov MY, Shoemaker BA, Garbuzynskiy SO, Fong JH, Panchenko AR, Galzitskaya OV. ComSin: database of protein structures in bound (complex) and unbound (single) states in relation to their intrinsic disorder. Nucleic Acids Res 2010; 38(Database issue):D283-7; PMID:19906708; http://dx.doi.org/10.1093/nar/gkp963
- Fong JH, Panchenko AR. Intrinsic disorder and protein multibinding in domain, terminal, and linker regions. Mol Biosyst 2010; 6(10):1821-8; PMID:20544079; http://dx.doi.org/10.1039/c005144f
- Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res 2013; 41(Web Server issue):W349-57; PMID:23748958; http://dx.doi.org/10.1093/nar/gkt381
- Mandell DJ, Chorny I, Groban ES, Wong SE, Levine E, Rapp CS, Jacobson MP. Strengths of hydrogen bonds involving phosphorylated amino acid side chains. J Am Chem Soc 2007; 129(4):820-7; PMID:17243818; http://dx.doi.org/10.1021/ja063019w
- Goodier JL, Cheung LE, Kazazian, Jr. HH. Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res 2013; 41(15):7401-19; PMID:23749060; http://dx.doi.org/10.1093/nar/gkt512
- Moldovan J, Moran J, Malik H. The zinc-finger antiviral protein ZAP inhibits LINE and alu retrotransposition. PLOS Genetics 2015; 11(5):e1005121; PMID:25951186; http://dx.doi.org/10.1371/journal.pgen.1005121
- Goodier J, Pereira GC, Cheung LE, Rose RJ, Kazazian HH Jr. The broad-spectrum antiviral protein ZAP restricts human retrotransposition. PLOS Genetics 2015; 11(5):e1005252; PMID:26001115; http://dx.doi.org/10.1371/journal.pgen.1005252
