ABSTRACT
MicroRNAs (miRNAs) are non-coding RNAs that regulate target gene expression at the post-transcriptional level and are supposed to be implicated in the control of adipogenesis. We aimed to identify miRNAs which are involved in the regulation of human adipogenesis and searched for their molecular targets.
Applying microarray-analysis we identified miR125b-5p as upregulated during human adipocyte differentiation, although its role during adipogenesis is unknown. We identified and characterized the matrix metalloproteinase 11 (MMP11) as a direct target of miR125b-5p by showing that miR125b-5p overexpression significantly reduces MMP11 luciferase activity and mutation of any single binding site was sufficient to abolish the miR125b-5p mediated inhibition of luciferase activity. MMP11 overexpression decreased fat accumulation, indicating that MMP11 acts as an anti-adipogenic regulator. In contrast, overexpression of miR125b-5p itself reduced adipogenesis.
In summary, we identified miR125b-5p as upregulated during human adipogenesis indicating that miR125b-5p may serve as a regulator of human adipocyte differentiation. We further show that miR125b-5p downregulates the anti-adipogenic MMP11, but directly inhibits adipogenesis itself. Taken together, these data implicate that miR125b-5p can affect human adipogenesis via MMP11 and probably additional targets.
Introduction
Adipocyte differentiation is a major mechanism in the accumulation of body fat mass.Citation1 Hence, dysregulation of adipogenesis may contribute to the development of obesity. Potential regulators of tissue development in general and adipogenesis specifically include miRNAs. Over the past few years, it has become clear that miRNAs are involved in critical biological processes, including development, metabolism, differentiation and proliferation, as well as apoptosis, and are thereby implicated in the development of different diseases including obesity.Citation2-4 MiRNAs are a family of 19-24 nucleotide small RNAs that downregulate gene expression at the post-transcriptional level by binding to the 3′untranslated regions (UTRs) of target genes.Citation5 Mature miRNA sequences are evolutionarily highly conservedCitation5 and exhibit high tissue specificity.Citation6 Computer-based predictions estimated that 2–3% of all genes encode miRNAs and 30% of all human genes may be targeted by miRNAs.Citation7 Although miRNA expression and function have been extensively investigated in neuronal, muscle and haematopoietic systems, little is known about their involvement in adipogenesis.Citation8,9 However, the finding that suppression of Drosha, an essential enzyme for miRNA maturation, abolished adipocyte differentiationCitation10 suggests that miRNAs affect adipogenesis.Citation11 So far, only few miRNAs have been investigated for their impact on adipogenesis in more detail. For example, miR143,Citation12 miR27bCitation13 and miR130Citation14 were shown to act as pro- or antiadipogenic regulators by suppressing PPARg, while Let-7 miRNA regulates adipocyte differentiation by targeting HMGA2 in 3T3-L1 cells.Citation15 There are supposedly many more miRNAs with an implication in adipogenesis.
The aim of this work was to identify miRNAs implicated in adipocyte differentiation and evaluate their relevance for human adipogenesis.
Results
MiR125b-5p is differentially expressed during human adipogenesis
To identify differentially expressed miRNAs, we differentiated human preadipocytes of the SGBS cell line into mature adipocytes, extracted RNA at different time points and searched for differentially regulated miRNAs via sequencing in 2 independent adipocyte differentiation courses.Citation16 Representative images at days 0, 4, 8 and 12 after adipogenic induction are shown in . The increasing expression of the adipogenic markers PPARg, FABP4 and ATGL () verified adipogenesis.
Figure 1. Inverse regulation of miR125b-5p and MMP11 during human adipogenesis. (A) Representative phase contrast images of cells at 0-12 d after adipogenic induction. (B) PPARG, (C) FABP4, (D) ATGL, (E) MMP11 (white bars), miR125b-5p (black bars) and miR125b-3p (gray bars) mRNA levels and (F) MMP11 protein levels during adipogenesis. MMP11 protein levels were normalized to total protein amount in each sample. The data are presented as mean+-SEM of these 2 independent adipocyte differentiation courses. (G) Homology comparison of predicted miR125b-5p binding sites in the 3′UTR region of human MMP11. Conserved miR125b-5p binding sites between different species are indicated by a bold sequence. For standardization, target gene expression was normalized to the mean of the 2 housekeeping genes: β-actin (ACTB), and TATAbox-binding protein (TBP). *, p < 0.05; ***, p < 0.001, ns, not significant.
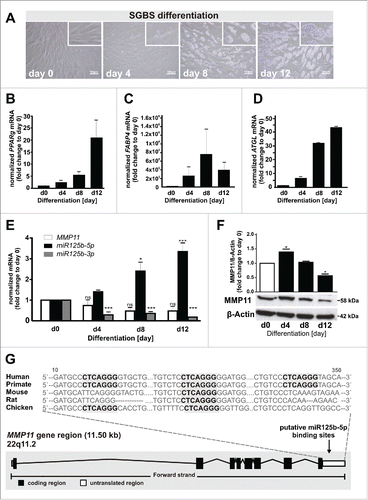
We identified 195 differentially expressed miRNAs during human adipogenesis. For further analyses we prioritized at least 2.5-fold upregulated miRNAs (p < 0.001) at day 12 compared to day 0 after induction of adipogenesis. Seven miRNAs fulfilled these criteria (miR378a-3p, miR-let7c, miR193a-5p, miR107, miR7d-5p, miR148a-3p and miR125b-5p). Twelve miRNAs were repressed to 40% at day 12 (miR21-5p, miR493-3p, miR409-3p, miR125b-1-3p, miR381, miR431-5p, miR155-5p, miR146a-5p, miR486-5p, miR21-3p, miR299-3p and miR100-3p), while the rest did not show differential expression. We selected miR125b-5p for further analysis, since this miRNA has so far not been investigated in the context of human adipogenesis in contrast to the other miRNAs.Citation15,17-22 We confirmed a 3.3-fold upregulation of the miR125b-5p strand and a 5.5-fold downregulation of the miR125b-3p strand during adipogenesis ().
