abstract
The recent approval of clincially effective immune checkpoint inhibitors illustrates the potential of cancer immunotherapy. A challenging task remains the identification of specific targets guiding immunotherapy. Facilitated by technical advances, the direct identification of physiologically relevant targets is enabled by analyzing the HLA ligandome of cancer cells. Since recent publications demonstrate the immunogenicity of ovarian cancer (OvCa), immunotherapies, including peptide-based cancer vaccines, represent a promising treatment approach. To identify vaccine peptides, we employed a combined strategy of HLA ligandomics in high-grade serous OvCa samples and immunogenicity analysis. Only few proteins were naturally presented as HLA ligands on all samples analyzed, including histone deacetylase (HDAC) 1 and 2. In vitro priming of CD8+ T cells demonstrated that two HDAC1/2-derived HLA ligands can induce T-cell responses, capable of killing HLA-matched tumor cells. High HDAC1 expression shown by immunohistochemistry in 136 high-grade serous OvCa patients associated with significantly reduced overall survival (OS), whereas patients with high numbers of CD3+ tumor-infiltrating lymphocytes (TILs) in the tumor epithelium and CD8+ TILs in the tumor stroma showed improved OS. However, correlating HDAC1 expression with TILs, high levels of TILs abrogated the impact of HDAC1 on OS. This study strengthens the role of HDAC1/2 as an important tumor antigen in OvCa, demonstrating its impact on OS in a large cohort of OvCa patients. We further identified two immunogenic HDAC1-derived peptides, which frequently induce multi-functional T-cell responses in many donors, suitable for future multi-peptide vaccine trials in OvCa patients.
Abbreviations
| HDAC | = | histone deacetylase |
| LC-MS/MS | = | liquid-chromatography coupled tandem mass spectrometry |
| OS | = | overall survival |
| OvCa | = | ovarian cancer |
| RCC | = | renal cell carcinoma |
| TAA | = | tumor-associated antigen |
| TMA | = | tissue microarray. |
Introduction
OvCa is the most lethal gynecological malignancy and the fifth leading cause of cancer death in women in the Western world. Citation1 Due to a lack of early detection methods and nonspecific symptoms, 75% of patients are diagnosed with stage III or IV disease. Although standard therapy, consisting of surgical tumor debulking followed by platinum-based chemotherapy Citation2 is able to induce primary clinical response, 70–80% of patients will relapse after initial treatment. In addition to stage and tumor-specific properties, interactions between the immune system and the tumor have an important impact on disease outcome. OvCa was one of the first tumor entities demonstrating an association between TILs and improved survival Citation3,4 as summarized in a large meta-analysis. Citation5 The significant impact of TILs supports current attempts to include the immune status of patients into general tumor classification Citation6 and to develop immunotherapies for the treatment of OvCa.
The encouraging clinical success of anti-CTLA4 antibody Ipilimumab in melanoma patients Citation7 started a new era in cancer immunotherapy. Meanwhile, several immune checkpoint inhibitors have been developed taking the breaks off a previously insufficient antitumor immune response. Anti-PD-L1 antibodies were demonstrated to be most effective in patients with pre-existing immune responses Citation8 implying that immune checkpoint inhibitors may profit from vaccine-induced tumor-specific immune responses. Immunotherapies already have shown clinical benefit in other solid tumors, especially melanoma and renal cell carcinoma (RCC). Citation9 In OvCa, antigen-specific approaches, such as peptide vaccinations targeting NY-ESO1 or Her2/neu resulted in immunological responses, Citation10,11 but failed to show clear clinical benefit. The limited clinical success might be, at least in part, due to suboptimal antigen selection according to gene expression data, having a distorted correlation to HLA presentation. Citation12 This implies that vaccination peptides should be selected according to their natural presentation on HLA molecules and their ability to induce T-cell responses. Considering this, we established a combined approach of HLA ligandomics and immunogenicity analysis, based on the identification of naturally presented HLA ligands on tumor samples, the selection of tumor-associated HLA ligands and subsequent immunogenicity testing by in vitro priming. Among others, our group has recently completed a clinical effective vaccination trial in RCC using tumor derived, naturally presented HLA-restricted peptides, confirming the feasibility of this approach. Citation13
The major objective of this current study was to perform HLA ligandome analysis of histologically defined high-grade serous OvCas Citation14 to identify tumor-associated peptides, tested for their immunogenic potential in subsequent experiments. In order to assess whether HLA ligandome analysis reflects protein expression, candidates were validated by immunohistochemistry and correlated with immune cell infiltration and OS in a large cohort of high-grade serous OvCa.
