Abstract
The detailed mechanism underlying adipocyte development and function remains to be fully understood. MicroRNAs are a class of non-coding regulators in adipocyte development and function in physiological and pathological conditions. Our recent study11 using adipose-specific dgcr8 knockout mice demonstrates that microRNA biogenesis has a depot-specific function in adipose and plays an important role in brown adipocyte feature maintenance. Interestingly, in a parallel study from Kahn's group, Mori et al observed similar phenotypes in adipose-specific Dicer knockout mice. This study also reveals that the expression of Dicer is downregulated in the adipose from patients with HIV-associated lipodystrophy (HALS), suggesting that dysregulation of microRNA biogenesis may play a role in the onset of HALS.
The past few years have seen an upsurge of interest in adipocyte biology, coincident with the onset of epidemic of obesity and associated metabolic diseases. Brown fat, unlike white fat, has a major function in energy expenditure by uncoupling oxidative phosphoration from ATP production, thereby receiving much attention for its therapeutic potential against many metabolic diseases.Citation1–4 There has been significant progression in understanding the regulatory mechanism underlying brown adipocyte differentiation and function. Many microRNAs, including miR-193b-365,Citation5 miR-133,Citation6–8 miR-155,Citation9 miR-196a,Citation10 etc., positively or negatively regulate brown adipogenesis. However, whether microRNAs are necessary for feature maintenance in mature brown adipocytes were poorly understood.
To address this question, our group ablated Dgcr8, an essential component in microRNA biogenesis, in mature adipocytes.Citation11 We bred mice carrying the Dgcr8-floxed allele with mice carrying the adipoQ-Cre transgene. Since AdipoQ-Cre is not expressed in preadipocytes but only expressed in later stage of differentiation, it will allow us to assess the function of microRNAs in mature adipocytes but not in adipogenesis.Citation12 The Dgcr8 knockout animals displayed an enlarged brown fat with a cream color, accompanied by decreased expression of UCP1 and other brown fat markers. Interestingly, another independent study from Kahn's group confirmed our findings using a different mouse model in which Dicer, a downstream component of Dgcr8 in microRNA biogenesis, was ablated in mature adipocytes, resulting in very similar phenotypes.Citation13 These two studies demonstrate that microRNAs are not only important for brown adipocyte differentiation but also for its functional maintenance.
As mentioned above, in both studies, target gene deletion was detected in mature adipocytes but not in preadipocytes. However, only based on these observations, we can't safely exclude the possibility that the loss of brown fat features in mature adipocytes is due to a defect during the course of adipogenesis, because the intermediates between these 2 stages could not be readily identified and isolated in vivo, precluding the possibility to directly examine whether the gene deletion can occur during adipogenesis. To address this issue, Mori's study employed another mouse model, inducible Ap2-Cre strain, to specifically delete Dicer in mature adipocytes.Citation13 After tamoxifen injection-induced Dicer deletion, these knockout animals manifest most of the aforementioned phenotypes, including larger brown fat, lower brown fat marker expression and impaired systemic insulin sensitivity. These data indicate that the defects observed in brown fat are indeed due to the absence of microRNAs in mature brown adipocytes.
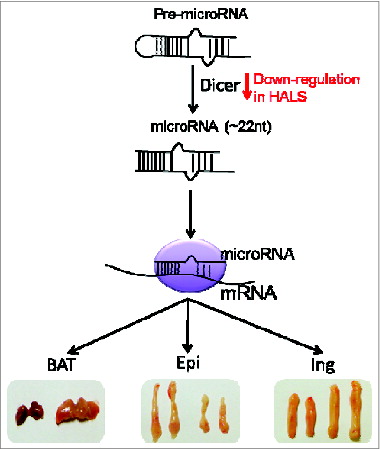
Consistent with the notion that distinct fat depot has different lineage origin.1415 both our and Mori et al's studies show that microRNAs have depot-specific functions (). Depletion of microRNAs in adipose results in enlarged cream color brown fat, bigger inguinal fat but smaller epididymal fat. Similar phenotypes and an impaired insulin signaling pathway of inguinal fat were observed in Mori et al's study. Thus, microRNAs seem to be essential for brown fat to maintain its energy expenditure features, for subcutaneous fat to keep insulin sensitivity, and for epididymal fat to retain lipids in this depot. However, these data complicate the interpretation of systemic insulin resistance developed in these mouse models. It remains to be determined which fat depot is the primary defective organ accounting for the whole body phenotype. A straightforward experiment is to block microRNA biogenesis specifically in brown fat using the UCP1-Cre strain, which will allow us to uncouple the contribution of brown fat from that of white fat.