Identification of MMP11 as a target of miR125b-5p
To identify putative targets of miR125b-5p we applied PicTar algorithmsCitation23 revealing 531 potential targets. We prioritized targets i) with a PicTar score higher than 5, ii) which were confirmed by other computer-based algorithms (TargetScanHuman), and iii) which have already been described in the context of adipogenesis. Applying this scoring strategy, the StAR-related lipid transfer domain protein 13 (STARD13) and the matrix metalloproteinase-11 (MMP11) ranked highest. Although, MMP11 mRNA expression appeared lower in mature adipocyte, the downregulation of MMP11 was not significant on mRNA level (), as well as STARD13 (data not shown) during adipogenesis. However, MMP11 protein levels were significantly down regulated (). In subsequent analyses, we were not able to verify an interaction between miR125b-5p and STARD13 with a reporter assay (data not shown), and thus we focused on MMP11. PicTar predictions revealed 3 putative miR125b-5p binding sites in the 3′UTR region of MMP11 (). Homology comparison across different species (human, primate, mouse, rat and chicken) revealed a high degree of sequence conservation, particularly for the second miR125b-5p binding site ().
Mir125b-5p directly represses MMP11 in adipocytes
To determine whether miR125b-5p targets MMP11 in human preadipocytes, we overexpressed or downregulated miR125b-5p using precursor or antagonizing molecules (pre-miR125b-5p or anti-miR125b-5p) (). By measuring miR125b-5p mRNA and protein levels after miR125b-5p overexpression, we confirmed a 10-fold up-regulation of miR125b-5p (), which was accompanied by an almost complete downregulation of MMP11 mRNA and protein levels; whereas miR125b-5p downregulation had no effect on MMP11 mRNA or protein levels ().
Figure 2. MiR125b-5p targets MMP11 by binding to the 3′UTR. (A) MiR125b-5p overexpression (pre-miR125b-5p) and downregulation (anti-miR125b-5p) efficiency was verified by qRT-PCR and normalized to scrambled control (pre-ntRNA, anti-ntRNA). (B) MMP11 mRNA and protein levels after overexpression or downregulation of miR125b-5p. (C) MMP11 mRNA expression after stratification in tertiles according to miR125b-5p mRNA levels in isolated human adipocytes of children. (D) Luciferase reporter plasmids carrying wild-type (WT) or mutant (MUT) miR125b-5p binding sites (gray boxes). MUT(1-3) indicate individual mutations of the single binding site; MUT(1.2.3), mutation of all binding sites. The insertion of the mutation is indicated in bold. (E) Relative luciferase activity after co-transfection of control or MMP11 with pre-miR125b-5p/pre-ntRNA or antimiR125b-5p/anti-ntRNA in SGBS preadipocytes. In addition, relative luciferase activity after cotransfection of control or mutated versions of miR125b-5p binding sites with pre-miR125b-5p/pre-ntRNA or anti-miR125b-5p/anti-ntRNA in SGBS preadipocytes. For standardization, target gene expression was normalized to the mean of the 2 housekeeping genes: β-actin (ACTB), and TATAbox-binding protein (TBP). *, p < 0.05; ***, p < 0.001.
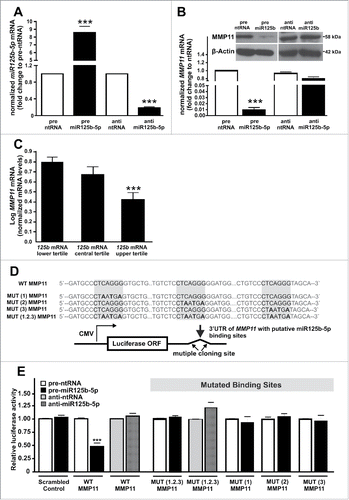
To investigate whether this co regulation is also detectable in vivo, we analyzed MMP11 and miR125b-5p expression in subcutaneous AT samples of 100 healthy children. To minimize the effect from cells of the stromal vascular fraction (macrophages, endothelial cells, etc.), we measured mRNA expression only in isolated adipocytes. In 49% of our samples we observed an undetectable MMP11 expression. Therefore, we decided to stratify AT samples into tertiles according to miR125b-5p expression (). We observed a twofold decrease in MMP11 expression in samples with high miR125b-5p compared to samples with low/absent miR125b-5p mRNA levels (). To assess whether this co regulation results from a direct interaction of miR125b-5p and MMP11, we cotransfected preadipocytes with luciferase reporter constructs containing wild-type (WT) or mutant (MUT) miR125b-5p binding sites together with pre-miR125b/pre-ntRNA or anti-miR125b/anti-ntRNA oligos (). Comparable transfection efficiency was ensured by normalizing all values to a renilla control. Luciferase activity of WT MMP11 was significantly reduced in the presence of miR125b-5p (), while luciferase activity did not change in the presence of anti-miR125b-5p. Mutation of all 3 miR125b-5p binding sites MUT (1.2.3) abolished the regulatory effect of miR125b-5p (), confirming that miR125b-5p directly interacts with MMP11 mRNA. We further assessed the relevance of the individual binding sites by selectively mutating each of the 3 distinct sites in the MMP11 3′UTR. The lack of any single binding site was sufficient to abolish miR125b-5p mediated inhibition of luciferase activity (), indicating that all sites are essential for the miR125b-5p/MMP11 interaction.