Results
HLA ligands from HDAC are frequently presented on OvCa tissue
In order to identify OvCa associated antigens for the development of targeted immunotherapies, we isolated HLA molecules by affinity chromatography from six samples of high-grade serous OvCa and analyzed the respective HLA ligands by liquid-chromatography tandem mass spectrometry (LC-MS/MS). For each tumor, we identified between 1194 and 2762 unique HLA ligands ( ) emanating from 1208 to 2513 different source proteins indicating that most of the peptides were unique for the corresponding antigen. Comparing the source proteins between the different samples revealed a great degree of heterogeneity. Only 45 proteins were represented in all analyzed tumors by at least one HLA ligand (Table S1) and predominantly consisted of ubiquitously expressed (non-tumor-associated) housekeeping proteins such as histone H2B, actin or HLA itself. Nevertheless, we also detected five well-described tumor-associated antigens (TAA): vimentin, ornithine decarboxylase 1, DNA-dependent protein kinase catalytic subunit (DN-PKC), β-catenin and histone deacetylase (HDAC) 1.
Table 1. HLA ligandome analysis and immunohistochemistry of OvCa samples.
Of these, HDAC1 was represented on the majority of tumors (4/6) by more than one particular HLA ligand culminating in three different coincidently presented HDAC1 ligands on one sample (OvCa41). Altogether, we identified four different peptides derived from HDAC1 (RMLPHAPGV, YTTDRVMTV, EYSKQMQRF and LPHAPGVQM), which potentially could also derive from the isoform HDAC2 due to the high conservation in the amino acid sequence, as well as on the functional level. One particular peptide, RMLPHAPGV, was detected in five of six samples.
Assigning the HLA restriction of the peptides, highest scores were yielded for RMLPHAPGV and YTTDRVMTV by HLA-A*02, EYSKQMQRF by HLA-A*24 and LPHAPGVQM by HLA-B*07 (Table S2). Notably, HLA-A*02-restricted RMLPHAPGV and YTTDRVMTV were also identified on HLA-A*02− samples implying further presentation on other HLA allotypes.
To assess the overlap of peptide profiles between benign and malignant ovarian tissues, we performed HLA ligandome analysis of six benign ovarian samples, resulting in comparable amounts in peptide yield between malignant and benign tissues (727-2244 unique HLA ligands per benign sample) (Table S3). Altogether 44 proteins were presented on all six analyzed benign ovaries with 14 proteins presented by both—maligant and benign—ovaries (Table S4).
Overlapping source proteins of the benign and maligant HLA ligandome were mainly non-tumor associated antigens like HLA class I or histone H2B. However, also previously described TAAs like vimentin were presented on all analyzed benign samples. HDAC1 was presented on two benign samples (RMLPHAPGV: OvN39; EYSKQMQRF: OvN38), although presentation frequency among benign samples was much lower compared to OvCa (2/6 versus 6/6).
HDAC derived HLA ligands are immunogenic
Frequent presentation on HLA class I molecules indicates the potential of HDAC1-derived peptides as suitable targets for immunotherapy. Immunogenicity of these peptides was subsequently investigated by in vitro priming of CD8+ T cells using aAPCs. After three stimulation intervals, HLA tetramer staining revealed that peptide-specific T cells were induced in five of seven (71%) healthy donors by the HLA-B*07-restricted peptide LPHAPGVQM and 13 of 16 (81%) donors by HLA-A*02-restricted peptide RMLPHAPGV ( ). Exemplary HLA tetramer staining results are shown for RMLPHAPGV in as well as for LPHAPGVQM in Fig. S1. To ensure peptide specificity and to exclude cross-reactivity, cross-staining of LPHAPGVQM-specific T cells with RMLPHAPGV-specific tetramer and vice versa was performed resulting in no observable cross-staining, respectively (data not shown). Tetramer staining after short-time stimulation of the same donors' PBMCs was negative (data not shown) demonstrating that the observed T-cell responses were mediated by in vitro primed naive T cells rather than by pre-existing memory T cells.