Figure 1. MicroRNA biogenesis has a depot-specific function in adipose. Our study demonstrated that microRNA biogenesis is essential in feature maintenance in BAT, and lipid accumulation and distribution in Epi and Ing white fat. Similar phenotypes were observed in Mori et al's study in which downregulation of Dice is connected with HALS.
To explore which microRNAs may account for the brown fat phenotype, we performed microRNA microarray and identified a set of microRNAs that are highly enriched in brown fat and upregulated during brown adipocyte differentiation. Some of these microRNAs, including miR-193b and miR-365.Citation5 have been reported before as regulators in brown adipocytes. We further identified miR-182 and miR-203 as novel regulators required for brown adipocyte differentiation. However, we were not able to rescue UCP1 and other marker expression in the knockout brown adipocytes using any of these microRNA mimics in primary cell culture. Interestingly, Mori et al rescued UCP1 expression by about 20-fold with miR-365 mimic in immortalized Dicer knockout brown adipocytes. The reason behind this discrepancy remains to be determined.
Mori's study further linked the abnormal fat redistribution with the HIV-associated lipodystrophy syndrome (HALS), the most common form of non-congenital lipodystrophy. HALS occurs in many HIV-infected patients with a prevalence rate varying widely from 11% to 83% according to international cross-sectional studies.Citation16,17 HALS patients often manifest disarrangement of adipose tissue with a depletion of subcutaneous fat, significantly worsen lipid profiles with elevated triglycerides, systemic insulin resistance and decreased adiponectin concentration. All these phenotypes are similar in both adipose Dicer and Dgcr8 knockout mice models. Interestingly, Mori et al demonstrated that Dicer expression levels are significantly decreased in dorsocervical and abdominal subcutaneous adipose from HALS patients, strongly suggesting that the decreased microRNA biogenesis is a contributing factor of HALS.
It remains unknown the mechanism by which Dicer expression levels decrease in HALS adipose. It is widely accepted that the main culprit for HALS is highly active antiretroviral therapy (HAART), which is administrated to decrease morbidity and prolong life expectancy, but also leads to lipodystrophy as a side effect.Citation18 The manifestations of lipodystrophy differ with respect to the choice of HAART drugs.Citation19 Nucleoside reverse transcriptase inhibitors (NRTIs) are commonly associated with lipoatrophy of extremities, while protease inhibitors are more linked to lipoaccumulation, hypertriglyceridemia, and systemic insulin resistances. Further studies need to be performed to determine whether and which HAART drugs lead to Dicer downregulation and whether other players in microRNA biogenesis such as Dgcr8 and Drosha are also downregulated.
Although these 2 knockout models are valuable for HALS studies, we should realize that the manifestations of HALS are complicated and there are some aspects that these mouse models do not resemble. For example, HALS is often characterized by a depletion of subcutaneous fat but an accumulation of central fat.Citation20 However, both mouse models manifest a smaller visceral fat and the Dgcr8 knockout mice even have a bigger inguinal fat. We should be aware of these limitations when these animals are used as HALS models.
