ABSTRACT
Individual cell types vary enormously in the amount of different organelles they contain. One such organelle is the mitochondrion. Understanding how mitochondrial levels are controlled is essential since so many disease states seem to involve mitochondrial function. The beige adipocyte is an inducible form of adipocyte that emerges in response to cold exposure and some other external stimuli. To perform its thermogenic function, its level of mitochondria increases dramatically. If the stimuli are removed the mitochondrial levels return to base line. Following the withdrawal of external stimuli, beige adipocytes directly acquire a white fat-like phenotype through mitophagy-mediated mitochondrial degradation. The beige cell is therefore a dynamic model for studying the mechanism of mitochondrial biogenesis and degradation.
A dynamic model of mitochondrial biogenesis and degradation
Mitochondria misregulation occurs in human diseases, such as obesity, diabetes and cancer.Citation1 The beige cell is a type of cold-inducible, mitochondrian-rich adipocyte and appears in the white adipose tissue (WAT).Citation2 Adult human do not have interscapular brown adipose tissue (BAT), but showed brown-adipose-tissue activity in the supraclavicular region by static positron emission tomography of 18F-fluorodeoxyglucose in combination with computed tomography (18F-FDG PET-CT).Citation3,Citation4 The BAT activity is significantly related with cold-challenge, body mass index and age, which are actually beige adipocytes.Citation5,Citation6 It can be distinguished from the classical interscapular brown adipose tissue (iBAT). Classical brown adipocytes develop prenatally from a Myf5, Pax7, Engrailed-1 positive progenitor population.Citation7–Citation9 Beige adipocytes postnatally develop from PDGFα/β, Ebf2, Sca-1, SMA positive progenitor population.Citation10-Citation13 Mitochondria biogenesis in white adipose tissue (WAT) can be activated by cold or β3 adrenergic agonist (β3-AR).Citation14 In the past decade, research interests were intensely focused on the transcriptional control of those mitochondrial-rich adipocyte. Transcription factors PRDM16, PGC-1α, et al, switch on beige adipose specific gene transcription and protein expression, thus promote mitochondria biogenesis in WAT.Citation15,Citation16 Recently, a new type of glycolytic beige adipocyte is found from β-adrenergic receptor less mouse under chronic cold adaptation, in which presented enhanced glucose oxidation.Citation17 The thermal stress induced progenitor cell plasticity in adipose tissue is still on the beginning of its discoveries.
Adipose tissue is able to sense environmental temperature and secretary factors.Citation18 Growth factors are regulating fat storage and fatty acid transport. Beige cell can be induced by inhibiting several cellular growth factors signaling pathways such as those controlled by VEGF-A, VEGF-B, PDGFRα and TGFβ pathway.Citation19-Citation21 Exercise training results in adaptations of mitochondria biogenesis in both skeletal muscle and subcutaneous adipose.Citation22-Citation24 At the epigenetic level, methylation or acetylation enzymes are important in sensing and regulating the differentiation of beige cell. For example, EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Beige cell can be induced by the external β3-AR agonist CL316,243 in WAT of wild type mice, but not in the adipose specific EMHT1 knockout mice.Citation25 The Sirtuin family is a class of stress responsive protein deacetylase and mono-ADP ribosyltransferase enzymes. The Chang lab has discovered that Sirt1 is playing a major role in high-fat-diet induced liver metabolic damage.Citation26 Cold-inducible Sirt6 regulates thermogenesis in both brown and beige fat.Citation27
Beige-to-white transition
The balance between energy-storage and energy-expenditure is always a systematic regulation process. When normal conditions are restored, the enriched mitochondria in beige cells disappear. After withdrawal of cold or β3-AR agonist stimuli, there always follows mitochondria degradation and the beige-to-white transition in WAT.Citation28 The mitochondria uncoupling protein UCP1 and the oxidative phosphorylation complex OXPHOS are specific markers of mitochondria formation during the transition to beige adipocytes. As the Kajimura lab reported recently, the expression of mitochondria marker UCP1 and OXPHOS were both rapidly decreased in the beige adipocyte after withdrawing stimuli.Citation29 During beige-to-white adipocyte transition, gene-annotation enrichment analysis found that the majority of the cluster I gene was related to mitochondria in cellular components, the electron transport chain and oxidation reduction related biological process.Citation14 Thus, the phenotype of mitochondria degradation right after withdrawal of stimuli is well established in mouse inguinal adipose tissue.
