ABSTRACT
Obesity has reached epidemic proportions, leading to severe associated pathologies such as insulin resistance, cardiovascular disease, cancer and type 2 diabetes. Adipose tissue has become crucial due to its involvement in the pathogenesis of obesity-induced insulin resistance, and traditionally white adipose tissue has captured the most attention. However in the last decade the presence and activity of heat-generating brown adipose tissue (BAT) in adult humans has been rediscovered. BAT decreases with age and in obese and diabetic patients. It has thus attracted strong scientific interest, and any strategy to increase its mass or activity might lead to new therapeutic approaches to obesity and associated metabolic diseases. In this review we highlight the mechanisms of fatty acid uptake, trafficking and oxidation in brown fat thermogenesis. We focus on BAT's morphological and functional characteristics and fatty acid synthesis, storage, oxidation and use as a source of energy.
Introduction
Importance of adipose tissue in obesity
Current life styles and continuous nutrient excess are increasing the incidence of obesity at an alarming rate, especially at younger ages. Worldwide there are more than 600 million obese subjects and, importantly, most of the world's population live in countries where overweight and obesity kills more people than underweight.Citation1 Very worrisome are the concurrent and parallel increases in the prevalence of pathologic conditions associated with obesity such as insulin resistance, cardiovascular and Alzheimer disease, cancer, and type 2 diabetes.
Over the last 2 decades the obesity epidemic has put a spotlight on the adipose tissue as a key player in the mechanisms involved in obesity-related disorders. Human fat consists of energy-storing white adipose tissue (WAT) and brown adipose tissue (BAT), which controls thermogenesis by dissipating energy to produce heat. In addition to adipocytes, adipose tissue is well vascularized and contains connective tissue and numerous immune cells such as macrophages, T and B cells, mast cells and neutrophils.Citation2 It has been demonstrated that obesity-induced insulin resistance is due to several factors: ectopic fat deposition,Citation3 increased inflammation and endoplasmic reticulum (ER) stress,Citation4,5 adipose tissue hypoxia and mitochondrial dysfunction,Citation6,7 and impaired adipocyte expansion and angiogenesis.Citation8-10 Fat is also an active endocrine tissue that secretes hormones such as leptin, adiponectin or resistin and inflammatory cytokines such as tumor necrosis factor α (TNFα), interleukin (IL)-6, IL-1β, etc. in response to several stimuli. Adipose tissue is therefore a complex and active organ controlling very important metabolic pathways such as energy expenditure, appetite, insulin sensitivity, endocrine and reproductive functions, inflammation and immunity.
Rediscovery of human active BAT
The fusion of positron-emission tomography (PET) and computed tomography (CT) images has allowed radiologists to retrieve both functional and structural information from a single image. In the course of using PET-CT to detect and stage tumors in humans, active BAT that increased after cold exposure was rediscovered.Citation11,12 Until that moment BAT was considered exclusive to rodents and human neonates. However, the breakthrough came in 2009, when 5 independent research groups used PET-CT to identify the presence and relevance of BAT in adult humans.Citation13-17 All showed major depots of metabolically active fat in the cervical-supravicular region. Furthermore, these depots expressed type 2 iodothyronine deiodinase (DIO2), the β3-adrenergic receptor, and the brown adipocyte-specific protein, uncoupling protein 1 (UCP1), which physiologically uncouples ATP production from mitochondrial respiration, thereby dissipating energy as heat.Citation18 The expression of these proteins indicated the potential responsiveness of human BAT to both hormonal and pharmacological stimuli. Here, we review the possibility that BAT could be induced to enhance its lipid-burning function even further and thus be an effective target to fight against obesity and associated metabolic disorders.
Brown adipose tissue characteristics
BAT localization and morphology
Our knowledge of BAT has been significantly influenced by studies in rodent models. There, BAT is situated at the interscapular, cervical, mediastinal and retroperitoneal regions.Citation19 While in infants BAT is mainly found in the interscapular area, in adult humans BAT is localized in a region extending from the anterior neck to the thorax.Citation20
In contrast to white adipocytes, which are unilocular, with polygonal morphology that optimizes their fat storing capacity, brown adipocytes are multiloculate and their color is due to their high mitochondrion content and vascular suply.Citation21 BAT thermogenesis takes place in its numerous, densely-packed mitochondria containing the BAT-specific inner membrane protein UCP1. Multilocular lipid stores provide a rapid source of fatty acids (FAs) for activated mitochondria. FAs released into the circulation by the WAT are also an important source of FAs for brown adipocytes. Thermogenesis is classified into: 1) Obligatory thermogenesis, which takes into account the standard metabolic rate (energy used for basic function of cells and organs) and the heat generated during food metabolism (digestion, absorption, processing and storing of energy); and 2) Adaptive thermogenesis or heat production in response to environmental temperature and diet. Adaptive thermogenesis can be further divided into: cold-induced shivering thermogenesis, which takes place in skeletal muscle; cold-induced non-shivering thermogenesis, which takes place mainly in brown fat; and diet-induced thermogenesis triggered by overfeeding, which also takes place in BAT.Citation22 Thus, BAT generates heat, with 2 main consequences: protection against cold exposure via non-shivering thermogenesis; and dissipation of the excess of energy from food. Therefore, BAT can be considered as an organ that burns off excess lipids, and further examination of this property may lead to the development of novel strategies against diet-induced obesity.
Molecular BAT signature: beige and brown adipocytes
Comprehensive research is being done to define the still under debate cellular heterogeneity of human fat.Citation23 At least 2 types of thermogenic adipocyte exist in rodents and humans: classical brown adipocytes and beige (also called brite) adipocytes. They have both anatomical and developmental differences. While brown adipocytes are mainly located in the above-mentioned BAT depots, beige adipocytes co-locate with white adipocytes in WAT near vascular and neural innervation and appear in response to certain stimuli, such as chronic cold exposure or β3-adrenergic signaling. In adult humans the ratio of brown to beige increases as one moves deeper within the neck and back.Citation20,24-27
BAT releases endocrine factors such as insulin-like growth factor I (IGF-1), IL-6 or fibroblast growth factor 21 (FGF21).Citation28 Brown adipocytes differ from white adipocytes due to their high expression of DIO2, the lipolytic regulator cell death-inducing DNA fragmentation factor-α-like effector A (CIDEA), and the transcription co-regulators PR domain-containing 16 (PRDM16) and peroxisome proliferator activated receptor gamma coactivator 1 α (PGC1α).Citation29,30 Beige and brown adipocytes have overlapping but distinct gene expression patterns.Citation31 Both express the main thermogenic and mitochondrial genes, including Ucp1. However, some surface markers such as CD137, TBX1 and TMEM26 seem to be specific to murine beige adipocytesCitation24,27 while other genes, like Zic1 and Lhx8, appear to specifically mark classic brown adipocytes.Citation20,32 Basal UCP1 expression and uncoupled respiration before hormonal stimulation are highest in brown fat cells and lower in beige cells, the lowest being found in white fat cells.Citation27 However, stimulation with a β3-adrenergic agonist elevates UCP1 expression in beige cells to levels seen in brown fat cells (fold-change compared to white cells).Citation27,33 This suggests that beige cells have a unique molecular signature with a dual role. They store energy in the absence of thermogenic stimuli but initiate heat production when appropriate signals are received.Citation27 White-to-beige conversion of adipocytes is a potential therapeutic approach to targeting obesity; however, the signals involved in this process still remain unclear.
Brown adipocytes arise from mesenchymal precursor cells common to the myogenic cell lineage and express myogenic factor 5 (Myf5).Citation34 Beige adipocytes derive from precursor cells that differ from those in classical BAT and are closer to the white adipocyte cell lineage. Thus, while brown adipocytes come from a Pax7+/Myf5+ lineage shared with skeletal muscle, white and beige adipocytes derive from Pax7−/Myf5− cells via distinct precursor cells. Beige adipocytes differentiate following activation by cold or other stimuli, and when the cold challenge is ceased, they become inactive, taking on the morphology of a white adipocyte.Citation35 However, the cell lineage and developmental origin of the adipose tissue is not so simple. Individual brown and white fats contain a mixture of adipocyte progenitor cells derived from Myf5+ and Myf5− lineages, with numbers varying depending on the depot location. In fact, beige adipocytes in the retroperitoneal WAT are Myf5+.Citation36 For further information about the developmental origin of white, beige and brown adipocytes see other excellent reviews.Citation34,37,38
At least 2 mechanisms have been postulated to occur during the browning process: transdifferentiation of white into beige adipocytes vs. de novo brown adipogenesis. The transdifferentiation process is the conversion of a differentiated somatic cell type into another one.Citation39 The transdifferentiation of white into beige adipocytes has been reported in several studies.Citation40-43 On the other hand, Lee et al. have shown that β3-adrenergic stimulation induces the proliferation and further differentiation of precursors in WAT.Citation44 Furthermore, Myf5+ precursors have also been reported to differentiate into white adipocytes.Citation36,45 Thus, whether the browning process arises from transdifferentiation or de novo brown adipogenesis is far from being fully understood. One could hypothesize that the 2 processes might take place simultaneously and to a different extend depending on the adipose depot or browning stimuli.
BAT activity in pathological conditions
Human studies showed that BAT was reduced in aging and in obese and diabetic patients, indicating that BAT participates in both cold-induced and diet-induced thermogenesis.Citation13 This significant discovery highlights that any strategy able to increase the mass or activity of BAT could potentially be a promising therapy for obese and diabetic patients. In contrast, enhanced BAT activation has been described as a negative effect on cancer cachexia.Citation46 In this study, mice with cachexia-inducing colorectal tumor showed increased BAT activity despite thermoneutrality, indicating that BAT activation may contribute to impaired energy balance in cancer cachexia. Hibernoma is another BAT pathological condition. A hibernoma is a benign tumor of BAT that up to date has no clear explanation of its cause. It is very rare in humans and it is successfully treated by complete surgical excision.Citation47,48 It has shown to express UCP1 and thus potentially contribute to whole-body energy balance.
Activators of thermogenesis
Despite some controversy, a large body of evidence indicates that browning entails the enhancement of thermogenesis within WAT, i.e. increased expression and activity of UCP1 in what are normally considered WAT depots.Citation49 Several factors have been described to activate the browning of the adipose tissue such as irisin,Citation50 natriuretic peptides,Citation51 bone morphogenetic protein 7 (BMP7)Citation52 and BMP8b,Citation53 norepinephrine,Citation54 meteorin-like,Citation55 bile acids,Citation56 adenosine,Citation57 or FGF21.Citation58 Interestingly, recent studies have shown activation of human BAT by the β3-adrenergic receptor agonist mirabegron.Citation59 β3-adrenergic receptor is expressed in humans on the surfaces of brown and white adipocytes and urinary bladder. Cypess et al. administered 200 mg of oral mirabegron, currently approved to treat overactive bladder, to healthy and young humans. Mirabegron acutely stimulated human BAT thermogenesis and increased resting metabolic rate. Further studies would be needed to explore the specificity of mirabegron's mechanism of action, possible adverse effects such as tachycardia, and the dose used, which was 4-fold higher than that prescribed for overactive bladder.
Although a large number of browning agents have been described (extensively reviewed elsewhere)Citation60,61 some studies showed that browning was a secondary consequence of enhanced heat loss, e.g. because of fur disruption in rodents.Citation49 The search for potential therapeutic browning agents to increase metabolism at thermoneutrality, to function through mechanisms other than those affecting heat loss and to finally decrease obesity should thus continue.
Fatty acid storage
FA synthesis, storage and metabolism are essential during thermogenesis because they are required for UCP1 proton transport activity in BAT.Citation62,63 Fundamentally, brown adipocytes have 2 mechanisms to obtain lipids: FA uptake via lipoproteins carriers and de novo FA synthesis, also known as lipogenesis.
Fatty acid uptake
While brown adipocytes synthesize FAs, the enzyme lipoprotein lipase (LPL), bound at the endothelial cell surface, is the major source of FAs in BAT.Citation64 After a meal, dietary lipids are transported by chylomicrons and very low density proteins (VLDL) via lymphatic vessels into the bloodstream. Once triglyceride (TG) rich-lipoproteins reach the bloodstream, LPL hydrolyzes them into free FAs (FFAs) and monoacylglycerol (MG) for BAT uptake. Indeed, BAT is an efficient modulator of triglyceridemia and it is contemplated as a major plasma lipid-clearing organ in rodents.Citation65-67 In fact, FA uptake under cold exposure is higher in BAT than in skeletal muscle.Citation65 Under cold exposure, the β3-adrenergic pathway enhances BAT FA flux and clearance via increased expression and activity of LPL.Citation65 However, the increase in LPL activity has also been shown to trigger adiposity and insulin resistance.Citation68 Adipocyte-specific LPL KO animals show an increase in FAs derived from lipogenesis and a decrease in polyunsaturated FAs, accompanied by an increase in the expression of lipogenic genes.Citation69 Glycosylphosphatidylinositol-anchored high density lipoprotein binding protein 1 (GPIHBP1) transports LPL across capillary endothelial cells, and GPIHBP1 KO mice show mislocated LPL in many tissues, including BAT,Citation70 decreased TG content and deficient lipolysis,Citation71 Administration of PPARγ agonists, such as rosiglitazone, in rodents increases BAT TG clearance and LPL activity, while lipogenesis is not increased. This suggests that under rosiglitazone treatment brown adipocytes metabolize FAs derived from TG hydrolyzed from lipoproteins or recycled from lipolysis.Citation72
Fatty acid transport
Once FAs are released by LPL, they are taken up into cells by plasmatic membrane receptors and transported for further utilization or storage.Citation65,73-75 The most important FA transporters in BAT are the following ():
Figure 1. FA uptake and lipogenesis in brown adipocytes. Schematic representation of FA uptake, transport, synthesis and storage in brown adipocytes, which provide substrate to mitochondria for thermogenesis. While brown adipocytes synthesize FAs, the enzyme lipoprotein lipase (LPL) is the major source of FAs in BAT. Once triglyceride (TG) rich-lipoproteins reach the bloodstream, LPL hydrolyzes them into FFAs for BAT uptake. FAs are sensed and taken up by FFAs 3 (FFA3) proteins, cluster of differentiation 36 (CD36) and/or FA transport proteins (FATPs). Inside the cytoplasm, FAs are transported by FA binding proteins (FABP). On the other hand, FAs can be synthesized by lipogenesis. This process takes place in the cytosol, and the first phase begins with the formation of malonyl-CoA from acetyl-CoA by the action acetyl-CoA caboxylase (ACC). Then, FA synthetase (FAS) catalyzes various reactions to finally generate palmitate, a 16-carbon saturated FA. In BAT, the last phases of lipogenesis are carried out by very long chain FA 3 (ELOVL3) and stearoyl-CoA desaturase 1 (SCD1). Once FAs are synthesized they can be esterified, becoming available for FAO or stored as TG in lipid droplets (LD). Blue arrows indicate enhanced processes or expression of proteins after cold stimulation and β3-adrenergic receptor activation.