MMP11 is a negative regulator of human adipogenesis
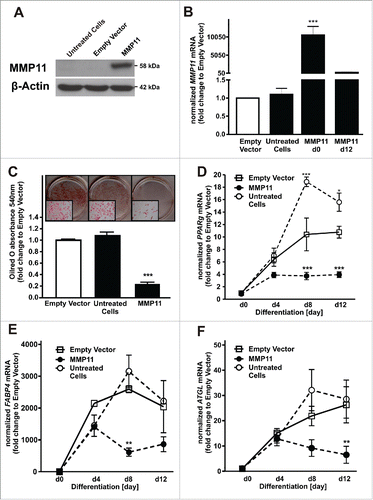
To evaluate the role of MMP11 itself during adipogenesis, we investigated the effect of MMP11 knockdown and overexpression on adipocyte differentiation. Efficient MMP11 knockdown was verified by decreased MMP11 mRNA at day 0 and 12 of adipogenesis and by decreased protein levels 48h after transfection of MMP11-specific siRNAs (). MMP11 knockdown had only a weak pro-adipogenic effect, indicated by Oilred-O-staining (). PPARg (), FABP4 () and ATGL () mRNA levels were not altered compared to a non-target scrambled siRNA control. In addition, we overexpressed MMP11 and investigated the effect on adipogenesis. Efficient MMP11 overexpression was verified by increased protein levels 48h after transfection () and significantly increased MMP11 mRNA expression at d0 of adipogenesis (). MMP11 overexpression significantly inhibited adipogenesis by 77% as shown by decreased Oilred-O absorbance () and decreased PPARg (), FABP4 () and ATGL () mRNA levels. Hence, MMP11 is a negative regulator of adipogenesis.
Figure 3. MMP11 knockdown only mildly affected adipogenesis. (A) SiRNA-mediated MMP11 knockdown efficiency was verified at mRNA level on day 0, 12 of adipogenesis and 48 h after transfection on protein level. (B) On day 12, Oilred-O absorbance was assessed to determine adipogenic capacity after MMP11 and PPARg knockdown. (C) The magnitude of PPARg, (D) FABP4, and (E) ATGL mRNA after transfection with siRNAs against control (white square), MMP11 (closed circle) or PPARg (open circle) were assessed by qRT-PCR analyses. For standardization, target gene expression was normalized to the mean of the 2 housekeeping genes: β-actin (ACTB), and TATAbox-binding protein (TBP). *, p < 0.05, **, p < 0.01; ***, p < 0.001.
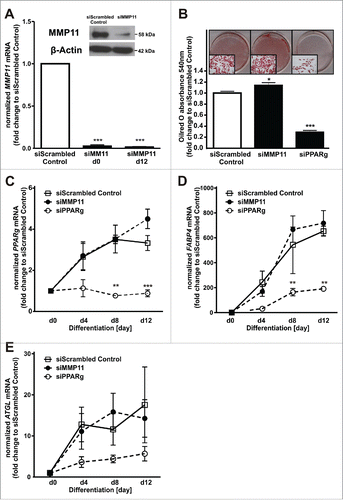
Figure 4. MMP11 acts as a negative regulator of adipogenesis. (A) MMP11 overexpression efficiency was measured on protein level and compared to cells transfected with empty vector or untreated cells. (B) MMP11 mRNA levels at day 0 and 12 of adipogenesis after MMP11 overexpression and compared to controls. (C) Effects of MMP11 overexpression on adipogenesis were measured by Oilred-O staining at day 12 of adipogenesis. (D) PPARg, (E) FABP4, and (F) ATGL mRNA levels after overexpression of MMP11 (closed circle) compared to an empty vector control (white square) or untreated cells (open circle). For standardization, target gene expression was normalized to the mean of the 2 housekeeping genes: β-actin (ACTB), and TATAbox-binding protein (TBP). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
MiR125b-5p overexpression inhibits adipogenesis
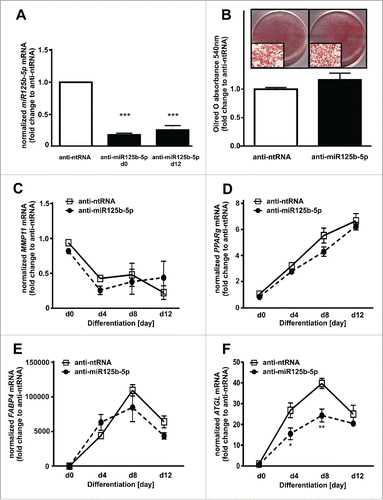
We finally evaluated the effect of miR125b-5p directly on adipogenesis. For this, we overexpressed miR125b-5p using precursor molecules (pre-miR125b-5p) and observed significantly increased levels of mature miR125b-5p at days 0 and 12 of adipogenesis (). MiR125b-5p overexpression led to a reduced fat accumulation as shown by a 57% lower frequency of differentiated adipocytes (). Moreover, miR125-5p overexpression resulted in a reduced MMP11 expression (), and was associated with significantly decreased PPARg mRNA expression (). Furthermore, FABP4 () and ATGL () mRNA levels were slightly reduced after miR125b-5p overexpression. We further assessed the relevance of miR125b-5p during adipogenesis by transfection of preadipocytes with antisense miR125b-5p oligos. MiR125b-5p knockdown was verified by 5-fold decreased miR125b-5p levels at day 0 and 12 of adipogenesis (). However, miR125b-5p knockdown did not influence adipogenesis, since we did not observe differences in Oilred-O absorbance (). In addition, we did not observe differences in MMP11 (), PPARg (), FABP4 (), and ATGL () mRNA levels.
Figure 5. MiR125b-5p act as a negative regulator of adipogenesis. (A) MiR125b-5p overexpression (pre-miR125b-5p) efficiency was assessed at day 0 and 12 of adipogenesis (black bars) and compared to pre-ntRNA control (white bar). (B) On day 12, Oilred-O absorbance was measured in order to investigate the effect of miR125b-5p on adipogenesis. (C) MMP11 mRNA levels after miR125b-5p (closed circle) overexpression compared to a pre-ntRNA (white square) control. (D) PPARg, (E) FABP4, and (F) ATGL mRNA levels after miR125b-5p (closed circle) overexpression compared to a pre-ntRNA (white square) control. For standardization, target gene expression was normalized to the mean of the 2 housekeeping genes: β-actin (ACTB), and TATAbox-binding protein (TBP). *, p < 0.05 **, p < 0.01, ***, p < 0.001.