The mitochondrial uncoupling protein UCP1 is an important marker for brown adipose tissue as well as beige adipose, and has been linked to thermogenesis. However beige fat thermogenesis is UCP1 dispensable. A recent study has discovered UCP1-independent thermogenesis pathways specifically in beige adipose. SERCA2b-mediated calcium cycling regulates UCP1-independent thermogenesis.Citation29 Enhanced Ca2+ cycling by activation of α1 – and/or β3-adrenergic receptors or the SERCA2b-RyR2 pathway stimulates UCP1-independent thermogenesis in beige adipocytes. The beige adipocytes of UCP1 deficient mice presented enhanced glycolysis, tricarboxylic acid metabolism and pyruvate dehydrogenase activity for ATP-dependent thermogenesis. In UCP1 knockout mice, beige cell can be highly induced by β3-AR agonist CL316,243, as well as mitochondrial degradation directly acquired after stimuli withdraw.Citation28 The oxygen consumption rate (OCR) of WAT decreased to wild type OCR level after 15 days, which presented a UCP1-independent manner in the beige-to-white transition. Thus, beige cell biogenesis and degradation of mitochondria is UCP1-independent.
The majority of studies about mitochondrial integrity are in mice for which the role of housing temperature in determining the relevance of any outcomes should be considered. To remove the temperature induced differences, a comparison in ambient temperature and in thermoneutrality (30°C) could be set up respectively for the animal study of mitochondria clearance in beige adipose tissue.Citation28
Mitophagy controls mitochondrial quantity and quality
Mitophagy is the degradation of mitochondria by autophagy.Citation30 To maintain the integrity and function of cells, it is important to eliminate damaged and aged mitochondria.Citation31-Citation33 During the beige-to-white transition, mitochondria are degraded in the adipose tissue by activation of autophagy.Citation14 Transcriptional regulators of mitochondrial biogenesis Pgc-1α, Nrf1/2, Tfam, et al. directly decrease in the early phase of beige-to-white transition.Citation34-Citation36 Changes in the autophagy and lysosome pathways were highly relevant in the gene enrichment analysis. Based on some sets of gene profiling data, autophagy related genes Atg5, Atg4b, Atg12, Atg16 were increased in the transition process.Citation37-Citation40 In others, autophagy related components and lysosomal enzyme related genes Cts, Arsg, Naga were also highly increased during the transition.Citation41-Citation43 The microtubule-associated protein 1A/1B-light chain 3 (LC3) is known to form a stable association with the membrane of autophagosomes.Citation44,Citation45 When GFP-LC3 mice are employed in the experiments, GFP-LC3 and mitochondria marker Tom20 co-localization indicates autophagosome formation. After 7 days of CL316,243 treatment to induce the beige phenotype, the number of GFP-LC3 punctate was significantly decreased in the beige adipose. 15 days after stopping the treatment, the autophagic flux was back to a natural level. LC3-II protein expression was consistent with the co-localization result. The degradation of LC3-II has indicated that autophagy/ mitophagy process is attenuated by external β3-AR stimuli in beige cell.