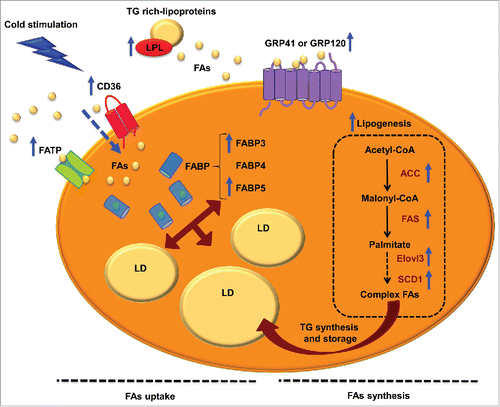
Cluster of differentiation 36 (CD36)
This integral membrane protein is expressed in BAT among other tissues.Citation73 CD36 belongs to the class B scavenger receptor family of cell surface proteins, whose main function is to translocate FAs, released by LPL activity, across the plasmatic membrane and thus provide a substrate for BAT thermogenesis.Citation65 Under cold exposure, CD36 expression and activity increase ().Citation65 However, CD36 is not a simple translocase; it is considered a lipid sensor and a regulator of FA uptake and transport in adipocytes.Citation76-78 CD36 KO mice die after 24 hours of cold exposure, which implicates CD36 in thermogenesis.Citation65 In addition, CD36 genetic variability has been associated with body weight differences in humans.Citation79
FA transport proteins (FATPs)
There are 6 isoforms of FATPs. FATP1 and 4 can be found specifically in BAT (extensively reviewed elsewhere).Citation80 These proteins translocate FAs into cells.Citation81 They display very long-chain acyl-CoA synthetase activity,Citation64,82 and their overexpression increases FA uptake.Citation83
G-protein-coupled receptors (GPCRs)
GPCRs comprise a family of proteins that respond to several ligands, and trigger a cascade of intracellular signaling (extensively reviewed elsewhere).Citation84 GPR41 (also known as FFA3) and GPR120 are activated by medium and long-chain FFA in BAT, and they are considered as sensors that maintain cell lipid homeostasis.Citation85 Interestingly, GRP120 mRNA expression increases under cold exposure (3).Citation86
Fatty acid binding proteins (FABPs)
Once in the cytoplasm, FFAs are minimally soluble. To prevent disruption of membrane or lipotoxicity, cells have soluble proteins that bind FFAs and transport them.Citation87 Brown adipocytes harbour 3 different isoforms: FABP3, FABP4 and FABP5.Citation88 FABP4, commonly known as adipocyte protein 2 (aP2), has been extensively used as a marker of adipocyte differentiation.Citation89 Although FABP4 is the most abundantly expressed isoform in BAT, only FABP3 and FABP5 are increased by cold exposure in rats.Citation88,90 Interestingly, FABP3 is overexpressed in mice with diet-induced obesity and in UCP1 KO mice, and it is associated with increased thermogenesis.Citation91 Thus, FFAs bind to FABPs present in brown adipocytes and are either stored or utilized to maintain thermogenesis ().
Lipogenesis
De novo FA synthesis or lipogenesis is the metabolic pathway that synthesizes FAs and ultimately induces TG synthesis.Citation92,93 Excellent studies on WAT report that glucose uptake, a preliminary step in de novo FA synthesis, is also involved in the regulation of lipogenesis.Citation94,95 Whether BAT contributes to this process is still unclear. A recent study examined the dynamics of de novo lipogenesis and lipolysis in classic brown, subcutaneous beige and classic white adipose tissues during chronic β3-adrenergic receptor stimulation.Citation96 Sustained β3-adrenergic stimulation increased de novo lipogenesis, TG turnover, and the expression of genes involved in FA synthesis and oxidation similarly in all adipose depots indicating that FA synthesis and FAO are tightly coupled during chronic β3-adrenergic stimulation.
Lipogenesis takes place in the cytosol and it can be summarized in 3 steps: synthesis of FAs from acetyl-CoA, elongation and desaturation. Lipogenesis begins with the carboxylation of acetyl-CoA to malonyl-CoA, the committed step catalyzed by acetyl-CoA carboxylase (ACC), which requires biotin cofactor.Citation97,98 Finally, FA synthetase (FAS), a multifunctional cytosolic protein, catalyzes different reactions to form palmitate, a 16-carbon saturated FA.Citation93 It has been shown that adipose-specific FAS KO mice have increased energy expenditure, which comes from the browning of subcutaneous WAT.Citation99
In the second phase of lipogenesis, FAs derived from the FAS enzymatic reaction, are elongated by membrane-bound enzymes mostly localized in the ER.Citation93 This process is induced by the elongation of very long chain FA (ELOVL) proteins, which have 7 members in mice and humans.Citation100 Among them, Elovl3 is expressed in BAT,Citation101 and its expression and activity are upregulated under cold conditions to re-establish the intracellular pool of TG and preserve lipid homeostasis.Citation102-104 ELOVL3 KO mice are resistant to diet-induced obesity, showing an increase in energy expenditure.Citation104
The final phase of lipogenesis is the desaturation of FAs. This process is catalyzed by desaturases, such as stearoyl-CoA desaturases (SCDs), which introduce double bonds at a specific position in a FA chain.Citation93,97 SCD1 is the predominant isoform in adipose tissue and liver, and its downregulation in liver prevents diet-induced obesity.Citation105,106 SCD1 KO mice show an increase in glucose uptake and glycogen metabolism, higher energy expenditure and basal thermogenesis in BAT.Citation107
Once FAs have been synthesized they can be esterified to be used for fatty acid oxidation (FAO) or stored as TG in lipid droplets. In BAT, the proper levels of TG are associated with thermogenic activity.Citation108 Since TG are composed of molecules of glycerol and 3 esterified FAs, TG synthesis depends on intracellular levels of glycerol-3-phosphate (G3P), the activated form of glycerol and first intermediary in TG synthesis. Thus, G3P levels are preserved under rigorous control in brown adipocytes.Citation66 It has been shown that rosiglitazone, a PPARγ agonist, increases BAT glycerolkinase activity, which phosphorylates glycerol generating G3P.Citation67 Since G3P comes from glucose, glucose metabolism plays a key role in BAT, as an important glucose-clearing organ, specifically under sympathetic activation.Citation65 In fact, G3P is a major substrate for BAT respiration.Citation109
In conclusion, coordinated FA uptake, transport and synthesis contribute to thermogenesis in BAT (). For this reason, the maintenance of the intracellular pool of TG to preserve lipid homeostasis in brown adipocytes is so important.Citation110,111 Indeed, any of these processes might be a potential target in the treatment of obesity-related disorders, such as insulin resistance and diabetes.
Fatty acid storage
Cells package the excess of intracellular lipids in a phylogenetically conserved organelle called lipid droplet, preventing the lipotoxicity of lipids and cholesterol in the cytoplasm.Citation112,113 Lipid droplets are composed of a neutral lipid core (cholesteryl ester and TGs) covered by a phospholipid monolayer, which contains proteins that regulate lipolysis. Among the lipid droplet membrane proteins found in brown adipocytes we will highlight fat storage-inducing transmembrane protein 2 (FITM2/FIT2), CIDEA and fat-specific protein 27 (FSP27 or CIDEC).
FITM2/FIT2 is strongly expressed in brown adipocytes and it determines the number of new lipid droplets formed in these cells.Citation114 FIT2 KO mice show few but larger lipid droplets in interscapular BAT without changes in cellular TG levels.Citation115 Thus, FIT2 is not essential for lipid droplet formation but it is required for normal storage of TG in vivo.
CIDEA is one of the 3 members of cell death-inducing DFF45-like effector (CIDE) family of proteins, which has emerged as an important regulator for various aspects of metabolism.Citation116 CIDEA is highly expressed in lipid droplet membranes and mitochondria of brown adipocytes. It is involved in the browning phenomenon and it is considered as a BAT differentiation marker.Citation117,118 CIDEA plays an inhibitory role during thermogenesis because it negatively modulates the activity of UCP1, being the first protein known to interact directly with an uncoupler protein.Citation119-121 Moreover, CIDEA mRNA and protein are down-regulated after cold exposureCitation121 and CIDEA-null mice are resistant to diet-induced obesity.Citation118
FSP27 also belongs to the CIDE family, and it is overexpressed during adipogenesis in BAT. It has been proposed as a novel lipid droplet protein that promotes TG storage and inhibits lipolysis, playing a key role in body energy homeostasis.Citation122,123 FSP27 interacts directly with another lipid droplet protein, perilipin 1, which is involved in lipolysis by indirect activation of the adipose triglyceride lipase (ATGL) at the lipid droplet surface.Citation123 Furthermore, FSP27 KO mice have larger lipid droplets and higher TG serum levels.Citation124
Thus, the above-mentioned proteins involved in FA storage contribute to the multilocular phenotype of brown adipocytes. Lipid droplets prevent lipotoxicity and provide FAs as substrates for mitochondrial thermogenesis. Therefore, the regulation and function of these proteins might be a target for enhancing BAT activity.
Fatty acids as a source of energy
Lipolysis
Intracellular lipolysis is the catabolic process that allows cells to obtain FAs and glycerol from the breakdown of TG stored in lipid droplets. Cells use these FAs and glycerol endogenously in times of metabolic need with the exception of WAT, which can also export them to circulation so they can reach other tissues in fasting or exercise periods.Citation125 In BAT, lipolysis is vital to its main physiological function, the cold response. To raise body temperature BAT dissipates energy as heat and mobilizes FAs from the breakdown of TGs stored in lipid droplets to mitochondria for thermogenesis.Citation108 Lipolysis can be classified in 2 types depending on the pH-optimum of action of the enzymes involved. Accordingly, there is neutral lipolysis, which relies on 3 key enzymes that work at a pH-optimum of 7, and acid lipolysis that depends on lysosomal degradation of TG by acidic lipases. Next we will focus on neutral lipolysis.
Neutral lipolysis
Neutral lipolysis takes place in the cytosol and it is the result of the action of 3 consecutive lipases that hydrolyze each ester bond of TG to obtain 3 FAs and glycerol. The 3 major lipases are ATGL, hormone sensitive lipase (HSL) and monoacylglycerol lipase (MGL).
ATGL/Desnutrin/calcium-independent phospholipase A2 ζ (iPLA2ζ) was discovered in 2004 by 3 independent laboratories.Citation126-128 It is strongly expressed in both WAT and BAT and performs the first step of TG lipolysis, the hydrolysis of TG into diacylglycerides (DG) and FAs ().Citation129 It exhibits high substrate specificity for TG and it is associated with lipid droplets.Citation126 ATGL regulation is complex and mRNA or protein levels of the enzyme do not always correlate with enzyme activity. This happens because ATGL has strong post translational regulation that often requires accessory proteins.Citation130 CGI-58 (Comparative gene identification-58) is a coactivator of ATGL that is necessary for full hydrolase activity.Citation131 On the other hand, G0S2 (G0/G1 switch gene 2) inactivates ATGL.Citation132 ATGL deficient mice accumulate TGs in all organs and have enlarged fat depots, especially BAT, which also displays defective thermogenesis.Citation133 Moreover, aP2-ATGL overexpressing mice display increased lipolysis and FAO in WAT and increased thermogenesis, resulting in higher energy expenditure and resistance to obesity.Citation134 Microarrays from ATGL KO BAT indicate a decrease in mRNA expression of genes involved in FAO and down-regulation of PPARα target genes.Citation135,136 In addition, a study using ATGL knock down brown adipocytes demonstrated that ATGL is required for the maximal induction of genes involved in FAO and mitochondrial electron transport.Citation137 All together, these results point to the crucial role of lipolysis and its first step, TG hydrolysis by ATGL, in thermogenesis.