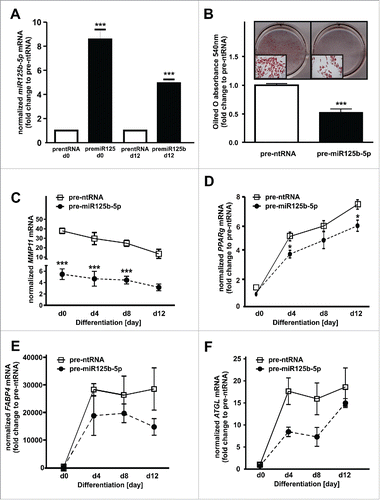
Figure 6. MiR125b-5p downregulation does not affect adipogenesis. (A) MiR125b-5p downregulation efficiency was assessed at day 0 and 12 of adipogenesis (black bars) and compared to pre-ntRNA control (white bar). (B) Oilred-O absorbance was measured at day 12 after adipogenic induction in order to investigate the effect of miR125b-5p on adipogenesis. (C) MMP11 mRNA levels after miR125b-5p (closed circle) downregulation compared to anti-ntRNA (white square) control. (D) PPARg, (E) FABP4, and (F) ATGL mRNA levels after miR125b-5p (closed circle) downregulation compared to an anti-ntRNA (white square) control. For standardization, target gene expression was normalized to the mean of the 2 housekeeping genes: β-actin (ACTB), and TATAbox-binding protein (TBP). *, p < 0.05 **, p < 0.01, ***, p < 0.001.
Discussion
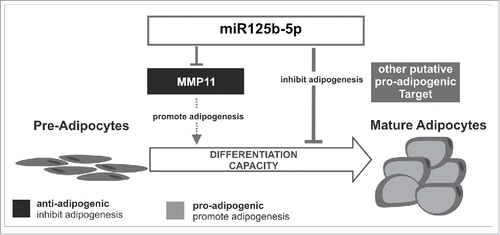
The aim of this study was to identify miRNAs which are involved in the regulation of human adipogenesis and to characterize their molecular targets and thereby address the functional relevance during adipocyte differentiation. We identified miR125b-5p as differentially up-regulated during human adipogenesis and showed that MMP11 is a direct target of miR125b-5p. Furthermore, MMP11 overexpression significantly inhibited adipocyte differentiation, suggesting that MMP11 itself is a negative regulator of human adipogenesis. In contrast, miR125b-5p-overexpression reduced fat accumulation, indicating that miR125b-5p itself has an additional antiadipogenic effect, potentially by the regulation of a so far unknown target which favors adipogenesis ().
Figure 7. Schematic overview of the findings. MiR125b-5p directly down-regulates MMP11, which in turn acts as negative regulator of human adipogenesis. Furthermore, miR125b-5p has an anti-adipogenic effect, most likely through the regulation of further unknown pro-adipogenic target(s), which may superimpose the effect of MMP11.
MiR125b-5p is induced during adipogenesis
We identified miR125b-5p as upregulated immediately after experimental induction of human adipogenesis, which is in line with other studies showing that miR125b was upregulated in subcutaneous AT from obese compared to non-obese subjectsCitation24 and in subcutaneous AT of patients with multiple symmetric lipomatosis.Citation25 Although miR125b has been identified as a brain-enriched miRNA,Citation26 these data indicate that miR125b-5p may have an important role during murine and human adipogenesis. It is, however, unknown at which level miR125b affects adipogenesis. In humans, the miR125b consists of 2 paralogs,Citation27 which are transcribed from 2 different loci located on chromosomes 11q23 (miR125b-1) and 21q21 (miR125b-2), coding for the same mature sequence.
MiR125b-5p decreases MMP11 protein levels by direct interaction
Using PicTar algorithms, we identified MMP11 as promising target genes for miR125b-5p. MMP11 (previously called stromelysin 3) is a member of the MMP family and is expressed in association with intense tissue remolding during embryogenesis, tissue involution, wound healing or metamorphosis.Citation28 In contrast to most MMPs, MMP11 is processed intracellularly and secreted as an active enzyme.Citation28,29 Downregulation of MMP11 mRNA and protein during adipogenesis in parallel with increasing miR125b-5p expression supports the hypothesis that miR125b-5p may be a direct regulator of MMP11. Furthermore, MMP11 expression was low in the situation of high miR125b-5p expression in subcutaneous AT samples from children, suggesting a counter-regulatory role of miR125b-5p on MMP11 in human AT. We confirmed this hypothesis by showing that overexpression of miR125b-5p significantly repressed MMP11. PicTar predictions revealed 3 potential binding sites for miR125b-5p in the 3′UTR of MMP11. Particularly the second miR125b-5p binding site showed a high degree of structural conservation across the different species, which lead to the assumption that this binding site could be critical for the miR125b-5p mediated downregulation of MMP11. Nonetheless, in our study all 3 binding sites were essential for the interaction and mutation of any single site abolished the regulatory effect of miR125b-5p on MMP11 expression.
MMP11 is a negative regulator of adipogenesis
Based on our findings of direct repression of MMP11 by miR125b-5p, we were interested whether MMP11 itself has an effect on adipogenesis. Particularly, MMP11 overexpression led to a significant decrease in fat accumulation and molecular adipogenesis markers, suggesting that MMP11 plays a role in adipocyte metabolism. These results are in line with earlier studies in mice showing significantly higher body weights in MMP11 deficient mice compared to control littermates, which was apparently attributed to better differentiation capacity of embryonic fibroblasts in MMP11 deficient mice.Citation30 Our observation of a rather weak effect on differentiation capacity after MMP11 knockdown may be explained by a generally low MMP11 expression in SGBS cells. Similar to our results, knockdown of other MMPs such as gelatinase B (MMP9) did not affect in vitro adipogenesis.Citation31 The mechanism and the timing by which MMP11 acts on adipogenesis are not fully clarified. It has been shown that MMP11 is not only able to reduce preadipocyte differentiation but also to revert mature adipocytes into preadipocytes.Citation32 Despite extensive research using various approaches, the specific substrate of MMP11 has not yet been found. It was suggested that MMP11 might have collagenolytic properties against the native α3 chain of collagen VI under normal and malignant conditions.Citation33 In addition, cancer cell-induced MMP11 expression by adjacent adipocytes led to their dedifferentiation,Citation34 suggesting that the event of adipocyte dedifferentiation might be the cleavage of collagen VI by MMP11. Apart from this assumption, results from our study indicate that MMP11 might play a role at early stages of adipogenesis, which is supported by the observation that MMP11 protein levels were downregulated at the end of adipogenesis. However, we cannot exclude that MMP11 could also play a role at later stages.