Mitophagy plays a major role in beige cell mitochondria clearance. In a recent study, we employed the genetic model of mt-Keima mouse, which specifically expresses Keima protein in the mitochondria.Citation29 The coral derived fluorescent protein Keima senses environmental pH value, giving green fluorescence in a regular cellular environment.Citation46,Citation47 In mitophagy, when the lysosome structure is formed and an acidic environment is generated, Keima protein gives a red fluorescence. The red to green fluorescence signal ratio can be precisely quantified by flow cytometry. The mt-Keima mouse presented a high level mitophagy-red/green ratio in white adipose tissue. After injecting mice for 7 days with the β3-AR agonist CL316,243 beige adipocyte biogenesis and attenuated mitophagy were induced in the inguinal adipose tissue and the red/green ratio shifted back to normal. After withdraw of the stimuli, the mitophagy signal gradually increases and recovers to regular level in 15–30 days. Thus using the mt-Keima mouse to measure the adipose tissue mitophagy stage or level reveals that the external β3-AR stimuli also induces beige cell biogenesis by attenuating the high levels of mitophagy that occur in normal WAT. Recently, Shirihai lab demonstrated that peridroplet mitochondria have higher pyruvate oxidation and ATP synthesis capacity, with unique structure and function that supports triacylglyceride synthesis.Citation48 It opens up a new area in mitochondria biology. Meanwhile, the mitophagy levels between peridroplet mitochondria and cytoplasmic mitochondria in different environmental or physiological conditions could be characterized by the mt-Keima model.
PINK-Parkin signaling is the key regulator of mitochondrial degradation.Citation49 Loss of mitochondrial membrane potential (depolarization) leads to PTEN-induced putative kinase 1 (PINK1) accumulation on the mitochondrial outer membrane.Citation50 PINK1 recruits Parkin, an E3 ubiquitin-protein ligase, which ubiquitinates proteins on the outer mitochondrial membrane and starts the autophagic degradation of dysfunctional mitochondria. At a cellular level, PINK1-Parkin mediated mitophagy can be visualized by FCCP or CCCP treatment in the Parkin over-expressed model.Citation51 CCCP is able to induce Parkin translocation to the mitochondria in mature beige adipocytes.Citation28 Accumulation of damaged mitochondria has been linked to Parkinson’s disease, Alzheimer’s disease, diabetes and age-related disorders, but the mechanism of accumulated mitochondria clearance in adipose tissue is still not clear.Citation52-Citation55 In a study of Drosophila, Parkin mutant flies significantly decreased the rate of mitochondrial protein turnover.Citation56 In the mouse adipose beige-to-white transition, Parkin also mediates the mitochondria clearance. After external β3-AR stimuli, beige cells can be induced in both wild type mice and Parkin deficient mice.Citation28 15 days after withdrawing the stimuli, beige cells disappeared in the wild type mouse, but still existed in the WAT of Parkin deficient mouse. When the oxygen consumption rate (OCR) was measured by Seahorse method the Parkin deficient mouse sustained a significant high level OCR than the wild type mouse in both basal and isoproterenol stimulated condition, which indicates that Parkin-dependent mitophagy is the key mechanism in beige cell mitochondrial clearance.
Control mitochondrial clearance by phosphorylating parkin
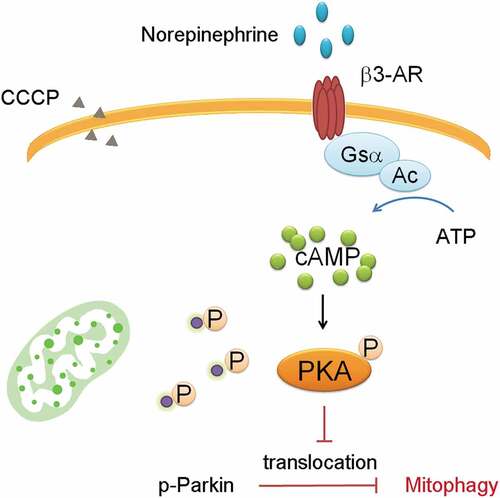
Parkin-mediated mitophagy plays a major role in beige cell mitochondrial clearance. In the CCCP induced-mitophagy beige cell model, Parkin translocation can directly be visualized. Norepinephrine can stimulate the cAMP signaling through β3-AR, activating PKA phosphorylation which phosphorylates downstream proteins.Citation6 Parkin translocation can be attenuated by pre-treatment of norepinephrine stimulated β3-AR signaling, presumably inhibiting mitophagy.Citation28 Pre-treatment with PKA inhibitors prevents the attenuated translocation when Parkin phosphorylation is preventing mitochondria degradation. In immunoprecipitation assay, norepinephrine induces Parkin phosphorylation which can also be blocked by the PKA inhibitors. The results suggest that Parkin recruitment into the mitochondria can be regulated by PKA signaling ().