Figure 1. Immunogenicity analysis of RMLPHAPGV-primed T cells. CD8+ T cells from healthy blood donors were primed in vitro using (A) aAPCs or (B+C) natural APCs. Numbers represent percentages of all T cells. (A) Exemplary tetramer staining of one donor primed with RMLPHAPGV-specific aAPCs. Tetramer staining was performed after three stimulations with aAPCs using control tetramer YLLPAIVHI and RMLPHAPGV tetramer. Numbers represent percentage of all T cells. This figure is representative of nine donors tested positive for RMLPHAPGV-specific T cells after in vitro expansion with aAPCs. (B) Exemplary intracellular IFNγ staining of one donor primed with RMLPHAPGV-presenting natural APCs. Staining was performed after five stimulations with natural APCs using control peptide YLLPAIVHI and RMLPHAPGV peptide. (C) T cells, which have been primed in vitro using natural APCs, (from panel B) were stained with irrelevant YLLPAIVHI and RMLPHAPGV tetramer.
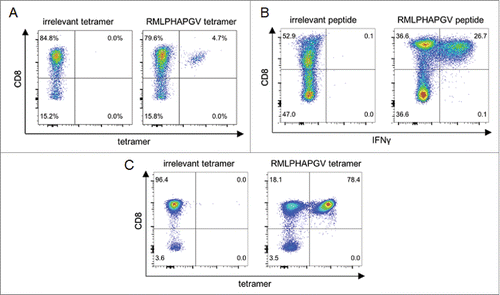
Table 2. Immunogenicity of HDAC-derived HLA ligands.
The immunogenic potential of RMLPHAPGV was confirmed by in vitro priming using peptide-loaded autologous DCs and B cells. After five stimulations intervals, RMLPHAPGV induced peptide-specific T-cell responses in 7 of 14 tested healthy donors demonstrated by IFNγ staining of CD8+ T cells ( ). In turn, staining with RMLPHAPGV tetramer in comparison to irrelevant tetramer revealed a peptide-specificity of 78.4% as shown for one exemplary donor ( ).
RMLPHAPGV-specific T cells are multi-functional and kill HLA-matched tumor cells
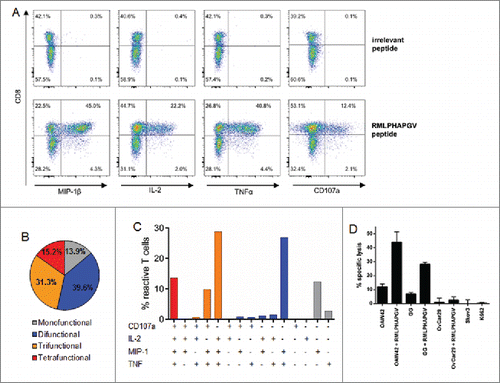
Following seven stimulations with peptide-loaded autologous DCs and B cells, multi-functionality of RMLPHAPGV-specific T cells was assessed by flow cytometric staining for additional activation markers MIP-1β, TNFα, IL-2 and CD107a ( ). 60.1% of RMLPHAPGV-specific T cells, indicated by HLA multimer staining, produced at least one activation marker. The combinations of different activation markers are depicted in . 91.2% of reactive T cells produced the pro-inflammatory chemokine ligand MIP-1β, 83.9% TNFα and 76.4% both activation markers at the same time. While 13.6% of all responding T cells expressed the four investigated activation markers simultaneously, only 16.9% were mono-functional, in the majority of cases expressing MIP-1β. CD107a, which is a surrogate marker for cytotoxicity, as well as IL-2, were almost exclusively secreted by T cells which also produced MIP-1β or TNFα. Incubation of RMLPHAPGV-specific T cells with HLA-A*02+ OvCa cell line OAW42 resulted in the secretion of CD107, but not of TNFα or IFNγ (Fig. S2).