Figure 2. Neutral lipolysis players and regulation in BAT. Neutral lipolysis allows cells to obtain 3 free fatty acids (FFAs) and glycerol from the hydrolysis of triglycerides (TG). Three enzymes control this process: adipose triglyceride lipase (ATGL), which hydrolyzes TG into diacylglycerol (DG), hormone sensitive lipase (HSL), which has high affinity for DG and converts them into monoacylglycerols (MG) and monoacylglycerol lipase (MGL), which finalizes the hydrolysis of MG into glycerol and FFA that are used as a fuel for thermogenesis. In basal state ATGL is inhibited by G0/G1 switch gene 2 (G0S2) and ATGL co-activator comparative gene identification-58 (CGI-58) is kidnapped by perilipin. In addition, HSL is located in the cytosol and thus unable to reach its substrates. Upon β3-adrenergic stimulation, adenyl cyclase (AC) increases cAMP levels that activate protein kinase A (PKA), which phosphorylates HSL promoting its translocation to the membrane of lipid droplets (LD). PKA also phosphorylates perilipin, which releases CGI-58 that can then fully activate ATGL. Phosphorylated perilipin also enhances HSL activity. On the other hand, insulin stimulation, through protein kinase B (PKB) activates phosphodiesterase 3B (PDE3B) which converts cAMP into AMP decreasing PKA activation and its lipolytic action. Figure insert: mouse models of the enzymes involved in neutral lipolysis. ATGL KO mice accumulate TGs and have enlarged BAT, which displays defective thermogenesis.Citation133 aP2-ATGL overexpressing mice show a reduction in TGs and increased thermogenesis.Citation134 HSL KO mice accumulate TGs and specially large amounts of DG leading to an enlarged BAT.Citation142
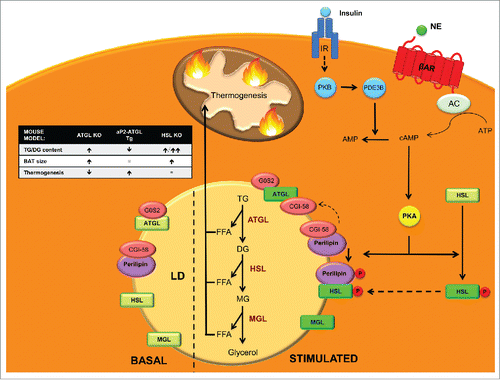
HSL performs the second step in TG lipolysis, hydrolyzing DG into MG and FAs ().Citation138 Similarly to ATGL, HSL mRNA and protein expression are highest in WAT and BAT.Citation139,140 Although DG are its preferred substrate, HSL can also hydrolyze TG, cholesterol esters, MGs and retinyl esters.Citation141 Before ATGL was known, HSL was believed to be the rate-limiting enzyme for TG hydrolysis. However, HSL-/- mice efficiently hydrolyze TG and accumulate large amounts of DG, indicating that, in vivo, HSL has a more important role as a DG than as a TG hydrolase.Citation142 Activation of HSL occurs in 2 steps: protein phosphorylation and binding to perilipins.Citation143 HSL has 5 putative phosphorylation sites and one of the most widely studied kinases that regulate its activity is cAMP-dependent Protein Kinase (PKA). Other kinases include AMPK, extracellular signal-regulated kinase (ERK), glycogen synthase kinase-4 and Ca2+/calmoduline dependent kinase II.Citation130
Glucagon, adrenaline or β3-adrenergic agonist stimulation, through adenyl cyclase (AC) activation increases cyclic AMP (cAMP) levels within the cell. This activates PKA that in turn phosphorylates HSL on serines 659 and 660, promoting its translocation from the cytoplasm to lipid droplets, where it acts on its substrates ().Citation144 Perilipins have an important role mediating this translocation, and PKA-mediated phosphorylation of perilipin is necessary for HSL translocation to lipid droplets and full induction of HSL activity. On the other hand, insulin activates cAMP phosphodiesterase, promoting cAMP hydrolysis, lowering PKA activity, HSL activation and lipolysis.Citation144 Mice lacking HSL display normal thermogenesisCitation145 and are not cold sensitive despite a lipolytic defect that results in brown adipocyte hypertrophy due to TG and DG accumulation. Apparently, during HSL deficiency sufficient amounts of FAs that are not HSL-dependent are mobilized for mitochondrial heat production. Later work on HSL null mice established that HSL KO mice are resistant to high-fat diet (HFD) effects due to an increase in energy expenditure.Citation146 This was linked to increased UCP1 and carnitine palmitoyltransferase (CPT) 1 expression levels in white adipocytes as well as an increase in white adipocyte mitochondrial size (see section 4.2.1 for more information). White adipocytes had increased basal O2 consumption and increased uncoupling. In addition, HSL is required to sustain normal expression levels of retinoblastoma protein (pRb) and receptor interacting protein 140 (RIP140), which both promote differentiation into the white, rather than the brown, adipocyte lineage. Thus, HSL may be involved in the determination of white versus brown adipocytes during adipocyte differentiation.Citation146
MGL specifically hydrolyzes MG derived from intracellular and extracellular TG hydrolysis and phospholipid hydrolysis into FAs and glycerol, culminating the lipolysis process (). It is ubiquitously expressed in tissues and localizes in cell membrane, cytoplasm and lipid droplets.Citation147 MGL has been implicated in the degradation of the bioactive MG 2-arachidonoyl glycerol, which is a potent endogenous agonist of cannabinoid receptors.Citation148
Upon lipolysis stimulation the most important mechanisms regulating lipolysis are: 1) Activation of ATGL by CGI-58; and 2) PKA-mediated phosphorylation of HSL and perilipin 1 (). In basal state, CGI-58 is bound and kidnaped by perilipin 1, and thus unable to activate ATGL. In addition, HSL is located in the cytosol far from its substrates. Upon hormonal stimulation, PKA phosphorylates HSL, promoting its translocation to the lipid droplet surface, where it hydrolyzes its substrates.Citation149,150 In addition, PKA phosphorylates perilipin 1, liberating CGI-58, which is then available to bind and activate ATGL.Citation151
The final result of lipolysis is the provision of energy to the cell in the form of FFAs and glycerol. Elevated levels of FFAs can be toxic for cells.Citation8 Brown adipocytes can prevent this lipotoxicity by matching this increment in FFAs with an increase in oxidative capacity. β3-adrenergic stimulation triggers lipase activation, resulting in a rise of lipolytic products that act as ligands of PPARα and PPARδ. PPAR activation promotes expression of oxidative genes such as UCP1 or PGC1α and as a result expands the oxidative capacity to match FA supply.Citation137 These findings highlight the importance of coupling lipolysis with increased oxidative capacity, which ultimately depends on the uptake of FAs to the mitochondria for FAO by carnitine acyltransferases.
Fatty acid oxidation
Consistent with its physiological role, BAT presents one of the highest FAO rates in the body.Citation152 Most of the oxidation of longchain fatty acids (LCFAs) to acetyl-CoA takes place in the mitochondrial matrix, although peroxisomal FAO has also been implicated in thermogenesis.
Mitochondrial fatty acid oxidation
Traditionally, research on the regulation of BAT thermogenesis has focused on the central role of UCP1 in maximizing rates of proton leak and heat production. In fact, FAs and HFD activate UCP1 and diet-induced thermogenesis.Citation153-155 However, studies by Yu et al.Citation156 support the hypothesis that additional systems and genes cooperate in the thermogenic system. These authors demonstrated that cold induces simultaneous FA synthesis and FAO in murine BAT. Similar conclusions were obtained by Mottillo et al.Citation96 In addition, it has recently been reported in primary brown adipocyte culture that intracellular FA levels are critical for thermogenesisCitation157 and that in rodents maximal BAT thermogenesis relies on the levels of LCFA pool, which activates entry to the mitochondria.Citation158 Acyl-CoA synthetase-1 (ACSL) mediates the conversion of FAs to acyl-CoA and specifically directs them toward mitochondrial FAO via the CPT1 system (). Experiments with ACSL KO mice showed that ACSL is required for cold thermogenesis.Citation159
Figure 3. Mitochondrial and peroxisomal fatty acid oxidation. Transport of long-chain fatty acids (LCFAs) from the cytosol to the mitochondrial matrix for FAO involves the activation to acyl-CoA by acyl-CoA synthetase-1 (ACSL), conversion of LCFA-CoA to LCFA-carnitines by carnitine palmitoyltransferase (CPT) 1, translocation across the inner mitochondrial membrane by the carnitine/acylcarnitine translocase (CACT) and reconversion to LCFA-CoA by CPT2. These acyl-CoAs are β-oxidized and render acetyl-CoA. The entry of acetyl-CoA to the tricarboxylic acid cycle generates NADH and FADH. These cofactors transfer the electrons to the electron transport chain, where the protons are transported to the mitochondrial intermembrane space to generate energy as ATP. UCP1 dissipates the proton gradient, releasing energy as heat. Very long chain fatty acids (VLFA) enter the peroxisome to be shortened by peroxisomal FAO. Shortened acyl-CoAs and acetyl-CoA are transported to the mitochondria to be completely oxidized.
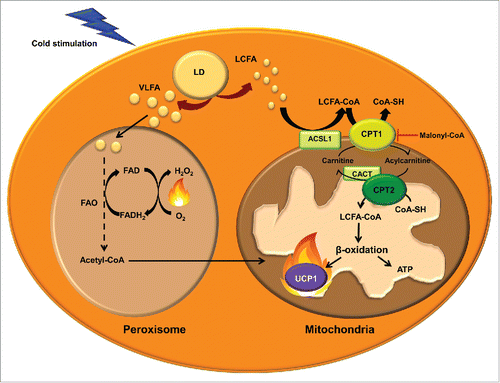
The CPT system permits the entry of LCFAs into the mitochondrial matrix, where they can undergo FAO. The first component of the system, CPT1 is located on the mitochondrial outer membrane (). This enzyme catalyzes the trans esterification of a fatty acyl group from CoA to carnitine, which is considered the rate-limiting step in the regulation of mitochondrial FAO of FAs.Citation160,161 Acylcarnitines are shuttled to the mitochondrial matrix by the transporter carnitine/acylcarnitine translocase (CACT). Once inside the mitochondria, acylcarnitines are converted to acyl-CoA by CPT2, located in the inner mitochondrial membrane, and can thus enter the FAO cycle. There are 3 isoforms of CPT1, denoted CPT1A,Citation162,163 CPT1BCitation164 and CPT1C.Citation165 They differ in their sequence, tissue distribution, intracellular location, kinetics and malonyl-CoA sensitivity. CPT1B is strongly expressed in BAT, skeletal muscle, heart, testis and, in some species, in WAT (human, rat and hamster)Citation108,166 while CPT1A is predominant in other tissues such as liver, kidney, lung, ovary, spleen, brain, intestine, mouse WAT and pancreas. CPT1C appears to be expressed exclusively in the neurons and testis. While CPT1A and B are located on the mitochondrial outer membrane and both isoforms are involved in the regulation of the flux of FAs into the mitochondria, CPT1C has been found on the ER membraneCitation167 and its function seems to be related with ceramide metabolism rather than FAO.Citation168,169
CPT1A and B isoforms have high sequence similarity but show important kinetic differences. In particular, they differ in their affinities for the substrate carnitine and their physiological inhibitor malonyl-CoA,Citation170 which is synthesized from acetyl-CoA by ACC and is degraded by malonyl-CoA decarboxylase.Citation171 CPT1B has higher affinity for the inhibitor malonyl-CoA and lower affinity for carnitine than CPT1A.Citation172,173 Doh et al.Citation152 examined the relation between the long-chain FAO rate and the CPT1A and CPT1B activity in different tissues. They found that all the tissues containing CPT1B showed a strong positive correlation between palmitate oxidation rate and the CPT1 activity and, among the tissues with CPT1B (heart, red and white gastrocnemius and BAT), BAT showed the highest palmitate oxidation and CPT1 activity. In addition, mice lacking CPT1B die during cold-exposure as a result of their inability to perform thermogenesis.Citation174 These observations indicate that CPT1B may play an important role in enhancing BAT thermogenesis. Indeed, inhibitors of CPT1 also inhibit mitochondrial respiration driven by LCFA in murine BAT.Citation158 Interestingly, BAT of diabetic rats showed decreased CPT1 activity and FAO.Citation175 Further, a recent study in adipose CPT2 KO mice demonstrated that FAO is required for both acute cold adaptation and the induction of thermogenic genes in BAT.Citation176 Taking into account the effect that this and other mitochondrial FAO alterationsCitation177 produce in thermogenesis and cold intolerance, it could be an appealing strategy to enhance FAO in BAT mitochondria to increase energy expenditure and fight against obesity-induced metabolic disorders. Studies enhancing FAO by CPT1 overexpression in the context of obesity have shown a decrease in TG content and an improvement in insulin sensitivity.Citation178-188 These results indicate that activation of FAO may provide the basis for the development of novel treatment options for obesity.Citation189
Peroxisomal fatty acid oxidation
Oxidation of very long chain FAs preferentially occurs in peroxisomes rather than in mitochondria ().Citation190 However, FAO in peroxisomes is not carried to completion. Since peroxisomes, unlike mitochondria, lack a citric-acid cycle and respiratory chain, shorter FAs can be shuttled to the mitochondria to be oxidized. During cold exposure peroxisomal FAO is also activated in BAT.Citation191 Furthermore, catalase protein, a marker of the quantity of peroxisomes, is dramatically increased in rat BAT under cold exposure.Citation192 However, the contribution of peroxisomal FAO to thermogenesis is not well established. Acetyl-CoAs produced by peroxisomal FAO may enter the mitochondria to fuel UCP1-mediated thermogenesis. Alternatively, a recent review by Lodhi et al.Citation193 suggests that peroxisomal FAO may contribute to adaptive thermogenesis independently of UCP1 by the generation of heat instead of ATP. Peroxisomes do not have a respiratory chain and the electrons from FADH2, obtained in the first step of peroxisomal FAO, are transferred by the flavoprotein acyl-CoA oxidase directly to O2 producing H2O2, and energy is released as heat.
In summary, mitochondrial and peroxisomal FAO are necessary for thermogenesis in BAT, and enhancing these catabolic processes is an unexplored strategy in our fight against the current obesity epidemic.
Future directions: BAT fat-burning power as a potential therapy against obesity
Despite the considerable current effort made worldwide, the prevalence of obesity and associated metabolic diseases is rising exponentially, jointly with their healthcare costs. The first line therapy is based on behavioral modifications, including healthy eating and exercise. However, this meets limited success when it comes to long-term maintenance of weight loss. Bariatric surgery achieves a sustained weight loss over the years, but its cost and associated dangers reduce its clinical indication to morbidly obese patients.Citation194 Moreover, the endocrine effects of bariatric surgery seem to be more important than the mechanically induced food restriction, which has led many researchers to assess obesity treatments based on the endocrine modifications derived from it.Citation195 Interestingly, bariatric surgery also leads to alterations in the microbiome.Citation196
Even though the list of potential anti-obesity drugs is increasing, the approval of new anti-obesity drugs has met relatively limited success. This is due to the history of withdrawals of anti-obesity drugs from the market due to serious adverse effects (i.e., dinitrophenol, fenfluramine, dexfenfluramine, phenylpropanolamine, sibutramine and rimonabant).Citation197,198 This has led US Food and Drug Administration (FDA) and European Medicines Agency (EMA) to make it harder for pharmaceutical corporations to market new anti-obesity drugs, especially in the case of the European regulator.Citation199 Today, the lipase inhibitor orlistat is approved by both FDA and EMA but it has shown limited long-term effectiveness.Citation200 In the US, the serotonergic lorcaserin is also approved,Citation201,202 but its European marketing authorization application has been withdrawn because of the lack of evidence regarding safety in tumorogenesis in long-term use.Citation203 Liraglutide, a previously approved antidiabetic drug, has recently been approved by both regulators for an anti-obesity indication.Citation204,205 Nonetheless, the fixed-dose combination of bupropion/naltrexone, which is described to act in the central nervous system (CNS) by increasing POMC neuron activity, has obtained marketing approval as an anti-obesity drug by the FDA,Citation206 but the EMA seems to be much more conservative regarding the approval of weight-management drug with effect on the CNS.Citation207
The mechanisms of action of obesity-management drugs are classified into 3 groups:Citation208 1) centrally acting medications impairing dietary intake (including bupropion/naltrexone and lorcaserin); 2) medications that act peripherally to impair dietary absorption (e.g., orlistat); and 3) medications that increase energy expenditure, whose effect is mediated by CNS. We propose that the increase in energy expenditure is a promising way to manage obesity, but only if this could be achieved via a direct effect on peripheral tissues without involving the CNS. Thus, some of the collateral effects, which caused other drugs to be withdrawn, would be overcome. Here we highlight an increase in FAO as a potential approach to enhance energy expenditure in peripheral tissues. Several studies enhancing FAO by CPT1 overexpression in the context of obesity have shown an improved metabolic phenotype.Citation178-187 Thus, an increasing body of evidence highlights FAO activation as a potential target to develop novel anti-obesity strategies.
