ABSTRACT
The long terminal repeat (LTR) and non-LTR retrotransposons comprise approximately half of the human genome, and we are only beginning to understand their influence on genome function and evolution. The LTR retrotransposon Ty1 is the most abundant mobile genetic element in the S. cerevisiae reference genome. Ty1 replicates via an RNA intermediate and shares several important structural and functional characteristics with retroviruses. However, unlike retroviruses Ty1 retrotransposition is not infectious. Retrotransposons integrations can cause mutations and genome instability. Despite the fact that S. cerevisiae lacks eukaryotic defense mechanisms such as RNAi, they maintain a relatively low copy number of the Ty1 retrotransposon in their genomes. A novel restriction factor derived from the C-terminal half of Gag (p22/p18) and encoded by internally initiated transcript inhibits retrotransposition in a dose-dependent manner. Therefore, Ty1 evolved a specific GAG organization and expression strategy to produce products both essential and antagonistic for retrotransposon movement. In this commentary we discuss our recent research aimed at defining steps of Ty1 replication influenced by p22/p18 with particular emphasis on the nucleic acid chaperone functions carried out by Gag and the restriction factor.
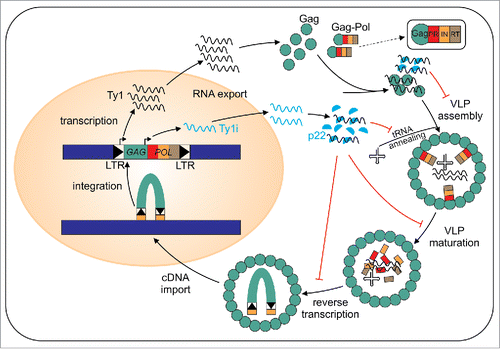
The Saccharomyces Ty1 element belongs to a widely disseminated group of retrotransposons that share pronounced structural and functional similarities with retroviruses. Ty1 is transcribed from LTR to LTR by RNA polymerase II, resulting in a genomic RNA that is exported to the cytoplasm and serves as a template for protein synthesis and replication.Citation1 Translation results in the synthesis of 2 primary gene products, Gag-p49, a structural protein and Gag-Pol. Gag-Pol is synthesized by a programmed +1 frameshift event that occurs at overlapping leucine codons present in GAG and POLCitation2 and contains Gag, protease (PR), integrase (IN) and reverse transcriptase (RT), and whose order is specific for the Ty1/copia superfamily. These enzymes are required for protein maturation, integration, and replication, respectively. Gag-p49, Gag-Pol and Ty1 RNA colocalize in the specific cytoplasmic foci termed retrosomes where they assemble into virus-like particles (VLPs).Citation3,4 During the VLP maturation Gag-p49 is cleaved to Gag-p45 (hereafter referred to as Gag). Encapsidated dimeric Ty1 RNA is reverse transcribed within VLPs.Citation5 Reverse transcription is initiated from the host-encoded tRNAiMet hybridized to the primer-binding site within Ty1 genomic RNA.Citation6 The process of Ty1 retrotransposition is completed by integration of a linear cDNA near genes transcribed by RNA polymerase III.Citation7
Ty1 is not infectious and retrotransposition may contribute to genetic diversity.Citation8 However, Ty1 insertions can also cause genome rearrangements and insertion mutations via homologous recombination between Ty1 copies or by integrating near or within genes, respectively.Citation9 Many eukaryotes utilize RNA interference to repress endogenous retroviruses. In addition, DNA-methylation has been widely observed in transposons, suggesting that this process may function to keep transposition in check.Citation10,11 However, S. cerevisiae lacks these cellular defense mechanisms,Citation12 yet most if not all characterized strains maintain a low copy number of Ty1.Citation13 The factor underlying this mechanism was just recently discovered.Citation14 The Garfinkel lab demonstrated that a self-encoded capsid derivative – p22 that can be considered an element encoded restriction factor is responsible for copy number control (CNC). CNC is defined as a decrease in transposition when additional elements are present in the genome. Genetic analysis of CNC region showed that mutations abrogating CNC map within GAG downstream of 2 internal AUG codons. The separation of function phenotype displayed by one of the GAG mutants suggested that an altered form of Gag confers CNC. Co-expression of p22 and Ty1 significantly decreased mobility [reviewed inCitation15]. Intriguingly, p22 is encoded by GAG but is not a product of Gag-p49 proteolytic cleavage or degradation. The transcript used for p22 synthesis is an internally-initiated (Ty1i) RNA that starts within GAG.Citation14 Our studies revealed that p22 synthesis can be initiated from either of 2 closely positioned AUG codons located downstream of the Ty1i start-sitesCitation16 and that the structure of the Ty1i 5′-UTR is important for efficient translation in vitro (Błaszczyk et al, unpublished). Similar to Gag-p49, p22 undergoes maturation by Ty1 protease to form AUG1p18 (translated from first start codon) and AUG2p18 (translated from second start codon).Citation16 Both AUG1p18 and AUG2p18 execute strong inhibition of Ty1 mobility that correlates with the level of their expression. In contrast, the proteins corresponding to N-terminal (NTR; 1-173 aa) or C-terminal (CTR; 174-401 aa) regions of Gag do not inhibit retrotransposition (). Thus, restriction does not result from simple Gag truncation but is specific to a ∼150 aa sequence within the C-terminal half of Gag. However, extension of the ∼150 aa protein by an additional ∼80 aa toward the Gag N-terminus abolishes restriction.Citation16 Together, our results suggest that Ty1 GAG evolved a specific organization and expression strategy to produce products both essential and antagonistic for retrotransposon movement.