MiR125b-5p acts as a direct repressor of adipogenesis
Considering that the miR125 family has been shown to play a crucial role in cell differentiation, proliferation and apoptosis,Citation35 we were interested whether miR125b-5p directly affects adipogenesis. Considering the downregulation of the anti-adipogenic MMP11, an increased adipogenesis after miR125b-5p overexpression may have been expected. However, overexpression of miR125b-5p significantly repressed adipogenesis. Potential explanations for this finding include that each miRNA can interact with hundreds of targets and hence miR125b-5p may regulate other pro-adipogenic target genes that superimpose the effect of MMP11.Citation36,37 In 3T3 preadipocytes, miR125b-5p affected adipogenesis and proliferation, at least partially through downregulating Smad4. In order to investigate if this is also true for human adipogenesis, we overexpressed miR125b-5p in preadipocytes and analyzed the global mRNA expression via Illumina BeadChip Microarray. However, Smad4 was not downregulated, indicating that it is no direct target of miR125b-5p during human adipogenesis.Citation38 Nonetheless, we identified CCAAT/enhancer-binding protein β (CEBPB), CCAAT/enhancer-binding protein delta (CEBPD), and early B-cell factor 1 EBF1 as significantly downregulated (data not shown) after miR125b-5p overexpression. However, we did not find miR125b-5p binding sites in these genes. The confirmation of further miR125b-5p targets with a pro-adipogenic effect is further complicated as we do not know if miR125b-5p regulated these targets itself or earlier steps of adipogenesis. First, it might be possible that miR125b-5p down regulates a target which has an activating effect on C/EBP-β or C/EBP-δ. At this point it is pure speculation what target this could be. For example C/EBP-ß expression depends on other genes that are also essential to adipogenesis, such as cAMP responsive element binding protein 1 (CREB1) and the early growth response protein 2 (EGR2).Citation39 Egr2 (also called Krox20) is induced early in 3T3-L1 adipogenesis and promotes C/EBP-β expression. Decreasing amounts of Krox20 reduces the ability of 3T3-L1 cells to differentiate.Citation40 However, PicTar algorithms did not reveal a putative miR125b-5p binding site for KROX20 as well as CREB1. Second, it might be possible that the unknown target is involved in early steps of adipogenesis, such as proliferation. Therefore, at this point we cannot conclusively answer the question concerning what pro-adipogenic target is involved in the miR125b-5p interaction.
Overall, our observations indicate that the miR125b-5p/MMP11 interaction may not represent a key regulation in the process of adipogenesis. Nevertheless, this interaction could be essential for other mechanisms. It has recently been shown that MMP11 is induced in AT by cancer cells as they invade their surrounding environment.Citation28,32 MMP11 negatively regulates adipogenesis which in turn leads to the accumulation of non-malignant peritumoral fibroblast-like cells favoring cancer cell survival and tumor progression.Citation32 Possibly, the miR125b-5p/MMP11 axis might play a role in the MMP11-mediated bi-directional crosstalk between invading cancer cells and adjacent adipocytes.
In conclusion, we identified miR125b-5p as a regulator of human adipogenesis. One of the targets of miR125b-5p is MMP11, which confers slightly pro-adipogenic effects. The direct anti-adipogenic effect of miR125b-5p implies that an additional target of miR125b-5p affects human adipogenesis.
Methods
Cell culture and adipocyte differentiation
We applied the human preadipocyte cell line Simpson-Golabi-Behmel syndrome (SGBS), which was a generous gift from Martin Wabitsch (Ulm, Germany).Citation16 Cells were cultured in Dulbecco's modified Eagle medium (DMEM/HAM F12) (Life Technologies, Karlsruhe, Germany) supplemented with 10%FCS, 33 µmol/l biotin, 17 µmol/l pantothenic acid. SGBS cells were differentiated into mature adipocytes as previously described.Citation41 To quantify differentiation capacity, differentiated adipocytes were fixed at day 12 post-induction in Roti-Histofix 4% (Carl Roth GmbH, Karlsruhe, Germany), washed with PBS and stained with Oilred-O-solution (0.3% in 60% isopropanol, Sigma, St. Louis, USA) for 15 min. After washing with PBS, Oilred-O-solution was extracted by incubation with isopropanol, and quantified at 540 nm using the FLUOstar OTIMA (BMG LABTECH, Offenburg, Germany).
Primary adipocytes of the Leipzig Adipose Tissue Childhood Cohort
Subcutaneous adipose tissue (AT) samples were obtained from the Leipzig Adipose Tissue Childhood Cohort as previously describedCitation42 encompassing 100 Caucasian children (0–18 years) undergoing elective orthopedic surgery (n = 77), herniotomy/orchidopexy (n = 10), or abdominal, thoracic or back surgery (n = 13). Children were free of severe diseases and medication potentially influencing AT biology. The following exclusion criteria were applied: diabetes, generalized inflammation, malignant disease, genetic syndromes, or permanently immobilized children. Written informed consent was obtained from all parents. The study was approved by the local ethics committee (265–08, 265–08-ff) and is registered in the National Clinical Trials database (NCT02208141).Citation42
RNA extraction and qRT-PCR analyses
Total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, USA) and reverse transcribed using miScript II RT Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. qRT-PCRs of MMP11, peroxisome proliferator-activated receptor gamma (PPARg), fatty acid binding protein 4 (FABP4) and adipose triglyceride lipase (ATGL) were performed with 2xqPCR MasterMix Plus Low ROX (Eurogentec, Seraing, Belgium), 900nM of each primer and 200nM probe using the ABI 7500 Real-Time PCR system (Applied Biosystems, Darmstadt, Germany). Mature miR125b-5p and miR125b-3p RNA were reverse transcribed using the miScript II RT Kit (Qiagen). For miR125b qRT-PCR primer (hs-miR-125b_1 and hsmiR-125b_1*_1, Qiagen) and the miScript SYBR Green PCR Kit (Qiagen) were used according to the manufacturer's instructions. For standardization, target gene expression was normalized to the mean of the 2 housekeeping genes: β-actin (ACTB), and TATAbox-binding protein (TBP).