Besides HALS, microRNA biogenesis pathway was also downregulated in aging. An earlier study from Mori et al showed that Dicer expression is significantly decreased in preadipocytes from elderly humans. Loss-of-dicer in C. elegans reduces their lifespan and tolerance to stress while overexpression of Dicer confers stress resistance.Citation21 Thus, the components of microRNA biogenesis, thought to play house-keeping role, are regulated in adipocytes at different physiology and pathological conditions. It is conceivable that besides the global regulation of microRNAs biogenesis, the expression of some individual microRNAs is also regulated through other mechanisms such as transcriptional control. How these 2 layers of regulation intertwine together in HALS and aging will need be elucidated in further studies.
In summary, our and Mori's studies have revealed a feature maintenance function of microRNA biogenesis in brown fat and also demonstrated that the role of microRNAs in adipose is depot-specific. In addition, both animal models manifest a disarrangement of adipose, worsen lipid profiles and systemic insulin resistance, partially resembling the phenotypes of HALS in which Dicer expression is downregulated in adipose. Determining the mechanism of Dicer downregulation in HALS and dissecting which microRNAs are functional in these pathological processes may lead to new therapeutic strategies to ameliorate HALS.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by Singapore NRF fellowship (NRF-2011NRF-NRFF 001–025) to L.S.
References
- Lee P, Swarbrick MM, Ho KKY. Brown adipose tissue in adult humans: a metabolic renaissance. Endocr Rev [Internet] 2013; 34:413-38; [cited 2013 Apr 5]; PMID:23550082; http://dx.doi.org/10.1210/er.2012-1081
- Lo KA, Sun L. Turning WAT into BAT: a review on regulators controlling the browning of white adipocytes. Biosci Rep [Internet] 2013; 33:711-9; PMID:23895241; http://dx.doi.org/10.1042/BSR20130046
- Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med [Internet] 2013; [cited 2013 Dec 13]; 19:1252-63; PMID:24100998; http://dx.doi.org/10.1038/nm.3361
- Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell [Internet] 2014; [cited 2014 Jul 11]; 156:20-44; PMID:24439368; http://dx.doi.org/10.1016/j.cell.2013.12.012
- Sun L, Xie H, Mori MA, Alexander R, Yuan B, Hattangadi SM, Liu Q, Kahn CR, Lodish HF. Mir193b-365 is essential for brown fat differentiation. Nat Cell Biol [Internet] 2011; [cited 2011 Jul 16]; 13:958-65; PMID:21743466; http://dx.doi.org/10.1038/ncb2286
- Trajkovski M, Ahmed K, Esau CC, Stoffel M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nat Cell Biol [Internet] 2012; [cited 2012 Nov 12]; 14:1-8; PMID:23143398; http://dx.doi.org/10.1038/ncb2612
- Liu W, Bi P, Shan T, Yang X, Yin H, Wang Y-X, Liu N, Rudnicki MA, Kuang S. miR-133a regulates adipocyte browning in vivo. PLoS Genet [Internet] 2013; [cited 2013 Jul 17]; 9:e1003626; PMID:23874225; http://dx.doi.org/10.1371/journal.pgen.1003626
- Yin H, Pasut A, Soleimani VD, Bentzinger CF, Antoun G, Thorn S, Seale P, Fernando P, Ijcken W, Van Grosveld F, et al. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting prdm16. Cell Metab [Internet] 2013; 17:210-24; PMID:23395168; http://dx.doi.org/10.1016/j.cmet.2013.01.004
- Chen Y, Siegel F, Kipschull S, Haas B, Fröhlich H, Meister G, Pfeifer A. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat Commun [Internet] 2013; [cited 2013 Apr 24]; 4:1769; PMID:23612310; http://dx.doi.org/10.1038/ncomms2742
- Mori M, Nakagami H, Rodriguez-Araujo G, Nimura K, Kaneda Y. Essential role for miR-196a in brown adipogenesis of white fat progenitor cells. PLoS Biol [Internet] 2012; [cited 2012 Apr 27]; 10:e1001314; PMID:22545021; http://dx.doi.org/10.1371/journal.pbio.1001314
- Kim HJ, Cho H, Alexander R, Patterson HC, Gu M, Lo KA, Xu D, Goh VJ, Nguyen LN, Chai X, et al. MicroRNAs are required for the feature maintenance and differentiation of brown adipocytes. Diabetes [Internet] 2014; [cited 2014 Nov 4]; 1-12; PMID:25008181; http://dx.doi.org/10.2337/db14-0466
- Lee KY, Russell SJ, Ussar S, Boucher J, Vernochet C, Mori MA, Smyth G, Rourk M, Cederquist C, Rosen ED, et al. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes [Internet] 2013; [cited 2013 Feb 28]; 62:864-74; PMID:23321074; http://dx.doi.org/10.2337/db12-1089
- Mori MA, Thomou T, Boucher J, Lee KY, Lallukka S, Kim JK, Torriani M, Yki-Järvinen H, Grinspoon SK, Cypess AM, et al. Altered miRNA processing disrupts brown/white adipocyte determination and associates with lipodystrophy. J Clin Invest [Internet] 2014 [cited 2014 Jul 2]; 124:3339-51; PMID:24983316; http://dx.doi.org/10.1172/JCI73468.