The pathogenesis of obesity is multifactorial and complex. However, the rediscovery of active BAT in adult humans and its relevance in metabolism has put a spotlight on this tissue as a potential target for therapies against obesity and metabolic diseases. A large number of activators of thermogenesis are increasingly being identified. This year, the β3-adrenergic receptor agonist mirabegron, the first compound able to stimulate human BAT thermogenesis and to increase resting metabolic rate, has been described.Citation59 Although further studies are needed to explore the specificity of its mechanism of action and potential adverse effects, mirabegron provides the first evidence of human BAT thermogenesis stimulation.
As a controller of thermogenesis BAT is a good modulator of triglyceridemia, an important consumer of glucose and the major plasma lipid-clearing organ in rodents. Furthermore, diabetes has been shown to decrease CPT1 activity and FAO in rat BAT.Citation175 Thus, strategies designed to enhance the fat-burning power of BAT and to increase lipid mobilization and oxidation could be very useful in the treatment of obesity and associated pathologies. Traditionally, most research has focused on the activation of BAT thermogenesis through UCP1. However, recent studies have shown that cold stimulates both FA synthesis and FAO in murine BAT.Citation96,156 Moreover, BAT peroxisomal FAO may generate heat independently of UCP1.Citation193 BAT transplantation is another strategy proving BAT's lipid-burning capacity in obesity. BAT transplantation to HFD-induced obese mice has shown a beneficial effect improving whole-body energy metabolism by increasing FAO-related genes such as PPARα, PGC1α, CPT1B and UCP1 in endogenous BAT.Citation209 Although the present review is focused on obesity, it is noteworthy to mention the role of BAT in atherosclerosis. Data showing both a positive and negative effect of BAT activation in the development of atherosclerosis have been reported.Citation210,211 On the one hand, Dong et al.Citation210 showed that sustained cold exposure accelerated the atherosclerotic plaque development by increasing plasma levels of small low-density lipoproteins (LDL) in apolipoprotein E (ApoE) and LDL receptor (LDLr) deficient atherosclerosis mouse models. On the other hand, Berbee et al.Citation211 reported that APOE*3-Leiden.CETP mice (a model for human-like lipoprotein metabolism) treated with Western diet, to induce hyperlipidaemia and atherosclerosis, plus CL316243 (a β3-adrenergic receptor agonist) had fewer atherosclerotic lesions. In this case BAT activation lead to enhanced uptake of FAs from TG-rich lipoproteins into BAT and increased hepatic clearance of cholesterol-enriched remnants and lower plasma cholesterol levels. The apparent opposite effects between the 2 studies could be explained by the different mouse model used. ApoE and LDL receptor are essential for hepatic clearance of TG-rich lipoprotein remnants. Thus, this pathway is blocked in ApoE (-/-) or LDLr (-/-) mice but not in APOE*3-Leiden.CETP mice. In fact, the antiatherogenic effect seen by Berbee et al. was blunted in ApoE (-/-) or LDLr (-/-) mice. Importantly, mice treated with the β3-receptor agonist lost weight and had increased FAO. This indicates that the beneficial effect of BAT activation on atherosclerosis could be the consequence of decreased obesity and enhanced FAO shedding light into FAO as a potential target to fight against obesity-induced metabolic disorders such as atherosclerosis.
At least 3 questions still need to be answered before increased BAT FAO can become an effective approach for obesity therapy. First, it is not known whether FAO enhancement might reach a limit in BAT, in which thermogenesis is tightly adjusted to the environmental temperature. Second, since increasing flux through FAO would only make sense together with a corresponding enhancement of energy demand,Citation212 the physiological relevance of this strategy might be questioned if the individual is at thermoneutrality. Third, secondary effects of BAT pharmacological activation may include excessive body temperature or increased food intake as a compensatory effect to re-establish energy balance. Increased BAT FAO may augment mitochondrial burning capacity through an increase in the number of mitochondria and/or the increased expression of UCPs, and thus dissipate the excess of energy as heat and ATP. These could well alleviate the mitochondrial pressure found in lipid overload states.
In summary, an increase in FAO and BAT mass and/or activity could indeed be one of the underlying protective mechanisms against obesity-induced metabolic abnormalities. Although more research is needed, we strongly believe that enhancing BAT thermogenic power through increased FAO may be available in the near future as a therapy to treat obesity and its associated severe diseases.
Abbreviations
| AC | = | adenyl cyclase |
| ACC | = | acetyl-CoA carboxylase |
| AMPK | = | AMP-dependent protein kinase |
| ATG | = | autophagy-related protein |
| ATGL | = | adipose triglyceride lipase |
| BAT | = | brown adipose tissue |
| BMP8b | = | bone morphogenetic protein 8b |
| cAMP | = | cyclic AMP |
| CIDEA | = | cell death-inducing DNA fragmentation factor-α-like effector A |
| CGI-58 | = | comparative gene identification-58 |
| CNS | = | central nervous system |
| CPT | = | carnitine palmitoyltransferase |
| DG | = | diacylglycerol |
| DIO2 | = | type 2 iodothyronine deiodinase |
| ELOVL | = | elongation of very long chain FA |
| ER | = | endoplasmic reticulum |
| FA | = | fatty acid |
| FAO | = | fatty acid oxidation |
| FFA | = | free fatty acids |
| FGF21 | = | fibroblast growth factor 21 |
| G0S2 | = | G0/G1 switch gene 2 |
| GPCRs | = | G-protein-coupled receptors |
| HFD | = | high-fat diet |
| HSL | = | hormone-sensitive lipase |
| IGF-1 | = | insulin-like growth factor I |
| IL-1β | = | interleukin-1β |
| IL-6 | = | interleukin-6 |
| KO | = | knockout |
| LAL | = | lysosomal acid lipase |
| MEFs | = | primary mouse fibroblasts |
| MG | = | monoacylglycerol |
| MGL | = | monoacylglycerol lipase |
| Myf5+ | = | myogenic factor 5-positive |
| iPLA2ζ | = | calcium-independent phospholipase A2 ζ |
| PGC1α | = | peroxisome proliferator activated receptor gamma coactivator 1 alpha |
| PKA | = | protein kinase A |
| PKB | = | protein kinase B |
| PRDM16 | = | PR domain-containing 16 |
| pRb | = | retinoblastoma protein |
| RIP140 | = | receptor interacting protein 140 |
| TG | = | triglyceride |
| TNFα | = | tumor necrosis factor α |
| UCP1 | = | uncoupling protein-1 |
| WAT | = | white adipose tissue |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank Robin Rycroft for valuable assistance in the preparation of the English manuscript.
Funding
This work was supported by the Ministry of Spain (SAF2013-45887-R to LH, SAF2014-52223-C2-1-R to DS (grant cofounded by Fondos Europeos de Desarrollo Regional de la Unión Europea (FEDER)) and doctoral fellowship to JFM), by the Centro de Investigación Biomédica en Red Fisiopatología de la Obesidad y la Nutrición (CIBEROBN) (Grant CB06/03/0001 to DS), by Generalitat de Catalunya (2014SGR465 to DS), and by the European Foundation for the Study of Diabetes (EFSD)/Janssen-Rising Star and L'Oréal-UNESCO “For Women in Science” research fellowships to LH. MW is a recipient of the Ciência sem Fronteiras-CNPq fellowship (237976/2012-9).
References
- World Health Organization. Obesity and overweight. Available from: http://www.who.int/mediacentre/factsheets/fs311/en/ (accessed 1 October 2015)
- Mathis D. Immunological goings-on in visceral adipose tissue. Cell Metab 2013; 17:851-9; PMID:23747244; http://dx.doi.org/10.1016/j.cmet.2013.05.008
- Samuel VT, Shulman GI. Mechanisms for insulin resistance: common threads and missing links. Cell 2012; 148:852-71; PMID:22385956; http://dx.doi.org/10.1016/j.cell.2012.02.017
- Shoelson SE, Herrero L, Naaz A. Obesity, inflammation, and insulin resistance. Gastroenterology 2007; 132:2169-80; PMID:17498510; http://dx.doi.org/10.1053/j.gastro.2007.03.059
- Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 2010; 140:900-17; PMID:20303879; http://dx.doi.org/10.1016/j.cell.2010.02.034
- Hosogai N, Fukuhara A, Oshima K, Miyata Y, Tanaka S, Segawa K, Furukawa S, Tochino Y, Komuro R, Matsuda M, et al. Adipose tissue hypoxia in obesity and its impact on adipocytokine dysregulation. Diabetes 2007; 56:901-11; PMID:17395738; http://dx.doi.org/10.2337/db06-0911
- Patti M-E, Corvera S. The role of mitochondria in the pathogenesis of type 2 diabetes. Endocr Rev 2010; 31:364-95; PMID:20156986; http://dx.doi.org/10.1210/er.2009-0027
- Virtue S, Vidal-Puig A. Adipose tissue expandability, lipotoxicity and the Metabolic Syndrome–an allostatic perspective. Biochim Biophys Acta 2010; 1801:338-49; PMID:20056169; http://dx.doi.org/10.1016/j.bbalip.2009.12.006
- Sun K, Kusminski CM, Scherer PE. Adipose tissue remodeling and obesity. J Clin Invest 2011; 121:2094-101; PMID:21633177; http://dx.doi.org/10.1172/JCI45887
- Sun K, Wernstedt Asterholm I, Kusminski CM, Bueno AC, Wang ZV, Pollard JW, Brekken RA, Scherer PE. Dichotomous effects of VEGF-A on adipose tissue dysfunction. Proc Natl Acad Sci U S A 2012; 109:5874-9; PMID:22451920; http://dx.doi.org/10.1073/pnas.1200447109
- Hany TF, Gharehpapagh E, Kamel EM, Buck A, Himms-Hagen J, Von Schulthess GK. Brown adipose tissue: a factor to consider in symmetrical tracer uptake in the neck and upper chest region. Eur J Nucl Med 2002; 29:1393-8; PMID:12271425; http://dx.doi.org/10.1007/s00259-002-0902-6
- Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab 2007; 293:E444-52; PMID:17473055; http://dx.doi.org/10.1152/ajpendo.00691.2006
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med 2009; 360:1509-17; PMID:19357406; http://dx.doi.org/10.1056/NEJMoa0810780
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, et al. Functional brown adipose tissue in healthy adults. N Engl J Med 2009; 360:1518-25; PMID:19357407; http://dx.doi.org/10.1056/NEJMoa0808949
- Zingaretti MC, Crosta F, Vitali A, Guerrieri M, Frontini A, Cannon B, Nedergaard J, Cinti S. The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue. Faseb J 2009; 23:3113-20; PMID:19417078; http://dx.doi.org/10.1096/fj.09-133546
- Saito M, Okamatsu-Ogura Y, Matsushita M, Watanabe K, Yoneshiro T, Nio-Kobayashi J, Iwanaga T, Miyagawa M, Kameya T, Nakada K, et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes 2009; 58:1526-31; PMID:19401428; http://dx.doi.org/10.2337/db09-0530
- Van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ. Cold-activated brown adipose tissue in healthy men. N Engl J Med 2009; 360:1500-8; PMID:19357405; http://dx.doi.org/10.1056/NEJMoa0808718
- Mailloux RJ, Harper M-E. Uncoupling proteins and the control of mitochondrial reactive oxygen species production. Free Radic Biol Med 2011; 51:1106-15; PMID:21762777; http://dx.doi.org/10.1016/j.freeradbiomed.2011.06.022
- Frontini A, Cinti S. Distribution and development of brown adipocytes in the murine and human adipose organ. Cell Metab 2010; 11:253-6; PMID:20374956; http://dx.doi.org/10.1016/j.cmet.2010.03.004
- Cypess AM, White AP, Vernochet C, Schulz TJ, Xue R, Sass CA, Huang TL, Roberts-Toler C, Weiner LS, Sze C, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat Med 2013; 19:635-9; PMID:23603815; http://dx.doi.org/10.1038/nm.3112
- Cinti S. Between brown and white: novel aspects of adipocyte differentiation. Ann Med 2011; 43:104-15; PMID:21254898; http://dx.doi.org/10.3109/07853890.2010.535557
- Tseng YH, Cypess AM, Kahn CR. Cellular bioenergetics as a target for obesity therapy. Nat Rev Drug Discov 2010; 9:465-82; PMID:20514071; http://dx.doi.org/10.1038/nrd3138
- Xue R, Lynes MD, Dreyfuss JM, Shamsi F, Schulz TJ, Zhang H, Huang TL, Townsend KL, Li Y, Takahashi H, et al. Clonal analyses and gene profiling identify genetic biomarkers of the thermogenic potential of human brown and white preadipocytes. Nat Med 2015; 21:760-8; PMID:26076036; http://dx.doi.org/10.1038/nm.3881
- Jespersen NZ, Larsen TJ, Peijs L, Daugaard S, Homøe P, Loft A, de Jong J, Mathur N, Cannon B, Nedergaard J, et al. A classical brown adipose tissue mRNA signature partly overlaps with brite in the supraclavicular region of adult humans. Cell Metab 2013; 17:798-805; PMID:23663743; http://dx.doi.org/10.1016/j.cmet.2013.04.011
- Lidell ME, Betz MJ, Dahlqvist Leinhard O, Heglind M, Elander L, Slawik M, Mussack T, Nilsson D, Romu T, Nuutila P, et al. Evidence for two types of brown adipose tissue in humans. Nat Med 2013; 19:631-4; PMID:23603813; http://dx.doi.org/10.1038/nm.3017