MMP11: F5′-CGGGCGCTGGGAGAA3′, R5′-TCACATCGCTCCATACCTTTAGG-3′, Probe 5-ACGGACCTCACCTACAGGATCCTTCGG-3´; PPARg: F5′-GATCCAGTGGTT GCAGATTACAA-3′, R5′-GAGGGAGTTGGAAGGCTCTTC-3′, Probe 5-TGACCTGAAACTTCAAGAGTACCAAAGTGCAA-3´; FABP4: F5′-GGAAAATCAACCACCATAAAGAGAA-3′, R5′-GGA AGTGACGCCTTTCATGAC-3′, Probe 5-CATTCCACCACCAGTTTATCATCCTCTCGT-3; ACTB: F5′-CGACGCGGCTACAGCTT-3′, R5′-CCTTAATGTCACGCACG ATTT-3′, Probe 5-ACCACCACGGCCGAGCGG-3; TBP: F5′-TTGTAAACTTGACCTAAAGACCATTGC-3′, R5′-TTCGTGGCTCTCTTATCCTCATG-3′, Probe 5-AACGCCGAATATAATCCCAAGCGGTTG-3. Expression analyses for ATGL were performed with TaqMan Gene Expression Assay (Hs00386101_m1, Applied Biosystem, Darmstadt, Germany). For RNA analyses of primary human adipocytes, AT samples were separated as previously described.Citation42
MMP11 and miR125b-5p overexpression and knockdown
Preadipocytes were transfected using the Neon Transfection System 100 μl Kit (Invitrogen) according to Bernhard et al.Citation43 For MMP11 overexpression, 1.25 µg of expression plasmid (Ambion, Abingdon, UK) were transfected and empty pCMV6-XL5 vector was used as a control. For miR125b-5p overexpression, 500 nmol of pre-miR125b-5p (Ambion) or a scrambled negative control (pre-ntRNA, Ambion) were transfected. For MMP11 and PPARg knockdown experiments, gene-specific ON-TARGETplus SMARTpool small interfering (si)RNAs and ON-TARGETplus control reagents (Dharmacon, Lafayette, LA, USA) were used at a final concentration of 500 nmol/l. The downregulation of miR125b-5p was carried out by transfection with antagonizing miR125b-5p (anti-miR125-5p, Ambion) or scrambled negative control RNA (anti-ntRNA, Ambion). After electroporation 350,000 cells were seeded in 6-well format and differentiated. Efficient knockdown was confirmed on day 0 and 12 post-induction by qRT-PCR.
Luciferase reporter assay
To generate MMP11 luciferase reporter constructs, fragments of putative miR125b-5p binding sites were cloned into the 3′UTR region using HindIII-SpeI restriction sites of the pMIR-report plasmid (Ambion, Carlsbad, USA) (). Correct insertion was confirmed by sequencing. For dual luciferase assays, 3×105 SGBS cells/ml were co-transfected with 0.2 µg of firefly luciferase reporter plasmid, 0.1 µg of a control vector containing Renilla luciferase (Promega, Fitchburg, Madison, USA), and 500nmol of pre-miR125b-5p (Ambion) or scrambled negative control RNA (pre-ntRNA, Ambion) 24h post-transfection. Luciferase activity was quantified using Dual Luciferase Assay System (Promega) according to the manufacturer's instructions. Putative miR125b-5p binding sites were mutated using site-directed mutagenesis primers.
MMP11 mutagenesis Primer (1): F5′-TGGCTTGGATGCCCTAATGAGTGCTGACCCCTGCC-3′, R5′-GGCAGGGGTCAGCACTCATTAGGGCATCCAAGCCA-3′; MMP11 Mutagenesis Primer (2): F5′-CTGAGCCCATGTCTCCTAATGAGGATGGGGTGGGGT-3′, R5′-ACCCCACCCCATCCTCATTAGGAGACATGGGCTCAG-3′; MMP11 Mutagenesis Primer (3): F5′-TCTCCATCCCTGTCCCTAATGATAGCACCATGGCAGGAC-3′, R5′-GTCCTGCCATGGTGCTATCATTAGGGACAGGG ATGGAGA-3′.
Western blot analysis of MMP11
Adipocytes were lysed in RIPA buffer (50 mM Tris-HCL pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 1 mM PMSF, 0.25% Na-deoxycholate, 1mM Na3VO4, 1 mM NaF, 1 µg/ml aprotinin, 1 µg/ml leupeptin, 1 µg/ml pepstatin), incubated for 10 min on ice, and centrifuged at 12,000xg for 10 min at 4°C. Samples (30 µg) were separated on 12% SDS-polyacrylamide gels and transferred to a polyvinylidene difluoride membrane. Membranes were blocked for 1h at room temperature with 5% non-fat-milk in Tris-buffered saline plus 0.1% Tween20, followed by overnight incubation at 4°C with polyclonal rabbit α MMP11 antibody (1:1000, M0806, Sigma) or monoclonal α β-Actin (1:5000, A5316, Sigma). Signals were detected using a horseradish peroxidase (HRP) conjugated secondary antibody, followed by incubation with Pierce Super Signal West Femto Trial Kit (Thermo Scientific).