- Gesta S, Tseng Y, Kahn CR. Developmental origin of fat : tracking obesity to its source. Cell 2007;131:242-56; PMID:17956727
- Long JZ, Svensson KJ, Tsai L, Zeng X, Roh HC, Kong X, Rao RR, Lou J, Lokurkar I, Baur W, et al. A smooth muscle-like origin for beige adipocytes. Cell Metab [Internet] 2014; [cited 2014 May 6]; 19:810-20; PMID:24709624; http://dx.doi.org/10.1016/j.cmet.2014.03.025
- Carr A, Samaras K, Thorisdottir A, Kaufmann GR, Chisholm DJ, Cooper DA. Diagnosis, prediction, and natural course of HIV-1 protease-inhibitor-associated lipodystrophy, hyperlipidaemia, and diabetes mellitus: A cohort study. Lancet 1999; 353:2093-9; PMID:10382692; http://dx.doi.org/10.1016/S0140-6736(98)08468-2
- Gervasoni C, Ridolfo AL, Trifirò G, Santambrogio S, Norbiato G, Musicco M, Clerici M, Galli M, Moroni M. Redistribution of body fat in HIV-infected women undergoing combined antiretroviral therapy. AIDS 1999; 13:465-71; PMID:10197374; http://dx.doi.org/10.1097/00002030-199903110-00004
- Tsiodras S, Mantzoros C, Hammer S, Samore M. Effects of protease inhibitors on hyperglycemia, hyperlipidemia, and lipodystrophy: a 5-year cohort study. Arch Intern Med [Internet] 2000; [cited 2014 Nov 4]; 160:2050-6; PMID:10888979; http://dx.doi.org/10.1001/archinte.160.13.2050
- Mallewa JE, Wilkins E, Vilar J, Mallewa M, Doran D, Back D, Pirmohamed M. HIV-associated lipodystrophy: A review of underlying mechanisms and therapeutic options. J Antimicrob Chemother 2008; 62:648-60; PMID:18565973; http://dx.doi.org/10.1093/jac/dkn251
- Anuurad E, Semrad A, Berglund L. Human immunodeficiency virus and highly active antiretroviral therapy-associated metabolic disorders and risk factors for cardiovascular disease. Metab Syndr Relat Disord [Internet] 2009; [cited 2014 Oct 6]; 7:401-10; PMID:19355810; http://dx.doi.org/10.1089/met.2008.0096
- Mori MA, Raghavan P, Thomou T, Boucher J, Robida-Stubbs S, Macotela Y, Russell SJ, Kirkland JL, Blackwell TK, Kahn CR. Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab [Internet] 2012; [cited 2012 Sep 4]; 16:336-47; PMID:22958919; http://dx.doi.org/10.1016/j.cmet.2012.07.017