- Sharp LZ, Shinoda K, Ohno H, Scheel DW, Tomoda E, Ruiz L, Hu H, Wang L, Pavlova Z, Gilsanz V, et al. Human BAT possesses molecular signatures that resemble beige/brite cells. PLoS One 2012; 7:e49452; PMID:23166672; http://dx.doi.org/10.1371/journal.pone.0049452
- Wu J, Boström P, Sparks LM, Ye L, Choi JH, Giang A-H, Khandekar M, Virtanen KA, Nuutila P, Schaart G, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 2012; 150:366-76; PMID:22796012; http://dx.doi.org/10.1016/j.cell.2012.05.016
- Villarroya J, Cereijo R, Villarroya F. An endocrine role for brown adipose tissue? Am J Physiol Endocrinol Metab 2013; 305:E567-72; PMID:23839524; http://dx.doi.org/10.1152/ajpendo.00250.2013
- Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell 2007; 131:242-56; PMID:17956727; http://dx.doi.org/10.1016/j.cell.2007.10.004
- Ortega FJ, Jilkova ZM, Moreno-Navarrete JM, Pavelka S, Rodriguez-Hermosa JI, Kopeck Ygrave J, Fernandez-Real JM. Type I iodothyronine 5'-deiodinase mRNA and activity is increased in adipose tissue of obese subjects. Int J Obes 2011; 36:320-4; PMID:21610697; http://dx.doi.org/10.1038/ijo.2011.101
- Peirce V, Carobbio S, Vidal-Puig A. The different shades of fat. Nature 2014; 510:76-83; PMID:24899307; http://dx.doi.org/10.1038/nature13477
- Waldén TB, Hansen IR, Timmons JA, Cannon B, Nedergaard J. Recruited vs. nonrecruited molecular signatures of brown, “brite,” and white adipose tissues. Am J Physiol Endocrinol Metab 2012; 302:E19-31; PMID:21828341; http://dx.doi.org/10.1152/ajpendo.00249.2011
- Bartelt A, Heeren J. Adipose tissue browning and metabolic health. Nat Rev Endocrinol 2014; 10:24-36; PMID:24146030; http://dx.doi.org/10.1038/nrendo.2013.204
- Rosenwald M, Wolfrum C. The origin and definition of brite versus white and classical brown adipocytes. Adipocyte 2014; 3:4-9; PMID:24575363; http://dx.doi.org/10.4161/adip.26232
- Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, Hamilton DL, Gimeno RE, Wahlestedt C, Baar K, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci U S A 2007; 104:4401-6; PMID:17360536; http://dx.doi.org/10.1073/pnas.0610615104
- Sanchez-Gurmaches J, Hung C-M, Sparks CA, Tang Y, Li H, Guertin DA. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metab 2012; 16:348-62; PMID:22940198; http://dx.doi.org/10.1016/j.cmet.2012.08.003
- Tseng Y-H, Cypess AM, Kahn CR. Cellular bioenergetics as a target for obesity therapy. Nat Rev Drug Discov 2010; 9:465-82; PMID:20514071; http://dx.doi.org/10.1038/nrd3138
- Kajimura S, Spiegelman BM, Seale P. Brown and beige fat: physiological roles beyond heat generation. Cell Metab 2015; 22:546-59; PMID:26445512; http://dx.doi.org/10.1016/j.cmet.2015.09.007
- Zhou Q, Melton DA. Extreme makeover: converting one cell into another. Cell Stem Cell 2008; 3:382-8; PMID:18940730; http://dx.doi.org/10.1016/j.stem.2008.09.015
- Barbatelli G, Murano I, Madsen L, Hao Q, Jimenez M, Kristiansen K, Giacobino JP, De Matteis R, Cinti S. The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation. Am J Physiol Endocrinol Metab 2010; 298:E1244-53; PMID:20354155; http://dx.doi.org/10.1152/ajpendo.00600.2009
- Barbatelli G, Morroni M, Vinesi P, Cinti S, Michetti F. S-100 protein in rat brown adipose tissue under different functional conditions: a morphological, immunocytochemical, and immunochemical study. Exp Cell Res 1993; 208:226-31; PMID:8359217; http://dx.doi.org/10.1006/excr.1993.1241
- Himms-Hagen J, Melnyk A, Zingaretti MC, Ceresi E, Barbatelli G, Cinti S. Multilocular fat cells in WAT of CL-316243-treated rats derive directly from white adipocytes. Am J Physiol Cell Physiol 2000; 279:C670-81; PMID:10942717
- Rosenwald M, Perdikari A, Rülicke T, Wolfrum C. Bi-directional interconversion of brite and white adipocytes. Nat Cell Biol 2013; 15:659-67; PMID:23624403; http://dx.doi.org/10.1038/ncb2740
- Lee Y-H, Petkova AP, Mottillo EP, Granneman JG. In vivo identification of bipotential adipocyte progenitors recruited by β3-adrenoceptor activation and high-fat feeding. Cell Metab 2012; 15:480-91; PMID:22482730; http://dx.doi.org/10.1016/j.cmet.2012.03.009]
- Schulz TJ, Huang P, Huang TL, Xue R, McDougall LE, Townsend KL, Cypess AM, Mishina Y, Gussoni E, Tseng Y-H. Brown-fat paucity due to impaired BMP signalling induces compensatory browning of white fat. Nature 2013; 495:379-83; PMID:23485971; http://dx.doi.org/10.1038/nature11943
- Tsoli M, Moore M, Burg D, Painter A, Taylor R, Lockie SH, Turner N, Warren A, Cooney G, Oldfield B, et al. Activation of thermogenesis in brown adipose tissue and dysregulated lipid metabolism associated with cancer cachexia in mice. Cancer Res 2012; 72:4372-82; PMID:22719069; http://dx.doi.org/10.1158/0008-5472.CAN-11-3536
- Vijgen GHEJ, Bouvy ND, Smidt M, Kooreman L, Schaart G, van Marken Lichtenbelt W. Hibernoma with metabolic impact? BMJ Case Rep 2012; 2012:bcr2012006325-bcr2012006325; PMID:22914232
- Radi Z, Bartholomew P, Elwell M, Vogel WM. Comparative pathophysiology, toxicology, and human cancer risk assessment of pharmaceutical-induced hibernoma. Toxicol Appl Pharmacol 2013; 273:456-63; PMID:24141031; http://dx.doi.org/10.1016/j.taap.2013.10.011
- Nedergaard J, Cannon B. The browning of white adipose tissue: some burning issues. Cell Metab 2014; 20:396-407; PMID:25127354; http://dx.doi.org/10.1016/j.cmet.2014.07.005
- Lo JC, Cinti S, Tu H, Kajimura S, Korde A, Choi JH, Wu J, Zingaretti MC, Rasbach KA, Vind BF, et al. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 2012; 481:463-8; PMID:22237023; http://dx.doi.org/10.1038/nature10777
- Bordicchia M, Liu D, Amri EZ, Ailhaud G, Dessi-Fulgheri P, Zhang C, Takahashi N, Sarzani R, Collins S. Cardiac natriuretic peptides act via p38 MAPK to induce the brown fat thermogenic program in mouse and human adipocytes. J Clin Invest 2012; 122:1022-36; PMID:22307324; http://dx.doi.org/10.1172/JCI59701
- Boon MR, van den Berg SAA, Wang Y, van den Bossche J, Karkampouna S, Bauwens M, De Saint-Hubert M, van der Horst G, Vukicevic S, de Winther MPJ, et al. BMP7 activates brown adipose tissue and reduces diet-induced obesity only at subthermoneutrality. PLoS One 2013; 8:e74083; PMID:24066098; http://dx.doi.org/10.1371/journal.pone.0074083
- Whittle AJ, Carobbio S, Martins L, Slawik M, Hondares E, Vazquez MJ, Morgan D, Csikasz RI, Gallego R, Rodriguez-Cuenca S, et al. BMP8B increases brown adipose tissue thermogenesis through both central and peripheral actions. Cell 2012; 149:871-85; PMID:22579288; http://dx.doi.org/10.1016/j.cell.2012.02.066
- Villarroya F, Vidal-Puig A. Beyond the sympathetic tone: the new brown fat activators. Cell Metab 2013; 17:638-43; PMID:23583169; http://dx.doi.org/10.1016/j.cmet.2013.02.020
- Rao RR, Long JZ, White JP, Svensson KJ, Lou J, Lokurkar I, Jedrychowski MP, Ruas JL, Wrann CD, Lo JC, et al. Meteorin-like is a hormone that regulates immune-adipose interactions to increase beige fat thermogenesis. Cell 2014; 157:1279-91; PMID:24906147; http://dx.doi.org/10.1016/j.cell.2014.03.065
- Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 2006; 439:484-9; PMID:16400329; http://dx.doi.org/10.1038/nature04330
- Gnad T, Scheibler S, von Kügelgen I, Scheele C, Kilić A, Glöde A, Hoffmann LS, Reverte-Salisa L, Horn P, Mutlu S, et al. Adenosine activates brown adipose tissue and recruits beige adipocytes via A2A receptors. Nature 2014; PMID:25317558; PMID:25317558
- Hondares E, Rosell M, Gonzalez FJ, Giralt M, Iglesias R, Villarroya F. Hepatic FGF21 expression is induced at birth via PPARalpha in response to milk intake and contributes to thermogenic activation of neonatal brown fat. Cell Metab 2010; 11:206-12; PMID:20197053; http://dx.doi.org/10.1016/j.cmet.2010.02.001
- Cypess AM, Weiner LS, Roberts-Toler C, Elía EF, Kessler SH, Kahn PA, English J, Chatman K, Trauger SA, Doria A, et al. Activation of human brown adipose tissue by a β3-adrenergic receptor agonist. Cell Metab 2015; 21:33-8; PMID:25565203; http://dx.doi.org/10.1016/j.cmet.2014.12.009
- Wu J, Cohen P, Spiegelman BM. Adaptive thermogenesis in adipocytes: is beige the new brown? Genes Dev 2013; 27:234-50; PMID:23388824; http://dx.doi.org/10.1101/gad.211649.112
- Bonet ML, Oliver P, Palou A. Pharmacological and nutritional agents promoting browning of white adipose tissue. Biochim Biophys Acta - Mol Cell Biol Lipids 2013; 1831:969-85; PMID:23246573; http://dx.doi.org/10.1016/j.bbalip.2012.12.002
- Divakaruni AS, Humphrey DM, Brand MD. Fatty acids change the conformation of uncoupling protein 1 (UCP1). J Biol Chem 2012; 287:36845-53; PMID:22952235; http://dx.doi.org/10.1074/jbc.M112.381780
- Fedorenko A, Lishko PV, Kirichok Y. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell 2012; 151:400-13; PMID:23063128; http://dx.doi.org/10.1016/j.cell.2012.09.010
- Bartelt A, Merkel M, Heeren J. A new, powerful player in lipoprotein metabolism: brown adipose tissue. J Mol Med (Berl) 2012; 90:887-93; PMID:22231746; http://dx.doi.org/10.1007/s00109-012-0858-3
- Bartelt A, Bruns OT, Reimer R, Hohenberg H, Ittrich H, Peldschus K, Kaul MG, Tromsdorf UI, Weller H, Waurisch C, et al. Brown adipose tissue activity controls triglyceride clearance. Nat Med 2011; 17:200-5; PMID:21258337; http://dx.doi.org/10.1038/nm.2297
- Festuccia WT, Blanchard P-G, Deshaies Y. Control of brown adipose tissue glucose and lipid metabolism by PPARγ. Front Endocrinol (Lausanne) 2011; 2:84; PMID:22654830
- Festuccia WT, Blanchard P-G, Turcotte V, Laplante M, Sariahmetoglu M, Brindley DN, Richard D, Deshaies Y. The PPARgamma agonist rosiglitazone enhances rat brown adipose tissue lipogenesis from glucose without altering glucose uptake. Am J Physiol Regul Integr Comp Physiol 2009; 296:R1327-35; PMID:19211718; http://dx.doi.org/10.1152/ajpregu.91012.2008
- Duivenvoorden I, Teusink B, Rensen PC, Romijn JA, Havekes LM, Voshol PJ. Apolipoprotein C3 deficiency results in diet-induced obesity and aggravated insulin resistance in mice. Diabetes 2005; 54:664-71; PMID:15734841; http://dx.doi.org/10.2337/diabetes.54.3.664
- Bartelt A, Weigelt C, Cherradi ML, Niemeier A, Tödter K, Heeren J, Scheja L. Effects of adipocyte lipoprotein lipase on de novo lipogenesis and white adipose tissue browning. Biochim Biophys Acta 2013; 1831:934-42; PMID:23228690; http://dx.doi.org/10.1016/j.bbalip.2012.11.011
- Davies BSJ, Beigneux AP, Barnes RH, Tu Y, Gin P, Weinstein MM, Nobumori C, Nyrén R, Goldberg I, Olivecrona G, et al. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab 2010; 12:42-52; PMID:20620994; http://dx.doi.org/10.1016/j.cmet.2010.04.016
- Weinstein MM, Goulbourne CN, Davies BSJ, Tu Y, Barnes RH, Watkins SM, Davis R, Reue K, Tontonoz P, Beigneux AP, et al. Reciprocal metabolic perturbations in the adipose tissue and liver of GPIHBP1-deficient mice. Arterioscler Thromb Vasc Biol 2012; 32:230-5; PMID:22173228; http://dx.doi.org/10.1161/ATVBAHA.111.241406
- Laplante M, Festuccia WT, Soucy G, Gélinas Y, Lalonde J, Deshaies Y. Involvement of adipose tissues in the early hypolipidemic action of PPARgamma agonism in the rat. Am J Physiol Regul Integr Comp Physiol 2007; 292:R1408-17; PMID:17170230; http://dx.doi.org/10.1152/ajpregu.00761.2006
- Glatz JFC, Luiken JJFP, Bonen A. Membrane fatty acid transporters as regulators of lipid metabolism: implications for metabolic disease. Physiol Rev 2010; 90:367-417; PMID:20086080; http://dx.doi.org/10.1152/physrev.00003.2009
- McArthur MJ, Atshaves BP, Frolov A, Foxworth WD, Kier AB, Schroeder F. Cellular uptake and intracellular trafficking of long chain fatty acids. J Lipid Res 1999; 40:1371-83; PMID:10428973
- Sasaki A, Sivaram P, Goldberg IJ. Lipoprotein lipase binding to adipocytes: evidence for the presence of a heparin-sensitive binding protein. Am J Physiol 1993; 265:E880-8; PMID:8279543
- Baillie AG, Coburn CT, Abumrad NA. Reversible binding of long-chain fatty acids to purified FAT, the adipose CD36 homolog. J Membr Biol 1996; 153:75-81; PMID:8694909; http://dx.doi.org/10.1007/s002329900111
- Jochen AL, Hays J, Mick G. Inhibitory effects of cerulenin on protein palmitoylation and insulin internalization in rat adipocytes. Biochim Biophys Acta 1995; 1259:65-72; PMID:7492617; http://dx.doi.org/10.1016/0005-2760(95)00147-5
- Martin C, Passilly-Degrace P, Gaillard D, Merlin J-F, Chevrot M, Besnard P. The lipid-sensor candidates CD36 and GPR120 are differentially regulated by dietary lipids in mouse taste buds: impact on spontaneous fat preference. PLoS One 2011; 6:e24014.