Figure 1. Schematic representation of Ty1 Gag and Gag-derived proteins. At the bottom is a prediction of ty1 gag disordered (yellow) and α-helical (red) regions (16).
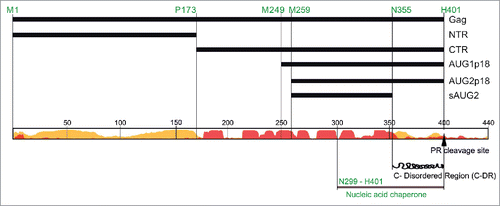
Ty1 Gag is not only the major structural component of the VLP but is a multifunctional regulator of Ty1 replication. Cristofari et al. demonstrated that a synthetic peptide corresponding to C-terminal 103 aa of Ty1 Gag displays nucleic acid chaperone (NAC) activity in vitro.Citation17 We showed that the CTR protein displays NAC activity similar to retroviral nucleocapsid proteins.Citation16,18 Therefore, the NAC activity of Ty1 Gag is most likely critical for RNA dimerization, packaging, annealing of the tRNA primer and strand-transfer during reverse transcription. Paradoxically, the NAC region is also present in p22/p18 even though Ty1 Gag and p22/p18 play opposing roles in retrotransposition.
Is there a reason for the NAC functionality to also be present in p22? We found that despite containing the nucleic acid chaperone domain, AUG1p18 and AUG2p18 show decreased NAC activity in comparison to the CTRCitation16 or Ty1 Gag (Pachulska-Wieczorek et al. unpublished). Moreover, AUG1p18 and AUG2p18 proteins display different properties even though they differ by only 10 residues located N-terminal to the NAC region (). Using an assay that mimics tRNA hybridization with the primer-binding site, AUG2p18 enhanced annealing to about 50% of the level measured for CTR, while the AUG1p18 enhanced annealing <20%. A similar trend was observed for protein-facilitated Ty1 RNA dimerization in vitro. Interestingly, AUG1p18 and the CTR behaved similarly in RNA binding analyses, since both bind Ty1 RNA with a low Kd even when electrostatic interactions were masked by increasing salt concentration. In contrast, AUG2p18 binding was sensitive to salt and thus less specific.
Despite the differences observed in the NAC activity and RNA binding specificity of the CTR and the restriction factors, hydroxyl radical footprinting revealed that they bind within the same regions of Ty1 RNA.Citation16 These results suggest that both forms of p18 and Gag compete for the same binding sites on Ty1 genomic RNA during the process of retrotransposition. A major binding site for the CTR and p18 was detected within the pseudoknotCitation16 present at the 5′-end of Ty1 RNA.Citation19,20 The Ty1 pseudoknot has unusual structural features since it encompasses a large RNA segment (>300 nt). We showed earlier that mutations disrupting pseudoknot formation interfere with retrotransposition,Citation19 indicating that it provides a critical biological function.Citation21 We found more than 50-fold decrease in transposition when the pseudoknot was destabilized in UC264AG mutant.Citation19 Analysis of the Ty1 RNA half-life, packaging of Ty1 RNAs into VLPs and cDNA accumulation suggest that the primary importance of the pseudoknot occurs during reverse transcription. However, the 2-fold decrease in cDNA accumulation cannot explain the 50-fold decrease in retrotransposition frequency.Citation19 Discovery of the p22/p18 restriction factor sheds new light on retrotransposition regulation, and in fact, may explain results obtained from mutational analysis of the pseudoknot. The fact that p18 binds Ty1 RNA in vitro within pseudoknot raises the possibility that mobility defects observed for RNA pseudoknot mutants might relate to interactions with p22/p18. Since p18 binds Ty1 RNA with less specificity than Gag, it is possible that p22/p18 can bind Ty1 RNA despite structural defects in the pseudoknot. Thus p22/p18 will limit the RNA available for interactions with Gag or the RT/IN heteromer, which is required for reverse transcription. Since p18 is also less active chaperone it could also decrease the efficiency or specificity of reverse transcription.