Figure 2. Multi-functionality and cytotoxicity of in vitro primed T cells reacting to RMLPHAPGV. (A) T cells show significant expression of TNFα, MIP-1β, IL-2 and CD107a after 6 h incubation with RMLPHAPGV. Numbers in the plot reflect percentage of reactive T cells for the respective marker. (B) Reactive T cells show different degrees of functionality. Only T cells expressing at least one activation marker (60.1% of all RMLPHAPGV-specific T cells) are displayed. 39.9% of RMLPHAPGV-tetramer-specific T cells did not react upon peptide stimulation. (C) Different combinations of activation markers are represented within the reactive T-cell population. (D) Percentage of specific lysis by antigen-specific T cells was determined by 51chromium release assay after 24 h coincubation with different RMLPHAPGV peptide-loaded or peptide-unloaded OvCa cell lines or HLA-negative K562 cells. Experiments were performed in triplicates with a 30:1 E:T ratio.
To investigate the cytotoxic potential in more detail, 51chromium release assays were performed using a RMLPHAPGV-primed T-cell population containing 78% RMLPHAPGV-specific T cells (assessed by HLA tetramer staining). After 24 h of co-incubation, RMLPHAPGV-specific T cells (E:T ratio 30:1) lysed HLA-A*02+ OvCa cell lines OAW42 (12.0%) and GG (4.6%) ( ) in an E:T ratio dependent manner (Fig. S3A), indicating that RMLPHAPGV is a naturally presented HLA ligand. Furthermore, HLA ligandome analysis identified RMLPHAPGV as a naturally presented HLA ligand on OAW42 cells (data not shown). Exogenous pulsing of HLA-A*02+ OvCa cell lines with RMLPHAPGV resulted in an increased peptide-specific killing of OAW42 and GG cells (44.0% and 26.9%). In contrast, HLA-A*02− OvCa cell lines OvCar29 (2.6%) and SKOV3 (0.0%) were not killed by RMLPHAPGV-specific T cells, even after pulsing with RMLPHAPGV (1.0%), confirming that the lysis mediated by RMLPHAPGV-recognizing T cells is both HLA-A*02- and peptide-dependent. The HLA− leukemia cell line K562 was not lysed (0.6%) after 24 h, excluding lysis by remaining NK cells. Similar results were obtained after 6 h of co-incubation although lysis of native OAW42 and GG cells did not reach significance due to shorter incubation times (Fig. S3B).
We screened for RMLPHAPGV-specific T cells among TILs from three OvCa patients by performing IFNγ-ELISpot assays (OvCa41: HLA-A*02, A*24, B*18, B*15, C*02, C*12; OvCa42: HLA-A*01, A*02, B*40, B*61, C*02, C*06; OvCa43: HLA-A*02, A*32, B*18, B*35, C*04, C*07). None of the tested donors showed a peptide-specific IFNγ response after stimulation with RMLPHAPGV but stimulation with the influenza virus peptide GILGFVFTL resulted in IFNγ secretion in one of three tested donors (OvCa41; Fig. S4). These results were confirmed by tetramer stainings.
The delayed and moderate lysis of native OvCa cells as well as the lack of RMLPHAPGV-specific TILs indicated that RMLPHAPGV-specific T cells have a low affinity to their cognate HLA-peptide complex. We performed immunohistochemical stainings of HDAC1 in thymus tissue samples to investigate whether HDAC1 is expressed by thymocytes which might cause the deletion of high-affinity RMLPHAPGV-specific T cells due to negative selection. Double staining of pan-cytokeratin (cytoplasmic) and HDAC1 (nuclear) demonstrated that HDAC1 is expressed by epithelial thymocytes (Fig. S5).