- Bokor S, Legry V, Meirhaeghe A, Ruiz JR, Mauro B, Widhalm K, Manios Y, Amouyel P, Moreno LA, Molnàr D, et al. Single-nucleotide polymorphism of CD36 locus and obesity in European adolescents. Obesity (Silver Spring) 2010; 18:1398-403; PMID:19893500; http://dx.doi.org/10.1038/oby.2009.412
- Gimeno RE. Fatty acid transport proteins. Curr Opin Lipidol 2007; 18:271-6; PMID:17495600; http://dx.doi.org/10.1097/MOL.0b013e3281338558
- Hagberg CE, Falkevall A, Wang X, Larsson E, Huusko J, Nilsson I, van Meeteren LA, Samen E, Lu L, Vanwildemeersch M, et al. Vascular endothelial growth factor B controls endothelial fatty acid uptake. Nature 2010; 464:917-21; PMID:20228789; http://dx.doi.org/10.1038/nature08945
- Stienstra R, Haim Y, Riahi Y, Netea M, Rudich A, Leibowitz G. Autophagy in adipose tissue and the beta cell: implications for obesity and diabetes. Diabetologia 2014; 57:1505-16; PMID:24795087; http://dx.doi.org/10.1007/s00125-014-3255-3
- Doege H, Stahl A. Protein-mediated fatty acid uptake: novel insights from in vivo models. Physiology (Bethesda) 2006; 21:259-68; PMID:16868315; http://dx.doi.org/10.1152/physiol.00014.2006
- Milligan G, Stoddart LA, Brown AJ. G protein-coupled receptors for free fatty acids. Cell Signal 2006; 18:1360-5; PMID:16716567; http://dx.doi.org/10.1016/j.cellsig.2006.03.011
- Ulven T. Short-chain free fatty acid receptors FFA2/GPR43 and FFA3/GPR41 as new potential therapeutic targets. Front Endocrinol (Lausanne) 2012; 3:111; PMID:23060857
- Rosell M, Kaforou M, Frontini A, Okolo A, Chan Y-W, Nikolopoulou E, Millership S, Fenech ME, MacIntyre D, Turner JO, et al. Brown and white adipose tissues: intrinsic differences in gene expression and response to cold exposure in mice. Am J Physiol Endocrinol Metab 2014; 306:E945-64; PMID:24549398; http://dx.doi.org/10.1152/ajpendo.00473.2013
- LaLonde JM, Bernlohr DA, Banaszak LJ. The up-and-down beta-barrel proteins. FASEB J 1994; 8:1240-7; PMID:8001736
- Yamamoto T, Yamamoto A, Watanabe M, Matsuo T, Yamazaki N, Kataoka M, Terada H, Shinohara Y. Classification of FABP isoforms and tissues based on quantitative evaluation of transcript levels of these isoforms in various rat tissues. Biotechnol Lett 2009; 31:1695-701; PMID:19565192; http://dx.doi.org/10.1007/s10529-009-0065-7
- Shan T, Liu W, Kuang S. Fatty acid binding protein 4 expression marks a population of adipocyte progenitors in white and brown adipose tissues. FASEB J 2013; 27:277-87; PMID:23047894; http://dx.doi.org/10.1096/fj.12-211516
- Nakamura Y, Sato T, Shiimura Y, Miura Y, Kojima M. FABP3 and brown adipocyte-characteristic mitochondrial fatty acid oxidation enzymes are induced in beige cells in a different pathway from UCP1. Biochem Biophys Res Commun 2013; 441:42-6; PMID:24129192; http://dx.doi.org/10.1016/j.bbrc.2013.10.014
- Yamashita H, Wang Z, Wang Y, Segawa M, Kusudo T, Kontani Y. Induction of fatty acid-binding protein 3 in brown adipose tissue correlates with increased demand for adaptive thermogenesis in rodents. Biochem Biophys Res Commun 2008; 377:632-5; PMID:18938135; http://dx.doi.org/10.1016/j.bbrc.2008.10.041
- Ameer F, Scandiuzzi L, Hasnain S, Kalbacher H, Zaidi N. De novo lipogenesis in health and disease. Metabolism 2014; 63:895-902; PMID:24814684; http://dx.doi.org/10.1016/j.metabol.2014.04.003
- Lodhi IJ, Wei X, Semenkovich CF. Lipoexpediency: de novo lipogenesis as a metabolic signal transmitter. Trends Endocrinol Metab 2011; 22:1-8; PMID:20889351; http://dx.doi.org/10.1016/j.tem.2010.09.002
- Herman MA, Peroni OD, Villoria J, Schön MR, Abumrad NA, Blüher M, Klein S, Kahn BB. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature 2012; 484:333-8; PMID:22466288; http://dx.doi.org/10.1038/nature10986
- Eissing L, Scherer T, Tödter K, Knippschild U, Greve JW, Buurman WA, Pinnschmidt HO, Rensen SS, Wolf AM, Bartelt A, et al. De novo lipogenesis in human fat and liver is linked to ChREBP-β and metabolic health. Nat Commun 2013; 4:1528; PMID:23443556; http://dx.doi.org/10.1038/ncomms2537
- Mottillo EP, Balasubramanian P, Lee Y-H, Weng C, Kershaw EE, Granneman JG. Coupling of lipolysis and de novo lipogenesis in brown, beige, and white adipose tissues during chronic β3-adrenergic receptor activation. J Lipid Res 2014; 55:2276-86; PMID:25193997; http://dx.doi.org/10.1194/jlr.M050005
- Berg JM, Tymoczko JL, Stryer L. Biochemistry. 7th edition. New York: WH Freeman; 2012.
- Bianchi A, Evans JL, Iverson AJ, Nordlund AC, Watts TD, Witters LA. Identification of an isozymic form of acetyl-CoA carboxylase. J Biol Chem 1990; 265:1502-9; PMID:1967254
- Lodhi IJ, Yin L, Jensen-Urstad APL, Funai K, Coleman T, Baird JH, El Ramahi MK, Razani B, Song H, Fu-Hsu F, et al. Inhibiting adipose tissue lipogenesis reprograms thermogenesis and PPARγ activation to decrease diet-induced obesity. Cell Metab 2012; 16:189-201; PMID:22863804; http://dx.doi.org/10.1016/j.cmet.2012.06.013
- Kihara A. Very long-chain fatty acids: elongation, physiology and related disorders. J Biochem 2012; 152:387-95; PMID:22984005; http://dx.doi.org/10.1093/jb/mvs105
- Tvrdik P, Westerberg R, Silve S, Asadi A, Jakobsson A, Cannon B, Loison G, Jacobsson A. Role of a new mammalian gene family in the biosynthesis of very long chain fatty acids and sphingolipids. J Cell Biol 2000; 149:707-18; PMID:10791983; http://dx.doi.org/10.1083/jcb.149.3.707
- Jakobsson A, Jörgensen JA, Jacobsson A. Differential regulation of fatty acid elongation enzymes in brown adipocytes implies a unique role for Elovl3 during increased fatty acid oxidation. Am J Physiol Endocrinol Metab 2005; 289:E517-26; PMID:15855229; http://dx.doi.org/10.1152/ajpendo.00045.2005
- Tvrdik P, Asadi A, Kozak LP, Nedergaard J, Cannon B, Jacobsson A. Cig30, a mouse member of a novel membrane protein gene family, is involved in the recruitment of brown adipose tissue. J Biol Chem 1997; 272:31738-46; PMID:9395518; http://dx.doi.org/10.1074/jbc.272.50.31738
- Zadravec D, Brolinson A, Fisher RM, Carneheim C, Csikasz RI, Bertrand-Michel J, Borén J, Guillou H, Rudling M, Jacobsson A. Ablation of the very-long-chain fatty acid elongase ELOVL3 in mice leads to constrained lipid storage and resistance to diet-induced obesity. FASEB J 2010; 24:4366-77; PMID:20605947; http://dx.doi.org/10.1096/fj.09-152298
- Miyazaki M, Flowers MT, Sampath H, Chu K, Otzelberger C, Liu X, Ntambi JM. Hepatic stearoyl-CoA desaturase-1 deficiency protects mice from carbohydrate-induced adiposity and hepatic steatosis. Cell Metab 2007; 6:484-96; PMID:18054317; http://dx.doi.org/10.1016/j.cmet.2007.10.014
- Ntambi JM, Miyazaki M, Stoehr JP, Lan H, Kendziorski CM, Yandell BS, Song Y, Cohen P, Friedman JM, Attie AD. Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc Natl Acad Sci U S A 2002; 99:11482-6; PMID:12177411; http://dx.doi.org/10.1073/pnas.132384699
- Rahman SM, Dobrzyn A, Lee S, Dobrzyn P, Miyazaki M, Ntambi JM. Stearoyl-CoA desaturase 1 de ciency increases insulin signaling and glycogen accumulation in brown adipose tissue. Am J Physiol Endocrinol Metab October 2005; 53706:381-7
- Townsend KL, Tseng Y-H. Brown fat fuel utilization and thermogenesis. Trends Endocrinol Metab 2014; 25:168-77; PMID:24389130; http://dx.doi.org/10.1016/j.tem.2013.12.004
- Orr AL, Quinlan CL, Perevoshchikova IV, Brand MD. A refined analysis of superoxide production by mitochondrial sn-glycerol 3-phosphate dehydrogenase. J Biol Chem 2012; 287:42921-35; PMID:23124204; http://dx.doi.org/10.1074/jbc.M112.397828
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev 2004; 84:277-359; PMID:14715917; http://dx.doi.org/10.1152/physrev.00015.2003
- Moura MAF, Festuccia WTL, Kawashita NH, Garófalo MAR, Brito SRC, Kettelhut IC, Migliorini RH. Brown adipose tissue glyceroneogenesis is activated in rats exposed to cold. Pflugers Arch 2005; 449:463-9; PMID:15688247; http://dx.doi.org/10.1007/s00424-004-1353-7
- Brasaemle DL, Wolins NE. Packaging of fat: an evolving model of lipid droplet assembly and expansion. J Biol Chem 2012; 287:2273-9; PMID:22090029; http://dx.doi.org/10.1074/jbc.R111.309088
- Walther TC, Farese RV. Lipid droplets and cellular lipid metabolism. Annu Rev Biochem 2012; 81:687-714; PMID:22524315; http://dx.doi.org/10.1146/annurev-biochem-061009-102430
- Beller M, Thiel K, Thul PJ, Jäckle H. Lipid droplets: a dynamic organelle moves into focus. FEBS Lett 2010; 584:2176-82; PMID:20303960; http://dx.doi.org/10.1016/j.febslet.2010.03.022
- Miranda DA, Kim J-H, Nguyen LN, Cheng W, Tan BC, Goh VJ, Tan JSY, Yaligar J, Kn BP, Velan SS, et al. Fat storage-inducing transmembrane protein 2 is required for normal fat storage in adipose tissue. J Biol Chem 2014; 289:9560-72; PMID:24519944; http://dx.doi.org/10.1074/jbc.M114.547687
- Gong J, Sun Z, Li P. CIDE proteins and metabolic disorders. Curr Opin Lipidol 2009; 20:121-6; PMID:19276890; http://dx.doi.org/10.1097/MOL.0b013e328328d0bb
- Cinti S. Transdifferentiation properties of adipocytes in the adipose organ. Am J Physiol Endocrinol Metab 2009; 297(5):E977-86; PMID:19458063; http://dx.doi.org/10.1152/ajpendo.00183.2009
- Zhou Z, Yon Toh S, Chen Z, Guo K, Ng CP, Ponniah S, Lin S-C, Hong W, Li P. Cidea-deficient mice have lean phenotype and are resistant to obesity. Nat Genet 2003; 35:49-56; PMID:12910269; http://dx.doi.org/10.1038/ng1225
- Li P. Cidea, brown fat and obesity. Mech Ageing Dev 2004; 125:337-8; PMID:15063110; http://dx.doi.org/10.1016/j.mad.2004.01.002
- Qi J, Gong J, Zhao T, Zhao J, Lam P, Ye J, Li JZ, Wu J, Zhou H-M, Li P. Downregulation of AMP-activated protein kinase by Cidea-mediated ubiquitination and degradation in brown adipose tissue. EMBO J 2008; 27:1537-48; PMID:18480843; http://dx.doi.org/10.1038/emboj.2008.92
- Shimizu T, Yokotani K. Acute cold exposure-induced down-regulation of CIDEA, cell death-inducing DNA fragmentation factor-alpha-like effector A, in rat interscapular brown adipose tissue by sympathetically activated beta3-adrenoreceptors. Biochem Biophys Res Commun 2009; 387:294-9; PMID:19577538; http://dx.doi.org/10.1016/j.bbrc.2009.06.147
- Grahn THM, Kaur R, Yin J, Schweiger M, Sharma VM, Lee M-J, Ido Y, Smas CM, Zechner R, Lass A, et al. Fat-specific protein 27 (FSP27) interacts with adipose triglyceride lipase (ATGL) to regulate lipolysis and insulin sensitivity in human adipocytes. J Biol Chem 2014; 289:12029-39; PMID:24627478; http://dx.doi.org/10.1074/jbc.M113.539890
- Puri V, Konda S, Ranjit S, Aouadi M, Chawla A, Chouinard M, Chakladar A, Czech MP. Fat-specific protein 27, a novel lipid droplet protein that enhances triglyceride storage. J Biol Chem 2007; 282:34213-8; PMID:17884815; http://dx.doi.org/10.1074/jbc.M707404200
- Toh SY, Gong J, Du G, Li JZ, Yang S, Ye J, Yao H, Zhang Y, Xue B, Li Q, et al. Up-regulation of mitochondrial activity and acquirement of brown adipose tissue-like property in the white adipose tissue of fsp27 deficient mice. PLoS One 2008; 3:e2890; PMID:18682832; http://dx.doi.org/10.1371/journal.pone.0002890
- Lafontan M, Langin D. Lipolysis and lipid mobilization in human adipose tissue. Prog Lipid Res 2009; 48:275-97; PMID:19464318; http://dx.doi.org/10.1016/j.plipres.2009.05.001
- Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, Lass A, Neuberger G, Eisenhaber F, Hermetter A, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 2004; 306:1383-6; PMID:15550674; http://dx.doi.org/10.1126/science.1100747
- Villena JA, Roy S, Sarkadi-Nagy E, Kim K-H, Sul HS. Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: ectopic expression of desnutrin increases triglyceride hydrolysis. J Biol Chem 2004; 279:47066-75; PMID:15337759; http://dx.doi.org/10.1074/jbc.M403855200
- Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem 2004; 279:48968-75; PMID:15364929; http://dx.doi.org/10.1074/jbc.M407841200
- Zechner R, Kienesberger PC, Haemmerle G, Zimmermann R, Lass A. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J Lipid Res 2009; 50:3-21; PMID:18952573; http://dx.doi.org/10.1194/jlr.R800031-JLR200
- Lass A, Zimmermann R, Oberer M, Zechner R. Lipolysis - a highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog Lipid Res 2011; 50:14-27; PMID:21087632; http://dx.doi.org/10.1016/j.plipres.2010.10.004
- Lass A, Zimmermann R, Haemmerle G, Riederer M, Schoiswohl G, Schweiger M, Kienesberger P, Strauss JG, Gorkiewicz G, Zechner R. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab 2006; 3:309-19; PMID:16679289; http://dx.doi.org/10.1016/j.cmet.2006.03.005
- Yang X, Lu X, Lombès M, Rha GB, Chi Y-I, Guerin TM, Smart EJ, Liu J. The G(0)/G(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab 2010; 11:194-205; PMID:20197052; http://dx.doi.org/10.1016/j.cmet.2010.02.003
- Haemmerle G, Lass A, Zimmermann R, Gorkiewicz G, Meyer C, Rozman J, Heldmaier G, Maier R, Theussl C, Eder S, et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 2006; 312:734-7; PMID:16675698; http://dx.doi.org/10.1126/science.1123965
- Ahmadian M, Duncan RE, Varady KA, Frasson D, Hellerstein MK, Birkenfeld AL, Samuel VT, Shulman GI, Wang Y, Kang C. Adipose overexpression of desnutrin promotes fatty acid use and attenuates diet-induced obesity. 2009; 58.