Recently, the Daumke and Wang labs have independently discovered that the structural plasticity of mitochondrial crista junctions is controlled by MIC60/Mitofilin.Citation57,Citation58 PINK1 phosphorylates inner mitochondrial membrane protein MIC60, which stabilizes MIC60 oligomerization. An earlier research was also focused on this pathway and found that PKA activation reduces PINK1 protein levels through phosphorylation of MIC60 and prevents the recruitment of Parkin to the mitochondria.Citation59 Nevertheless, the function of MIC60/Mitofilin in beige cell mitochondria clearance is still unclear. Temporal inhibition of the MIC60-Parkin axis in the adipose tissue could be a novel approach to retain thermogenic beige adipocytes.
Conclusions
A powerful tool to understand how mitochondrial levels are controlled is the beige adipocyte, an inducible regulator of thermogenesis. In the white to beige transition, mitochondrial levels are increased. In the beige to white transition, they are lowered. In both cases, the change in mitochondrial level is due to both expression of mitochondrial genes and the control of mitophagy. The uncoupling marker UCP1 does not play an important role in this regulation. Using single cell measurements of RNA levels, a complete description of the control of mitochondrial levels in adipocytes should soon be available. Such data should provide insights into how mitochondrial levels are regulated and into metabolism associated human diseases.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Ackowledgements
The author is grateful to S. Kajimura and R. B. Kelly (UCSF) for their scientific discussions.
Additional information
Funding
References
- Sung H, Siegel RL, Torre LA, Pearson-Stuttard J, Islami F, Fedewa SA, Goding Sauer A, Shuval K, Gapstur SM, Jacobs EJ, et al. Global patterns in excess body weight and the associated cancer burden. CA Cancer J Clin. 2018. doi:10.3322/caac.21499.
- Sponton CH, Kajimura S. Multifaceted roles of beige fat in energy homeostasis beyond UCP1. Endocrinology. 2018;159(7):2545–2553. doi:10.1210/en.2018-00371.
- Ong FJ, Ahmed BA, Oreskovich SM, Blondin DP, Haq T, Konyer NB, Noseworthy MD, Haman F, Carpentier AC, Morrison KM, et al. Recent advances in the detection of brown adipose tissue in adult humans: a review. Clin Sci (Lond). 2018;132(10):1039–1054. doi:10.1042/CS20170276.
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360(15):1509–1517. doi:10.1056/NEJMoa0810780.
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360(15):1500–1508. doi:10.1056/NEJMoa0808718.
- Shinoda K, Luijten IH, Hasegawa Y, Hong H, Sonne SB, Kim M, Xue R, Chondronikola M, Cypess AM, Tseng YH, et al. Genetic and functional characterization of clonally derived adult human brown adipocytes. Nat Med. 2015;21(4):389–394. doi:10.1038/nm.3819.
- Sanchez-Gurmaches J, Hung CM, Sparks CA, Tang Y, Li H, Guertin DA. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metab. 2012;16(3):348–362. doi:10.1016/j.cmet.2012.08.003.
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102(6):777–786. doi:10.1016/S0092-8674(00)00066-0.
- Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev Biol. 2006;296(1):164–176. doi:10.1016/j.ydbio.2006.04.449.
- Lee YH, Petkova AP, Mottillo EP, Granneman JG. In vivo identification of bipotential adipocyte progenitors recruited by beta3-adrenoceptor activation and high-fat feeding. Cell Metab. 2012;15(4):480–491. doi:10.1016/j.cmet.2012.03.009.
- Stine RR, Shapira SN, Lim HW, Ishibashi J, Harms M, Won KJ, Seale P. EBF2 promotes the recruitment of beige adipocytes in white adipose tissue. Mol Metab. 2015;5(1):57–65. doi:10.1016/j.molmet.2015.11.001.
- Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci USA. 2011;108(1):143–148. doi:10.1073/pnas.1010929108.
- Berry DC, Jiang Y, Graff JM. Mouse strains to study cold-inducible beige progenitors and beige adipocyte formation and function. Nat Commun. 2016;7:10184. doi:10.1038/ncomms10184.