The pseudoknot structure is absent from in vitro transcribed full-length Ty1 RNA,Citation20 suggesting that in vivo protein-mediated RNA chaperone activity may be involved in pseudoknot folding process. This hypothesis is in accordance with generally accepted role of nucleic acid chaperone proteins in the RNA folding process. We demonstrated that p18 proteins and a derivative incapable of RNA binding (; sAUG2) decrease the CTR chaperone activity. Therefore, one aspect of p18 restriction may involve inhibition of Gag NAC functions by p18/Gag interactions. If Gag NAC function helps stabilize the pseudoknot, p18 could interfere with its formation during restriction of Ty1 movement.
Additional CTR and p18 binding sites within Ty1 RNA are adjacent to the structural motifs important for tRNAiMet hybridization, cyclization and dimerization.Citation16,20 Assuming that Gag and p22/p18 proteins compete for those binding sites and since p22/p18 displays compromised NAC activity, retrotransposition may be targeted at all steps requiring nucleoprotein interactions mediated by Gag (). This idea is supported by findings that p22/p18 disrupts retrosome formation, VLP assembly and inhibits reverse transcription within the VLPs that are able to form.Citation14 If Ty1 and retroviral particle assembly are similar,Citation22 then RNA dimerization may be a prerequisite for VLP assembly, and p22/p18 could inhibit this process since it antagonizes Ty1 RNA dimerization in vitro.Citation16 Additional support comes from the analysis of a mutant lacking XRN1, which encodes a 5′-3′ exonuclease involved in RNA decay. This mutant produces more Ty1i RNA and p18Citation14 and fails to package Ty1 RNA into VLPs.Citation23 Furthermore, the low activity in tRNA annealing displayed by p18 could contribute to the block in reverse transcription in addition to the decreased level of IN/RT heteromer.Citation14
Figure 2. Ty1 replication cycle with the steps inhibited by p22/p18 highlighted. p22/p18 is shown in blue.
A recent study from the Garfinkel lab found that mutations in GAG confer resistance to the p22 restriction factor.Citation24 Most of the mutations mapped within the Gag region important for VLP assembly but they did not interfere with p22 binding to Gag.Citation24 Thus, productive Gag/Ty1 RNA interactions were rescued from p22 restriction in those mutants. Other characterized resistant mutants contained substitutions in the NAC region important for RNA binding but their NAC functions remain to be studied.
In summary, our work suggests that it may be difficult to define a single action of p22/p18 that is most destructive to Ty1 movement. Instead, both strong and weaker defects mediated by p22/p18 contribute to the severe inhibition of retrotransposition during CNC. Discovery of the Gag-derived p22/p18 restriction factor also answers a long-standing question concerning how Ty1 transposition is repressed and sets up a new paradigm for host-parasite interactions.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank an anonymous Reviewer for constructive comments.
Funding
This work was supported by the National Science Center Poland [2011/01/D/NZ1/03478, 2012/06/A/ST6/00384], the Foundation for Polish Science [HOMING PLUS/2012-6/12], the Ministry of Science and Higher Education Poland [0492/IP1/2013/72], and the National Institutes of Health [GM095622].
References
- Curcio MJ, Lutz S, Lesage P. The Ty1 LTR-retrotransposon of budding yeast. Microbiol Spectr 2015; 3:1-35; PMID:25893143
- Belcourt MF, Farabaugh PJ. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell 1990; 62:339-52; PMID:2164889; http://dx.doi.org/10.1016/0092-8674(90)90371-K
- Malagon F, Jensen TH. The T body, a new cytoplasmic RNA granule in Saccharomyces cerevisiae. Mol Cell Biol 2008; 28:6022-32; PMID:18678648; http://dx.doi.org/10.1128/MCB.00684-08
- Malagon F, Jensen TH. T-body formation precedes virus-like particle maturation in S. cerevisiae. RNA Biol 2011; 8:184-9; PMID:21358276; http://dx.doi.org/10.4161/rna.8.2.14822