- Pinent M, Hackl H, Burkard TR, Prokesch A, Papak C, Scheideler M, Hämmerle G, Zechner R, Trajanoski Z, Strauss JG. Differential transcriptional modulation of biological processes in adipocyte triglyceride lipase and hormone-sensitive lipase-deficient mice. Genomics 2008; 92:26-32; PMID:18572100; http://dx.doi.org/10.1016/j.ygeno.2008.03.010
- Ahmadian M, Abbott MJ, Tang T, Hudak CSS, Kim Y, Bruss M, Hellerstein MK, Lee H-Y, Samuel VT, Shulman GI, et al. Desnutrin/ATGL is regulated by AMPK and is required for a brown adipose phenotype. Cell Metab 2011; 13:739-48; PMID:21641555; http://dx.doi.org/10.1016/j.cmet.2011.05.002
- Mottillo EP, Bloch AE, Leff T, Granneman JG. Lipolytic products activate peroxisome proliferator-activated receptor (PPAR) α and δ in brown adipocytes to match fatty acid oxidation with supply. J Biol Chem 2012; 287:25038-48; PMID:22685301; http://dx.doi.org/10.1074/jbc.M112.374041
- Vaughan M, Berger J, Steinberg D. Hormone-sensitive lipase and tissue hormone-sensitive lipase and monoglyceride activities in adipose tissue. J Biol Chem 1964; 239:401-9; PMID:14169138
- Holm C, Osterlund T, Laurell H, Contreras JA. Molecular mechanisms regulating hormone-sensitive lipase and lipolysis. Annu Rev Nutr 2000; 20:365-93; PMID:10940339; http://dx.doi.org/10.1146/annurev.nutr.20.1.365
- Holm C, Fredrikson G, Cannon B, Belfrage P. Hormone-sensitive lipase in brown adipose tissue: identification and effect of cold exposure. Biosci Rep 1987; 7:897-904; PMID:3329536; http://dx.doi.org/10.1007/BF01119481
- Fredrikson G, Belfrage P. Positional specificity of hormone-sensitive lipase from rat adipose tissue. J Biol Chem 1983; 258:14253-6; PMID:6643478
- Haemmerle G, Zimmermann R, Hayn M, Theussl C, Waeg G, Wagner E, Sattler W, Magin TM, Wagner EF, Zechner R. Hormone-sensitive lipase deficiency in mice causes diglyceride accumulation in adipose tissue, muscle, and testis. J Biol Chem 2002; 277:4806-15; PMID:11717312; http://dx.doi.org/10.1074/jbc.M110355200
- Wang H, Hu L, Dalen K, Dorward H, Marcinkiewicz A, Russell D, Gong D, Londos C, Yamaguchi T, Holm C, et al. Activation of hormone-sensitive lipase requires two steps, protein phosphorylation and binding to the PAT-1 domain of lipid droplet coat proteins. J Biol Chem 2009; 284:32116-25; PMID:19717842; http://dx.doi.org/10.1074/jbc.M109.006726
- Holm C. Molecular mechanisms regulating hormone-sensitive lipase and lipolysis. Biochem Soc Trans 2003; 31:1120; PMID:14641008; http://dx.doi.org/10.1042/bst0311120
- Osuga J -I, Ishibashi S, Oka T, Yagyu H, Tozawa R, Fujimoto A, Shionoiri F, Yahagi N, Kraemer FB, Tsutsumi O, et al. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc Natl Acad Sci 2000; 97:787-92; PMID:10639158; http://dx.doi.org/10.1073/pnas.97.2.787
- Ström K, Hansson O, Lucas S, Nevsten P, Fernandez C, Klint C, Movérare-Skrtic S, Sundler F, Ohlsson C, Holm C. Attainment of brown adipocyte features in white adipocytes of hormone-sensitive lipase null mice. PLoS One 2008; 3:e1793; PMID:18335062; http://dx.doi.org/10.1371/journal.pone.0001793
- Sakurada T, Noma A. Subcellular localization and some properties of monoacylglycerol lipase in rat adipocytes. J Biochem 1981; 90:1413-9; PMID:7338512
- Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, Madeo F. FAT SIGNALS–lipases and lipolysis in lipid metabolism and signaling. Cell Metab 2012; 15:279-91; PMID:22405066; http://dx.doi.org/10.1016/j.cmet.2011.12.018
- Egan JJ, Greenberg AS, Chang M, Wek SA, Moos MC, Londos C. Mechanism of hormone-stimulated lipolysis in adipocytes: Translocation of hormone-sensitive lipase to the lipid storage droplet. 1992; 89:8537-41; PMID:1528859
- Clifford GM. Translocation of Hormone-sensitive Lipase and Perilipin upon Lipolytic Stimulation of Rat Adipocytes. J Biol Chem 2000; 275:5011-5; PMID:10671541; http://dx.doi.org/10.1074/jbc.275.7.5011
- Granneman JG, Moore H-PH, Granneman RL, Greenberg AS, Obin MS, Zhu Z. Analysis of lipolytic protein trafficking and interactions in adipocytes. J Biol Chem 2007; 282:5726-35; PMID:17189257; http://dx.doi.org/10.1074/jbc.M610580200
- Doh K-O, Kim Y-W, Park S-Y, Lee S-K, Park JS, Kim J-Y. Interrelation between long-chain fatty acid oxidation rate and carnitine palmitoyltransferase 1 activity with different isoforms in rat tissues. Life Sci 2005; 77:435-43; PMID:15894012; http://dx.doi.org/10.1016/j.lfs.2004.11.032
- Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab 2009; 9:203-9; PMID:19187776; http://dx.doi.org/10.1016/j.cmet.2008.12.014
- Ueta CB, Fernandes GW, Capelo LP, Fonseca TL, Maculan FD, Gouveia CHA, Brum PC, Christoffolete MA, Aoki MS, Lancellotti CL, et al. β(1) Adrenergic receptor is key to cold- and diet-induced thermogenesis in mice. J Endocrinol 2012; 214:359-65; PMID:22728333; http://dx.doi.org/10.1530/JOE-12-0155
- Rothwell NJ, Stock MJ. A role for brown adipose tissue in diet-induced thermogenesis. Nature 1979; 281:31-5; PMID:551265; http://dx.doi.org/10.1038/281031a0
- Yu XX, Lewin DA, Forrest W, Adams SH. Cold elicits the simultaneous induction of fatty acid synthesis and beta-oxidation in murine brown adipose tissue: prediction from differential gene expression and confirmation in vivo. FASEB J 2002; 16:155-68; PMID:11818363; http://dx.doi.org/10.1096/fj.01-0568com
- Li Y, Fromme T, Schweizer S, Schöttl T, Klingenspor M. Taking control over intracellular fatty acid levels is essential for the analysis of thermogenic function in cultured primary brown and brite / beige adipocytes. EMBO Rep 2014; 15:1069-76.
- Bukowiecki LJ, Follea N, Lupien J, Paradis A. Metabolic relationships between lipolysis and respiration in rat brown. Respiration 1981; 256:12840-8
- Ellis JM, Li LO, Wu P-C, Koves TR, Ilkayeva O, Stevens RD, Watkins SM, Muoio DM, Coleman RA. Adipose acyl-CoA synthetase-1 directs fatty acids toward beta-oxidation and is required for cold thermogenesis. Cell Metab 2010; 12:53-64; PMID:20620995; http://dx.doi.org/10.1016/j.cmet.2010.05.012
- McGarry JD, Foster DW. Regulation of hepatic fatty acid oxidation and ketone body production. Annu Rev Biochem 1980; 49:395-420; PMID:6157353; http://dx.doi.org/10.1146/annurev.bi.49.070180.002143
- McGarry JD, Woeltje KF, Kuwajima M, Foster DW. Regulation of ketogenesis and the renaissance of carnitine palmitoyltransferase. Diabetes Metab Rev 1989; 5:271-84; PMID:2656156; http://dx.doi.org/10.1002/dmr.5610050305
- Curtis DE, Sukumaran S, Shao X, Parameswara V, Rashid S, Corte A, Smith AR, Ren J, Esser V, Hammer RE, et al. Article Molecular Mechanisms of Hepatic Steatosis and Insulin Resistance in the AGPAT2-Deficient Mouse Model of Congenital Generalized Lipodystrophy. 2009; 165-76.