- Altshuler-Keylin S, Shinoda K, Hasegawa Y, Ikeda K, Hong H, Kang Q, Yang Y, Perera RM, Debnath J, Kajimura S. Beige adipocyte maintenance is regulated by autophagy-induced mitochondrial clearance. Cell Metab. 2016;24(3):402–419. doi:10.1016/j.cmet.2016.08.002.
- Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460(7259):1154–1158. doi:10.1038/nature08262.
- Hasegawa Y, Ikeda K, Chen Y, Alba DL, Stifler D, Shinoda K, Hosono T, Maretich P, Yang Y, Ishigaki Y, et al. Repression of adipose tissue fibrosis through a PRDM16-GTF2IRD1 complex improves systemic glucose homeostasis. Cell Metab. 2018;27(1):180–194.e6. doi:10.1016/j.cmet.2017.12.005.
- Chen Y, Ikeda K, Yoneshiro T, Scaramozza A, Tajima K, Wang Q, Kim K, Brack A, Kajimura S. Thermal stress induces glycolytic beige fat formation via a myogenic state. Nature. 2018. doi:10.1038/s41586-018-0801-z.
- Lu X, Ji Y, Zhang L, Zhang Y, Zhang S, An Y, Liu P, Zheng Y. Resistance to obesity by repression of VEGF gene expression through induction of brown-like adipocyte differentiation. Endocrinology. 2012;153(7):3123–3132. doi:10.1210/en.2012-1151.
- Jin H, Li D, Wang X, Jia J, Chen Y, Yao Y, Zhao C, Lu X, Zhang S, Togo J, et al. VEGF and VEGFB play balancing roles in adipose differentiation, gene expression, and function. Endocrinology. 2018;159(5):2036–2049. doi:10.1210/en.2017-03246.
- Gao Z, Daquinag AC, Su F, Snyder B, Kolonin MG. PDGFRalpha/PDGFRbeta signaling balance modulates progenitor cell differentiation into white and beige adipocytes. Development. 2018;145:1. doi:10.1242/dev.158527.
- Qian SW, Tang Y, Li X, Liu Y, Zhang YY, Huang HY, Xue RD, Yu HY, Guo L, Gao HD, et al. BMP4-mediated brown fat-like changes in white adipose tissue alter glucose and energy homeostasis. Proc Natl Acad Sci U S A. 2013;110(9):E798–807. doi:10.1073/pnas.1215236110.
- Tiano JP, Springer DA, Rane SG. SMAD3 negatively regulates serum irisin and skeletal muscle FNDC5 and peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1alpha) during exercise. J Biol Chem. 2015;290(12):7671–7684. doi:10.1074/jbc.M114.617399.
- Haczeyni F, Barn V, Mridha AR, Yeh MM, Estevez E, Febbraio MA, Nolan CJ, Bell-Anderson KS, Teoh NC, Farrell GC. Exercise improves adipose function and inflammation and ameliorates fatty liver disease in obese diabetic mice. Obesity (Silver Spring, Md). 2018;23(9):1845–1855. doi:10.1002/oby.21170.
- Aldiss P, Betts J, Sale C, Pope M, Budge H, Symonds ME. Exercise-induced ‘browning’ of adipose tissues. Metabolism. 2018;81:63–70. doi:10.1016/j.metabol.2017.11.009.
- Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature. 2015;504(7478):163–167. doi:10.1038/nature12652.
- Zhang Y, Geng C, Liu X, Li M, Gao M, Liu X, Fang F, Chang Y. Celastrol ameliorates liver metabolic damage caused by a high-fat diet through Sirt1. Mol Metab. 2016;6(1):138–147. doi:10.1016/j.molmet.2016.11.002.
- Yao L, Cui X, Chen Q, Yang X, Fang F, Zhang J, Liu G, Jin W, Chang Y. Cold-Inducible SIRT6 regulates thermogenesis of brown and beige fat. Cell Rep. 2017;20(3):641–654. doi:10.1016/j.celrep.2017.06.069.