TILs overcome the poor prognostic impact of HDAC1 overexpression
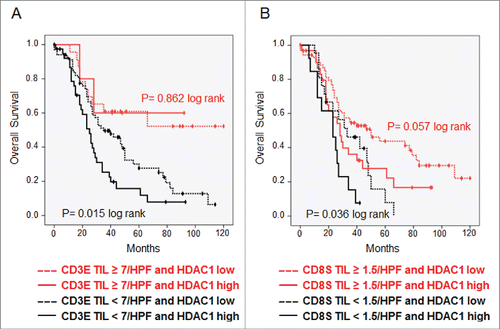
To investigate whether HDAC1 represents a tumor antigen of general importance in OvCa, immunohistochemistry was performed on tissue microarrays (TMAs) of 136 patients with histologically defined high-grade serous OvCa. Compared to surrounding normal tissue, 45 (33.1%) of the cases showed strong overexpression of HDAC1 (score ≥ 9), further characterized by a very diffuse nuclear expression, which was associated with poor OS (median 24 vs. 42 mo, p = 0.011, log rank) ( ). We further analyzed the TIL subtypes in the same tumor samples to investigate a possible association between the presence of TILs and HDAC1 overexpression. TIL subset analysis demonstrated that median OS was significantly longer for patients with high CD3E (66 vs. 29 mo, p = 0.002) ( ) and high CD8S (35 vs. 26 mo, p = 0.002) ( ), but not for CD3S (p = 0.225) and CD8E (p = 0.376) (data not shown). At the same time, the infiltration of TILs did not correlate with HDAC1 overexpression (data not shown). However, combining TIL subgroup analysis and HDAC1 expression data revealed that the intratumoral immune response reduced the prognostic effect of HDAC1. With high numbers of CD3E, strong HDAC1 overexpression failed to show a prognostic impact, whereas it was maintained in the group with low CD3E (OS median 25 vs. 35 mo, p = 0.015, log rank) ( ) with similar but less significant effects for CD8S ( ).
Figure 3. Immunohistochemistry of high-grade serous OvCa. Immunohistochemical stainings on TMAs with 136 high-grade serous OvCa samples were performed. (A) Expression of HDAC1: Top: Immunohistochemical staining for HDAC1 of two exemplary OvCa samples with high HDAC1 expression (score 12, left) and low HDAC1 expression (score 3, right). Bottom: Impact of HDAC1 on OS of high-grade serous OvCa stage III and IV (Kaplan Meier analysis, low expression: score < 9, n = 94; high expression score ≥ 9, n = 45). (B) Intraepithelial CD3+ TILs: Top: Immunohistochemical staining for CD3+ TILs of two exemplary OvCa samples with lower infiltration (left) and higher infiltration (right) of TILs (epithelial compartment is highlighted in yellow). Bottom: Impact of epithelial CD3+ TILs on OS of high-grade serous OvCa stage II to IV (Kaplan Meier analysis, low: < 7 cells/HPF, n = 108; high: ≥ 7 cells/HPF, n = 28). (C) Intrastromal CD8+ TILs: Top: Immunohistochemical staining for CD8+ TILs of two exemplary OvCa samples with lower infiltration (left) and higher infiltration (right) of TILs (stromal compartment is highlighted in green). Bottom: Impact of stromal CD8+ TILs on OS of high-grade serous OvCa stage II to IV (Kaplan Meier analysis, low: < 1.5 cells/HPF, n = 91; high: ≥ 1.5 cells/HPF, n = 45).
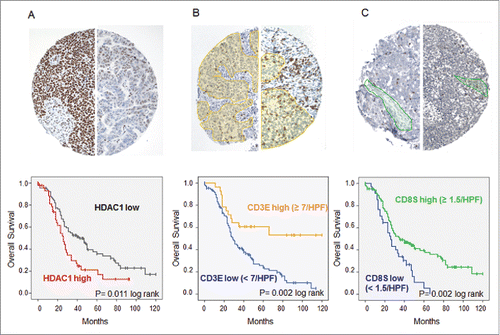
Figure 4. TILs overcome prognostic effect of HDAC1. Kaplan–Meier analysis was performed to investigate the survival effect of HDAC1 in high-grade serous OvCa stage II–IV with. (A) Low and high numbers of intraepithelial CD3+ TILs. (B) Low and high numbers of stromal CD8+ TILs.
We finally performed a multivariate analysis (Cox regression) of survival to characterize HDAC1 overexpression as well as the infiltration of CD3E and CD8S as independent prognostic markers, further considering the tumor resection status, tumor stage and platinum resistance. The multivariate analysis revealed HDAC1, CD3E TILs and platinum resistance as independent prognostic parameters of OS in our cohort (Table S5).