- Feng YX, Moore SP, Garfinkel DJ, Rein A. The genomic RNA in Ty1 virus-like particles is dimeric. J Virol 2000; 74:10819-21; PMID:11044130; http://dx.doi.org/10.1128/JVI.74.22.10819-10821.2000
- Keeney JB, Chapman KB, Lauermann V, Voytas DF, Aström SU, von Pawel-Rammingen U, Bystrom A, Boeke JD. Multiple molecular determinants for retrotransposition in a primer tRNA. Mol Cell Biol 1995; 15:217-26; PMID:7528326; http://dx.doi.org/10.1128/MCB.15.1.217
- Devine SE, Boeke JD. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev 1996; 10:620-33; PMID:8598291; http://dx.doi.org/10.1101/gad.10.5.620
- Huang CR, Burns KH, Boeke JD. Active transposition in genomes. Annu Rev Genet 2012; 46:651-75; PMID:23145912; http://dx.doi.org/10.1146/annurev-genet-110711-155616
- Babatz TD, Burns KH. Functional impact of the human mobilome. Curr Opin Genet Dev 2013; 23:264-70; PMID:23523050; http://dx.doi.org/10.1016/j.gde.2013.02.007
- Kazazian HH, Jr. Mobile elements: drivers of genome evolution. Science 2004; 303:1626-32; PMID:15016989; http://dx.doi.org/10.1126/science.1089670
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 2007; 8:272-85; PMID:17363976; http://dx.doi.org/10.1038/nrg2072
- Drinnenberg IA, Fink GR, Bartel DP. Compatibility with killer explains the rise of RNAi-deficient fungi. Science 2011; 333:1592; PMID:21921191; http://dx.doi.org/10.1126/science.1209575
- Garfinkel DJ, Nyswaner K, Wang J, Cho JY. Post-transcriptional cosuppression of Ty1 retrotransposition. Genetics 2003; 165:83-99; PMID:14504219
- Saha A, Mitchell JA, Nishida Y, Hildreth JE, Ariberre JA, Gilbert WA, Garfinkel DJ. A trans-dominant form of Gag restricts Ty1 retrotransposition and mediates copy number control. J Virol 2015; 89:3922-38; PMID:25609815; http://dx.doi.org/10.1128/JVI.03060-14
- Garfinkel DJ, Tucker JM, Saha A, Nishida Y, Pachulska-Wieczorek K, Blaszczyk L, Purzycka KJ. A self-encoded capsid derivative restricts Ty1 retrotransposition in Saccharomyces. Curr Genet 2015; PMID:26650614.
- Nishida Y, Pachulska-Wieczorek K, Blaszczyk L, Saha A, Gumna J, Garfinkel DJ, Purzycka KJ. Ty1 retrovirus-like element Gag contains overlapping restriction factor and nucleic acid chaperone functions. Nucleic Acids Res 2015; 43:7414-31; PMID:26160887; http://dx.doi.org/10.1093/nar/gkv695
- Cristofari G, Ficheux D, Darlix JL. The GAG-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J Biol Chem 2000; 275:19210-7; PMID:10766747; http://dx.doi.org/10.1074/jbc.M001371200
- Pachulska-Wieczorek K, Stefaniak AK, Purzycka KJ. Similarities and differences in the nucleic acid chaperone activity of HIV-2 and HIV-1 nucleocapsid proteins in vitro. Retrovirology 2014; 11:54; PMID:24992971; http://dx.doi.org/10.1186/1742-4690-11-54
- Huang Q, Purzycka KJ, Lusvarghi S, Li D, Legrice SF, Boeke JD. Retrotransposon Ty1 RNA contains a 5′-terminal long-range pseudoknot required for efficient reverse transcription. RNA 2013; 19:320-32; PMID:23329695; http://dx.doi.org/10.1261/rna.035535.112
- Purzycka KJ, Legiewicz M, Matsuda E, Eizentstat LD, Lusvarghi S, Saha A, Le Grice SF, Garfinkel DJ. Exploring Ty1 retrotransposon RNA structure within virus-like particles. Nucleic Acids Res 2013; 41:463-73; PMID:23093595; http://dx.doi.org/10.1093/nar/gks983
- Purzycka KJ, Garfinkel DJ, Boeke JD, Le Grice SF. Influence of RNA structural elements on Ty1 retrotransposition. Mob Genet Elements 2013; 3:e25060; PMID:23914314; http://dx.doi.org/10.4161/mge.25060
- Johnson SF, Telesnitsky A. Retroviral RNA dimerization and packaging: the what, how, when, where, and why. PLoS Pathog 2010; 6:e1001007; PMID:20949075; http://dx.doi.org/10.1371/journal.ppat.1001007
- Dutko JA, Kenny AE, Gamache ER, Curcio MJ. 5′ to 3′ mRNA decay factors colocalize with Ty1 gag and human APOBEC3G and promote Ty1 retrotransposition. J Virol 2010; 84:5052-66; PMID:20219921; http://dx.doi.org/10.1128/JVI.02477-09
- Tucker JM, Larango ME, Wachsmuth LP, Kannan N, Garfinkel DJ. The Ty1 Retrotransposon Restriction Factor p22 Targets Gag. PLoS Genet 2015; 11:e1005571; PMID:26451601; http://dx.doi.org/10.1371/journal.pgen.1005571