- Lu X, Altshuler-Keylin S, Wang Q, Chen Y, Henrique Sponton C, Ikeda K, Maretich P, Yoneshiro T, Kajimura S. Mitophagy controls beige adipocyte maintenance through a Parkin-dependent and UCP1-independent mechanism. Sci Signal. 2018;11(527):pii: eaap8526. doi:10.1126/scisignal.aap8526.
- Ikeda K, Kang Q, Yoneshiro T, Camporez JP, Maki H, Homma M, Shinoda K, Chen Y, Lu X, Maretich P, et al. UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat Med. 2017;23(12):1454–1465. doi:10.1038/nm.4429.
- Pickles S, Vigié P, Youle RJ. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr Biol. 2018;28(4):R170–R185. doi:10.1016/j.cub.2018.01.004.
- Murrow L, Malhotra R, Debnath J. ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat Cell Biol. 2015;17(3):300–310. doi:10.1038/ncb3112.
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524(7565):309–314. doi:10.1038/nature14893.
- Chouchani ET, Kazak L, Jedrychowski MP, Lu GZ, Erickson BK, Szpyt J, Pierce KA, Laznik-Bogoslavski D, Vetrivelan R, Clish CB, et al. Mitochondrial ROS regulate thermogenic energy expenditure and sulfenylation of UCP1. Nature. 2016;532(7597):112–116. doi:10.1038/nature17399.
- Chang JS, Ghosh S, Newman S, Salbaum JM. A map of the PGC-1alpha- and NT-PGC-1alpha-regulated transcriptional network in brown adipose tissue. Sci Rep. 2018;8(1):7876. doi:10.1038/s41598-018-26244-4.
- Bhide S, Trujillo AS, O’Connor MT, Young GH, Cryderman DE, Chandran S, Nikravesh M, Wallrath LL, Melkani GC. Increasing autophagy and blocking Nrf2 suppress laminopathy-induced age-dependent cardiac dysfunction and shortened lifespan. Aging Cell. 2018;17(3):e12747. doi:10.1111/acel.12747.
- Masand R, Paulo E, Wu D, Wang Y, Swaney DL, Jimenez-Morales D, Krogan NJ, Wang B. Proteome imbalance of mitochondrial electron transport chain in brown adipocytes leads to metabolic benefits. Cell Metab. 2018;27(3):616–629 e614. doi:10.1016/j.cmet.2018.01.018.
- Kimmey JM, Huynh JP, Weiss LA, Park S, Kambal A, Debnath J, Virgin HW, Stallings CL. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature. 2015;528(7583):565–569. doi:10.1038/nature16451.
- Pengo N, Agrotis A, Prak K, Jones J, Ketteler R. A reversible phospho-switch mediated by ULK1 regulates the activity of autophagy protease ATG4B. Nat Commun. 2017;8(1):294. doi:10.1038/s41467-017-00303-2.
- Malhotra R, Warne JP, Salas E, Xu AW, Debnath J. Loss of Atg12, but not Atg5, in pro-opiomelanocortin neurons exacerbates diet-induced obesity. Autophagy. 2015;11(1):145–154. doi:10.1080/15548627.2014.998917.
- Varga K, Nagy P, Arsikin Csordas K, Kovacs AL, Hegedus K, Juhasz G. Loss of Atg16 delays the alcohol-induced sedation response via regulation of Corazonin neuropeptide production in Drosophila. Sci Rep. 2016;6:34641. doi:10.1038/srep34641.
- Frese MA, Schulz S, Dierks T. Arylsulfatase G, a novel lysosomal sulfatase. J Biol Chem. 2008;283(17):11388–11395. doi:10.1074/jbc.M709917200.
- Hsin MC, Hsieh YH, Wang PH, Ko JL, Hsin IL, Yang SF. Hispolon suppresses metastasis via autophagic degradation of cathepsin S in cervical cancer cells. Cell Death Dis. 2017;8(10):e3089. doi:10.1038/cddis.2017.459.