Bioinformatic and sequencing analysis
Putative miR125b binding sites in the human MMP11 3′UTR were identified with PicTar (MDC, Berlin, Germany) and TargetScanHuman (Whitehead Institute for Biomedical Research, Cambridge, UK) database. For the identification of differentially expressed miRNAs, we differentiated SGBS preadipocytes and analyzed the global miRNA expression profile via high-throughput sequencing in 2 independent differentiation experiments. For this, 500 ng of RNA were used in the small RNA protocol with the TruSeq™ Small RNA sample prepkit v2 (Illumina) according to the instructions of the manufacturer. Data analysis was performed according to Stokowy et al.Citation44 Microarray data were confirmed by qRT-PCR.
Statistical analyses
Data are presented as mean ± SEM of at least 3 independent experiments measured in duplicates. Differences were analyzed by Student's t-test, one-way ANOVA with post-hoc Dunnett's test or 2-way ANOVA with Bonferroni correction for multiple comparisons. Statistical analysis of the data was performed using GraphPad Prism 5 software (GraphPad Software Inc., La Jolla, USA).
Table 1. Characteristics of the Childhood Adipose Tissue Cohort (n = 100) after stratification in tertiles according to miR125b-5p mRNA levels.
Abbreviations
| miRNA | = | microRNAs |
| MMP11 | = | matrix metalloproteinase 11 |
| SGBS preadipocytes | = | Simpson-Golabi-Behmel syndrome preadipocytes |
| UTRs | = | untranslated regions |
Disclosure of potential conflicts of interest
The authors do not claim any conflict of interest or competing financial interests.
Acknowledgments
High-Throughput Sequencing was conducted at the Core Unit DNA Technologies of the IZKF Leipzig at the Faculty of Medicine of the University Leipzig (Project Z03).
All authors are responsible for reported research and have participated in the concept and design, analysis and interpretation of data, drafting or revising, and have approved this manuscript as submitted.
Funding
This work was supported by grants from the German Research Council (DFG) for the Clinical Research Center “Obesity Mechanisms” CRC1052/1 C05 and the Federal Ministry of Education and Research (BMBF), Germany, FKZ: 01EO1001 (IFB AdiposityDiseases ADI K7-10 and ADI K7-11 to A.K.).
References
- Gregoire FM, Smas CM, Sul HS. Understanding adipocyte differentiation. Physiol Rev 1998; 78:783-809; PMID:9674695
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development 2005; 132:4653-62; PMID:16224045; http://dx.doi.org/10.1242/dev.02073
- Krützfeldt J, Stoffel M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab 2006; 4:9-12; PMID:16814728; http://dx.doi.org/10.1016/j.cmet.2006.05.009
- Lee CT, Risom T, Strauss WM. MicroRNAs in mammalian development. Birth Defects Res C Embryo Today 2006; 78:129-39; http://dx.doi.org/10.1002/bdrc.20072
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97; PMID:14744438; http://dx.doi.org/10.1016/S0092-8674(04)00045-5
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science 2005; 309:1519-24; PMID:16141061; http://dx.doi.org/10.1126/science.1111444
- Berezikov E, Guryev V, van de BJ, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 2005; 120:21-4; PMID:15652478; http://dx.doi.org/10.1016/j.cell.2004.12.031
- Gao FB. Posttranscriptional control of neuronal development by microRNA networks. Trends Neurosci 2008; 31:20-6; PMID:18054394; http://dx.doi.org/10.1016/j.tins.2007.10.004
- Callis TE, Deng Z, Chen JF, Wang DZ. Muscling through the microRNA world. Exp Biol Med (Maywood) 2008; 233:131-8
- Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ, Li X. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad Sci U S A 2008; 105:2889-94; PMID:18287052; http://dx.doi.org/10.1073/pnas.0800178105
- Romao JM, Jin W, Dodson MV, Hausman GJ, Moore SS, Guan LL. MicroRNA regulation in mammalian adipogenesis. Exp Biol Med (Maywood) 2011; 236:997-1004
- Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, et al. MicroRNA-143 regulates adipocyte differentiation. JBiolChem 2004; 279:52361-5
- Karbiener M, Fischer C, Nowitsch S, Opriessnig P, Papak C, Ailhaud G, Dani C, Amri EZ, Scheideler M. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem Biophys Res Commun 2009; 390:247-51
- Lee EK, Lee MJ, Abdelmohsen K, Kim W, Kim MM, Srikantan S, Martindale JL, Hutchison ER, Kim HH, Marasa BS, et al. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol 2011; 31:626-38
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 2007; 315:1576-9; PMID:17322030; http://dx.doi.org/10.1126/science.1137999
- Wabitsch M, Brenner RE, Melzner I, Braun M, Moller P, Heinze E, Debatin KM, Hauner H. Characterization of a human preadipocyte cell strain with high capacity for adipose differentiation. Int J Obes Relat Metab Disord 2001; 25:8-15; http://dx.doi.org/10.1038/sj.ijo.0801520
- Carrer M, Liu N, Grueter CE, Williams AH, Frisard MI, Hulver MW, Bassel-Duby R, Olson EN. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc Natl Acad Sci U S A 2012; 109:15330-5; http://dx.doi.org/10.1073/pnas.1207605109
- Meerson A, Traurig M, Ossowski V, Fleming JM, Mullins M, Baier LJ. Human adipose microRNA-221 is upregulated in obesity and affects fat metabolism downstream of leptin and TNF-α. Diabetologia 2013; 56:1971-9; PMID:23756832; http://dx.doi.org/10.1007/s00125-013-2950-9
- Nielsen LB, Wang C, Sorensen K, Bang-Berthelsen CH, Hansen L, Andersen ML, Hougaard P, Juul A, Zhang CY, Pociot F, et al. Circulating levels of microRNA from children with newly diagnosed type 1 diabetes and healthy controls: evidence that miR-25 associates to residual β-cell function and glycaemic control during disease progression. Exp Diabetes Res 2012; 2012:896362. Epub;%2012 Jul 5.:896362.