- Esser V, Britton CH, Weis BC, Foster DW, McGarry JD. Cloning, sequencing, and expression of a cDNA encoding rat liver carnitine palmitoyltransferase I: Direct evidence that a single polypeptide is involved in inhibitor interaction and catalytic function. J Biol Chem 1993; 268:5817-22; PMID:8449948
- Yamazaki N, Shinohara Y, Shima A, Terada H. High expression of a novel carnitine palmitoyltransferase I like protein in rat brown adipose tissue and heart: isolation and characterization of its cDNA clone. FEBS Lett 1995; 363:41-5; PMID:7729550; http://dx.doi.org/10.1016/0014-5793(95)00277-G
- Price N, van der Leij F, Jackson V, Corstorphine C, Thomson R, Sorensen A, Zammit V. A novel brain-expressed protein related to carnitine palmitoyltransferase I. Genomics 2002; 80:433-42; PMID:12376098; http://dx.doi.org/10.1006/geno.2002.6845
- Brown NF, Hill JK, Esser V, Kirkland JL, Corkey BE, Foster DW, McGarry JD. Mouse white adipocytes and 3T3-L1 cells display an anomalous pattern of carnitine palmitoyltransferase (CPT) I isoform expression during differentiation. Inter-tissue and inter-species expression of CPT I and CPT II enzymes. Biochem J 1997; 327 ( Pt 1):225-31; PMID:9355756; http://dx.doi.org/10.1042/bj3270225
- Sierra AY, Gratacós E, Carrasco P, Clotet J, Ureña J, Serra D, Asins G, Hegardt FG, Casals N. CPT1c is localized in endoplasmic reticulum of neurons and has carnitine palmitoyltransferase activity. J Biol Chem 2008; 283:6878-85; PMID:18192268; http://dx.doi.org/10.1074/jbc.M707965200
- Gao S, Zhu G, Gao X, Wu D, Carrasco P, Casals N, Hegardt FG, Moran TH, Lopaschuk GD. Important roles of brain-specific carnitine palmitoyltransferase and ceramide metabolism in leptin hypothalamic control of feeding. Proc Natl Acad Sci U S A 2011; 108:9691-6; PMID:21593415; http://dx.doi.org/10.1073/pnas.1103267108
- Ramírez S, Martins L, Jacas J, Carrasco P, Pozo M, Clotet J, Serra D, Hegardt FG, Diéguez C, López M, et al. Hypothalamic ceramide levels regulated by CPT1C mediate the orexigenic effect of ghrelin. Diabetes 2013; 62:2329-37; PMID:23493572; http://dx.doi.org/10.2337/db12-1451
- Ramsay RR, Gandour RD, Van Der Leij FR. Molecular enzymology of carnitine transfer and transport. Biochim Biophys Acta - Protein Struct Mol Enzymol 2001; 1546:21-43; PMID:11257506; http://dx.doi.org/10.1016/S0167-4838(01)00147-9
- Alam A, Saggerson E. Malonyl-CoA and the regulation of fatty acid oxidation in soleus muscle. Biochem J 1998; 241:233-41; PMID:9693125; http://dx.doi.org/10.1042/bj3340233
- Esser V, Brown NF, Cowan AT, Foster DW, Mcgarry JD, Lett HF. Expression of a cDNA Isolated from Rat Brown Adipose Tissue and Heart Identifies the Product as the Muscle Isoform of Carnitine Palmitoyltransferase I ( M-CPT I ) isolated from rat brown adipose tissue ( BAT ) by. Biochemistry 1996; 271:6972-7
- Morillas M, Gómez-Puertas P, Bentebibel A, Sellés E, Casals N, Valencia A, Hegardt FG, Asins G, Serra D. Identification of conserved amino acid residues in rat liver carnitine palmitoyltransferase I critical for malonyl-CoA inhibition: Mutation of methionine 593 abolishes malonyl-CoA inhibition. J Biol Chem 2003; 278:9058-63; PMID:12499375; http://dx.doi.org/10.1074/jbc.M209999200
- Ji S, You Y, Kerner J, Hoppel CL, Schoeb TR, Chick WSH, Hamm DA, Sharer JD, Wood PA. Homozygous carnitine palmitoyltransferase 1b (muscle isoform) deficiency is lethal in the mouse. Mol Genet Metab 2008; 93:314-22; PMID:18023382; http://dx.doi.org/10.1016/j.ymgme.2007.10.006
- Jamal Z, Saggerson ED. Changes in brown-adipose-tissue mitochondrial processes in streptozotocin-diabetes. Biochem J 1988; 252:293-6; PMID:3421907; http://dx.doi.org/10.1042/bj2520293
- Lee J, Ellis JM, Wolfgang MJ. Adipose fatty acid oxidation is required for thermogenesis and potentiates oxidative stress-induced inflammation. Cell Rep 2015; 10:266-79; PMID:25578732; http://dx.doi.org/10.1016/j.celrep.2014.12.023
- Schuler AM, Gower BA, Matern D, Rinaldo P, Vockley J, Wood PA. Synergistic heterozygosity in mice with inherited enzyme deficiencies of mitochondrial fatty acid beta-oxidation. Mol Genet Metab 2005; 85:7-11; PMID:15862275; http://dx.doi.org/10.1016/j.ymgme.2004.09.006
- Bruce CR, Hoy AJ, Turner N, Watt MJ, Allen TL, Carpenter K, Cooney GJ, Febbraio MA, Kraegen EW. Overexpression of carnitine palmitoyltransferase-1 in skeletal muscle is sufficient to enhance fatty acid oxidation and improve high-fat diet-induced insulin resistance. Diabetes 2009; 58:550-8; PMID:19073774; http://dx.doi.org/10.2337/db08-1078
- Perdomo G, Commerford SR, Richard A-MT, Adams SH, Corkey BE, O'Doherty RM, Brown NF. Increased beta-oxidation in muscle cells enhances insulin-stimulated glucose metabolism and protects against fatty acid-induced insulin resistance despite intramyocellular lipid accumulation. J Biol Chem 2004; 279:27177-86; PMID:15105415; http://dx.doi.org/10.1074/jbc.M403566200
- Stefanovic-Racic M, Perdomo G, Mantell BS, Sipula IJ, Brown NF, O'Doherty RM. A moderate increase in carnitine palmitoyltransferase 1a activity is sufficient to substantially reduce hepatic triglyceride levels. Am J Physiol Endocrinol Metab 2008; 294:E969-77; PMID:18349115; http://dx.doi.org/10.1152/ajpendo.00497.2007
- Orellana-Gavaldà JM, Herrero L, Malandrino MI, Pañeda A, Sol Rodríguez-Peña M, Petry H, Asins G, Van Deventer S, Hegardt FG, Serra D, et al. Molecular therapy for obesity and diabetes based on a long-term increase in hepatic fatty-acid oxidation. Hepatology 2011; 53:821-32; PMID:21319201; http://dx.doi.org/10.1002/hep.24140
- Monsenego J, Mansouri A, Akkaoui M, Lenoir V, Esnous C, Fauveau V, Tavernier V, Girard J, Prip-Buus C, Monsénégo J, et al. Enhancing liver mitochondrial fatty acid oxidation capacity in obese mice improves insulin sensitivity independently of hepatic steatosis. J Hepatol 2012; 56:632-9; PMID:22037024; http://dx.doi.org/10.1016/j.jhep.2011.10.008
- Sebastián D, Herrero L, Serra D, Asins G, Hegardt FG, Sebastian D, Herrero L, Serra D, Asins G, Hegardt FG. CPT I overexpression protects L6E9 muscle cells from fatty acid-induced insulin resistance. Am J Physiol Endocrinol Metab 2007; 292:E677-86; PMID:17062841; http://dx.doi.org/10.1152/ajpendo.00360.2006
- Namgaladze D, Lips S, Leiker TJ, Murphy RC, Ekroos K, Ferreiros N, Geisslinger G, Brüne B. Inhibition of macrophage fatty acid β-oxidation exacerbates palmitate-induced inflammatory and endoplasmic reticulum stress responses. Diabetologia 2014; 57:1067-77; PMID:24488024; http://dx.doi.org/10.1007/s00125-014-3173-4
- Gao X, Li K, Hui X, Kong X, Sweeney G, Wang Y, Xu A, Teng M, Liu P, Wu D. Carnitine palmitoyltransferase 1A prevents fatty acid-induced adipocyte dysfunction through suppression of c-Jun N-terminal kinase. Biochem J 2011; 435:723-32; PMID:21348853; http://dx.doi.org/10.1042/BJ20101680
- Malandrino MI, Fucho R, Weber M, Calderon-Dominguez M, Mir JF, Valcarcel L, Escoté X, Gómez-Serrano M, Peral B, Salvadó L, et al. Enhanced fatty acid oxidation in adipocytes and macrophages reduces lipid-induced triglyceride accumulation and inflammation. Am J Physiol Endocrinol Metab 2015; 308:E756-69; PMID:25714670; http://dx.doi.org/10.1152/ajpendo.00362.2014
- Herrero L, Rubí B, Sebastián D, Serra D, Asins G, Maechler P, Prentki M, Hegardt FG. Alteration of the malonyl-CoA/carnitine palmitoyltransferase I interaction in the beta-cell impairs glucose-induced insulin secretion. Diabetes 2005; 54:462-71; PMID:15677504; http://dx.doi.org/10.2337/diabetes.54.2.462
- Mera P, Mir JF, Fabriàs G, Casas J, Costa ASH, Malandrino MI, Fernández-López J-A, Remesar X, Gao S, Chohnan S, et al. Long-term increased carnitine palmitoyltransferase 1A expression in ventromedial hypotalamus causes hyperphagia and alters the hypothalamic lipidomic profile. PLoS One 2014; 9:e97195; PMID:24819600; http://dx.doi.org/10.1371/journal.pone.0097195
- Serra D, Mera P, Malandrino MI, Mir JF, Herrero L. Mitochondrial fatty acid oxidation in obesity. Antioxid Redox Signal 2013; 19:269-84; PMID:22900819; http://dx.doi.org/10.1089/ars.2012.4875
- Singh I, Moser AE, Goldfischer S, Moser HW. Lignoceric acid is oxidized in the peroxisome: implications for the Zellweger cerebro-hepato-renal syndrome and adrenoleukodystrophy. Proc Natl Acad Sci U S A 1984; 81:4203-7; PMID:6588384; http://dx.doi.org/10.1073/pnas.81.13.4203
- Nedergaard J, Alexson S, Cannon B. Cold adaptation in the rat: increased brown fat peroxisomal beta-oxidation relative to maximal mitochondrial oxidative capacity. Am J Physiol Cell Physiol 1980; 239:C208-16
- Ahlabo I, Barnard T. Observations on peroxisomes in brown adipose tissue of the rat. J Histochem Cytochem 1971; 19:670-5; PMID:4107748; http://dx.doi.org/10.1177/19.11.670
- Lodhi IJ, Semenkovich CF. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab 2014; 19:380-92; PMID:24508507; http://dx.doi.org/10.1016/j.cmet.2014.01.002
- Schneider BE, Mun EC. Surgical management of morbid obesity. Diabetes Care 2005; 28:475-80; PMID:15677820; http://dx.doi.org/10.2337/diacare.28.2.475
- Mumphrey MB, Patterson LM, Zheng H, Berthoud H-R. Roux-en-Y gastric bypass surgery increases number but not density of CCK-, GLP-1-, 5-HT-, and neurotensin-expressing enteroendocrine cells in rats. Neurogastroenterol Motil 2013; 25:e70-9; PMID:23095091; http://dx.doi.org/10.1111/nmo.12034
- Tremaroli V, Karlsson F, Werling M, Ståhlman M, Kovatcheva-Datchary P, Olbers T, Fändriks L, le Roux CW, Nielsen J, Bäckhed F. Roux-en-Y gastric bypass and vertical banded gastroplasty induce long-term changes on the human Gut microbiome contributing to fat mass regulation. Cell Metab 2015; 22:228-38; PMID:26244932; http://dx.doi.org/10.1016/j.cmet.2015.07.009
- Hurt RT, Edakkanambeth Varayil J, Ebbert JO. New pharmacological treatments for the management of obesity. Curr Gastroenterol Rep 2014; 16:394; PMID:24828101; http://dx.doi.org/10.1007/s11894-014-0394-0
- Jackson VM, Price DA, Carpino PA. Investigational drugs in Phase II clinical trials for the treatment of obesity: implications for future development of novel therapies. Expert Opin Investig Drugs 2014; 23:1055-66; PMID:25000213; http://dx.doi.org/10.1517/13543784.2014.918952
- EMA. Guideline on clinical evaluation of medicinal products used in weight control. London: 2014. Available from: http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2014/07/WC500170278.pdf
- Derosa G, Maffioli P. Anti-obesity drugs: a review about their effects and their safety. Expert Opin Drug Saf 2012; 11:459-71; PMID:22439841; http://dx.doi.org/10.1517/14740338.2012.675326
- O'Neil PM, Smith SR, Weissman NJ, Fidler MC, Sanchez M, Zhang J, Raether B, Anderson CM, Shanahan WR. Randomized Placebo-Controlled Clinical Trial of Lorcaserin for Weight Loss in Type 2 Diabetes Mellitus: The BLOOM-DM Study. Obes (Silver Spring) 2012; 20:1426-36; PMID:22421927; http://dx.doi.org/10.1038/oby.2012.66
- Khan A, Raza S, Khan Y, Aksoy T, Khan M, Weinberger Y, Goldman J. Current updates in the medical management of obesity. Recent Pat Endocr Metab Immune Drug Discov 2012; 6:117-28; PMID:22435392; http://dx.doi.org/10.2174/187221412800604644
- EMA. Withdrawal of the marketing authorisation application for Belviq (lorcaserin). London: 2013. Available from: http://www.ema.europa.eu/docs/en_GB/document_library/Medicine_QA/2013/05/WC500143811.pdf
- FDA. Liraglutide 3.0 mg for Weight Management NDA 206-321 Briefing Document Endocrinologic and Metabolic Drug Advisory Committee. Silver Spring, MD: 2014. Available from: http://www.fda.gov/downloads/advisorycommittees/committeesmeetingmaterials/drugs/endocrinologicandmetabolicdrugsadvisorycommittee/ucm413318.pdf
- EMA. Saxenda recommended for approval in weight management in adults Medicine to be used in addition to reduced-calorie diet and physical activity. London: 2015. Available from: http://www.ema.europa.eu/docs/en_GB/document_library/Press_release/2015/01/WC500180857.pdf
- Jeon WS, Park CY. Antiobesity pharmacotherapy for patients with type 2 diabetes: focus on long-term management. Endocrinol Metab (Seoul, Korea) 2014; 29:410-7; PMID:25559569; http://dx.doi.org/10.3803/EnM.2014.29.4.410
- Caixàs A, Albert L, Capel I, Rigla M. Naltrexone sustained-release/bupropion sustained-release for the management of obesity: review of the data to date. Drug Des Devel Ther 2014; 8:1419-27; PMID:25258511; http://dx.doi.org/10.2147/DDDT.S55587
- Hamdy O, Uwaifo GI, Oral EA. Obesity Medication. Medscape 2015; Available from: http://emedicine.medscape.com/article/123702-medication
- Zhu Z, Spicer EG, Gavini CK, Goudjo-Ako AJ, Novak CM, Shi H. Enhanced sympathetic activity in mice with brown adipose tissue transplantation (transBATation). Physiol Behav 2014; 125:21-9; PMID:24291381; http://dx.doi.org/10.1016/j.physbeh.2013.11.008
- Dong M, Yang X, Lim S, Cao Z, Honek J, Lu H, Zhang C, Seki T, Hosaka K, Wahlberg E, et al. Cold exposure promotes atherosclerotic plaque growth and instability via UCP1-dependent lipolysis. Cell Metab 2013; 18:118-29; PMID:23823482; http://dx.doi.org/10.1016/j.cmet.2013.06.003
- Berbée JFP, Boon MR, Khedoe PPSJ, Bartelt A, Schlein C, Worthmann A, Kooijman S, Hoeke G, Mol IM, John C, et al. Brown fat activation reduces hypercholesterolaemia and protects from atherosclerosis development. Nat Commun 2015; 6:6356; PMID:25754609; http://dx.doi.org/10.1038/ncomms7356
- Muoio DM, Neufer PD. Lipid-induced mitochondrial stress and insulin action in muscle. Cell Metab 2012; 15:595-605; PMID:22560212; http://dx.doi.org/10.1016/j.cmet.2012.04.010