- Stara V, Navarova J, Ujhazy E, Gasparova Z. Progressive increase of lysosomal enzyme activities in hippocampus associated with reduction of population spike in a rat model of neurodegeneration. Neuro Endocrinol Lett. 2016;37:(Suppl1):111–117.
- Cunha LD, Yang M, Carter R, Guy C, Harris L, Crawford JC, Quarato G, Boada-Romero E, Kalkavan H, Johnson MDL, et al. LC3-Associated phagocytosis in myeloid cells promotes tumor immune tolerance. Cell. 2018;pii: S0092-8674(18)31122-X. doi:10.1016/j.cell.2018.08.061.
- Xu Q, Mariman ECM, Roumans NJT, Vink RG, Goossens GH, Blaak EE, Jocken JWE. Adipose tissue autophagy related gene expression is associated with glucometabolic status in human obesity. Adipocyte. 2018;7(1):12–19. doi:10.1080/21623945.2017.1394537.
- Sun N, Yun J, Liu J, Malide D, Liu C, Rovira II, Holmstrom KM, Fergusson MM, Yoo YH, Combs CA, et al. Measuring in vivo mitophagy. Mol Cell. 2015;60(4):685–696. doi:10.1016/j.molcel.2015.10.009.
- Sun N, Malide D, Liu J, Rovira II, Combs CA, Finkel T. A fluorescence-based imaging method to measure in vitro and in vivo mitophagy using mt-Keima. Nat Protoc. 2017;12(8):1576–1587. doi:10.1038/nprot.2017.060.
- Benador IY, Veliova M, Mahdaviani K, Petcherski A, Wikstrom JD, Assali EA, Acin-Perez R, Shum M, Oliveira MF, Cinti S, et al. Mitochondria bound to lipid droplets have unique bioenergetics, composition, and dynamics that support lipid droplet expansion. Cell Metab. 2018;27(4):869–885 e866. doi:10.1016/j.cmet.2018.03.003.
- Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ, Pallanck LJ. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc Natl Acad Sci USA. 2013;110(16):6400–6405. doi:10.1073/pnas.1221132110.
- Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510(7503):162–166. doi:10.1038/nature13392.
- Hasson SA, Kane LA, Yamano K, Huang CH, Sliter DA, Buehler E, Wang C, Heman-Ackah SM, Hessa T, Guha R, et al. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature. 2013;504(7479):291–295. doi:10.1038/nature12748.
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–608. doi:10.1038/33416.
- Herst PM, Rowe MR, Carson GM, Berridge MV. Functional Mitochondria in Health and Disease. Front Endocrinol (Lausanne). 2017;8:296. doi:10.3389/fendo.2017.00296.
- Stanford KI, Goodyear LJ. Muscle-Adipose tissue cross talk. Cold Spring Harb Perspect Med. 2018;8(8):pii: a029801. doi:10.1101/cshperspect.a029801.
- Ikeda K, Maretich P, Kajimura S. The common and distinct features of brown and beige adipocytes. Trends Endocrinol Metab. 2018;29(3):191–200. doi:10.1016/j.tem.2018.01.001.
- Zviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci USA. 2010;107(11):5018–5023. doi:10.1073/pnas.0913485107.
- Tsai PI, Papakyrikos AM, Hsieh CH, Wang X. Drosophila MIC60/mitofilin conducts dual roles in mitochondrial motility and crista structure. Mol Biol Cell. 2018;28(24):3471–3479. doi:10.1091/mbc.e17-03-0177.
- Tsai PI, Lin CH, Hsieh CH, Papakyrikos AM, Kim MJ, Napolioni V, Schoor C, Couthouis J, Wu RM, Wszolek ZK, et al. PINK1 phosphorylates MIC60/Mitofilin to control structural plasticity of mitochondrial crista junctions. Mol Cell. 2018;69(5):744–756 e746. doi:10.1016/j.molcel.2018.01.026.
- Akabane S, Uno M, Tani N, Shimazaki S, Ebara N, Kato H, Kosako H, Oka T. PKA regulates PINK1 stability and parkin recruitment to damaged mitochondria through phosphorylation of MIC60. Mol Cell. 2016;62(3):371–384. doi:10.1016/j.molcel.2016.03.037.