- Ortega FJ, Mercader JM, Catalan V, Moreno-Navarrete JM, Pueyo N, Sabater M, Gomez-Ambrosi J, Anglada R, Fernandez-Formoso JA, Ricart W, et al. Targeting the circulating microRNA signature of obesity. Clin Chem 2013; 59:781-92
- Sun T, Fu M, Bookout AL, Kliewer SA, Mangelsdorf DJ. MicroRNA let-7 regulates 3T3-L1 adipogenesis. Mol Endocrinol 2009; 23:925-31; PMID:19324969; http://dx.doi.org/10.1210/me.2008-0298
- Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011; 474:649-53; PMID:21654750; http://dx.doi.org/10.1038/nature10112
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet 2005; 37:495-500
- Ortega FJ, Moreno-Navarrete JM, Pardo G, Sabater M, Hummel M, Ferrer A, Rodriguez-Hermosa JI, Ruiz B, Ricart W, Peral B, et al. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoSOne 2010; 5:e9022; http://dx.doi.org/10.1371/journal.pone.0009022
- Chen K, He H, Xie Y, Zhao L, Zhao S, Wan X, Yang W, Mo Z. miR-125a-3p and miR-483-5p promote adipogenesis via suppressing the RhoA/ROCK1/ERK1/2 pathway in multiple symmetric lipomatosis. SciRep 2015; 5:11909.
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 2004; 5:R13; PMID:15003116; http://dx.doi.org/10.1186/gb-2004-5-3-r13
- Shaham L, Binder V, Gefen N, Borkhardt A, Izraeli S. MiR-125 in normal and malignant hematopoiesis. Leukemia 2012; 26:2011-8; PMID:22456625; http://dx.doi.org/10.1038/leu.2012.90
- Rio MC. From a unique cell to metastasis is a long way to go: clues to stromelysin-3 participation. Biochimie 2005; 87:299-306; PMID:15781316; http://dx.doi.org/10.1016/j.biochi.2004.11.016
- Amano T, Fu L, Sahu S, Markey M, Shi YB. Substrate specificity of Xenopus matrix metalloproteinase stromelysin-3. Int J Mol Med 2004; 14:233-9
- Lijnen HR, Van HB, Frederix L, Rio MC, Collen D. Adipocyte hypertrophy in stromelysin-3 deficient mice with nutritionally induced obesity. Thromb Haemost 2002; 87:530-5
- Bauters D, Van Hul M, Lijnen HR. Gelatinase B (MMP-9) gene silencing does not affect murine preadipocyte differentiation. Adipocyte 2014; 3:50-3; PMID:24575369; http://dx.doi.org/10.4161/adip.26966
- Andarawewa KL, Motrescu ER, Chenard MP, Gansmuller A, Stoll I, Tomasetto C, Rio MC. Stromelysin-3 is a potent negative regulator of adipogenesis participating to cancer cell-adipocyte interaction/crosstalk at the tumor invasive front. Cancer Res 2005; 65:10862-71; PMID:16322233; http://dx.doi.org/10.1158/0008-5472.CAN-05-1231
- Motrescu ER, Blaise S, Etique N, Messaddeq N, Chenard MP, Stoll I, Tomasetto C, Rio MC. Matrix metalloproteinase-11/stromelysin-3 exhibits collagenolytic function against collagen VI under normal and malignant conditions. Oncogene 2008; 27:6347-55; PMID:18622425; http://dx.doi.org/10.1038/onc.2008.218
- Motrescu ER, Rio MC. Cancer cells, adipocytes and matrix metalloproteinase 11: a vicious tumor progression cycle. Biol Chem 2008; 389:1037-41
- Sun YM, Lin KY, Chen YQ. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol 2013; 6:6.
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature 2008; 455:64-71; PMID:18668037; http://dx.doi.org/10.1038/nature07242
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008; 455:58-63; PMID:18668040; http://dx.doi.org/10.1038/nature07228
- Ouyang D, Ye Y, Guo D, Yu X, Chen J, Qi J, Tan X, Zhang Y, Ma Y, Li Y. MicroRNA-125b-5p inhibits proliferation and promotes adipogenic differentiation in 3T3-L1 preadipocytes. Acta Biochim Biophys Sin (Shanghai) 2015; 47:355-61; PMID:25918183; http://dx.doi.org/10.1093/abbs/gmv024
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 2006; 7:885-96; PMID:17139329; http://dx.doi.org/10.1038/nrm2066
- Chen Z, Torrens JI, Anand A, Spiegelman BM, Friedman JM. Krox20 stimulates adipogenesis via C/EBPbeta-dependent and -independent mechanisms. Cell Metab 2005; 1:93-106; PMID:16054051; http://dx.doi.org/10.1016/j.cmet.2004.12.009
- Friebe D, Loffler D, Schonberg M, Bernhard F, Buttner P, Landgraf K, Kiess W, Korner A. Impact of metabolic regulators on the expression of the obesity associated genes FTO and NAMPT in human preadipocytes and adipocytes. PLoS One 2011; 6:e19526; http://dx.doi.org/10.1371/journal.pone.0019526
- Landgraf K, Rockstroh D, Wagner IV, Weise S, Tauscher R, Schwartze JT, Löffler D, Bühligen U, Wojan M, Till H, et al. Evidence of early alterations in adipose tissue biology and function and its association with obesity-related inflammation and insulin resistance in children. Diabetes 2015; 64(4); 1249–61.
- Bernhard F, Landgraf K, Klöting N, Berthold A, Büttner P, Friebe D, Kiess W, Kovacs P, Blüher M, Körner A. Functional relevance of genes implicated by obesity genome-wide association study signals for human adipocyte biology. Diabetologia 2013; 56:311-22; PMID:23229156; http://dx.doi.org/10.1007/s00125-012-2773-0
- Stokowy T, Eszlinger M, Wierniak M, Fujarewicz K, Jarz BB, Paschke R, Krohn K. Analysis options for high-throughput sequencing in miRNA expression profiling. BMCResNotes 2014; 7:144.