Discussion
The recent approval of immune checkpoint inhibitors Citation7 demonstrates the clinical effectiveness of cancer immunotherapy. A challenging task, however, remains the identification of physiologically relevant targets to guide tumor-specific immune responses, for example by peptide vaccination. We established a combined approach of HLA ligandomics and immunogenicity analysis to identify tumor-associated HLA ligands presented in a large number of patients. In this study, HLA ligandome analysis was performed for several primary serous OvCa samples revealing that only few proteins are presented on all analyzed tumors, including mainly house-keeping genes but also few TAAs such as HDAC1. HDAC1 mediated deacetylation of histones leads to increased cell proliferation, cell migration, angiogenesis and invasion by reducing the transcription of tumor suppressor genes. Citation15,16 HDAC1/2-derived peptides were represented on the majority of tumors by more than one HLA allotype simultanously, indicating a frequent processing and presentation and fostering our attempts to investigate HDAC1 in more detail. We were able to identify four different HDAC1/2-derived peptides: EYSKQMQRF (HLA-A*24), LPHAPGVQM (HLA-B*07), RMLPHAPGV and YTTDRVMTV (both HLA-A*02). Interestingly, both A*02-restricted peptides were additionally identified on HLA-A*02− patients suggesting further presentation by other HLA allotypes. We hypothesize that these peptides could be presented by HLA-E which is supposed to present a peptide pool similar to that of HLA-A*02:01 in the absence of dominant leader peptides. Citation17
We compared the HLA ligandome of malignant and benign ovarian samples, demonstrating that about one-third of abundantly presented proteins on benign samples were also presented on all OvCa samples, most of them being non-tumor-associated housekeeping proteins. HDAC1-derived peptides RMLPHAPGV and EYSKQMQRF were each presented on one benign sample. However, the presentation frequency is much higher on the malignant ovarian samples. Apart from this, targeting benign residual ovarian tissue in OvCa patients would be acceptable as the complete ovaries are resected during surgery.
In vitro priming of CD8+ T cells from healthy blood donors demonstrated that LPHAPGVQM and RMLPHAPGV are immunogenic in a high proportion of donors and capable of lysing HDAC1+ OvCa cell lines. By analyzing different activation markers, we could demonstrate the multi-functionality of RMLPHAPGV-specific T cells which has been previously associated with an improved control of viral infections and malignancies. Citation18,19 However, RMLPHAPGV-specific T cells were not detected among TILs in OvCa patients, although the samples size of three patients was too small for significant statements. Pre-existing RMLPHAPGV-specific memory T cells would provide a proof of principle of this approach. However, immune escape mechanisms in the tumor and tumor-draining lymph nodes might have suppressed the in vivo priming of RMLPHAPGV-specific T cells. Furthermore, frequencies below detection limit and low T-cell receptor affinities toward their corresponding HLA-peptide complex could explain the lack of RMLPHAPGV-specific T-cells, requiring an exogenous booster such as a peptide vaccination for their activation. This hypothesis is supported by the moderate lysis of target cells in the cytotoxicity assays employed here. The lack of high-affinity T cells might be explained by negative selection in the thymus as we could demonstrate HDAC1 expression in thymic epithelial cells.
Previous peptide vaccinations targeting for example NY-ESO1 Citation10 or p53 Citation20 resulted in immunological responses but failed to show clear clinical benefit. To improve peptide-based immunotherapies, several issues such as inadequate co-stimulation or immunosuppressive microenvironment have to be addressed. The lack of efficacy might be, at least partly, due to an inadequate antigen selection. Most previous studies relied on gene expression analysis, lacking evidence of natural HLA presentation. For example, HLA ligands derived from NY-ESO-1, a highly tumor-specific antigen expressed by 43% of OvCa patients, Citation21 have not been detected in OvCa samples so far. The distorted relationship between RNA expression levels and HLA presentation has been previously described Citation12 supporting our attempts to identify vaccination peptides based on the HLA ligandome. The feasibility and safety of multi-peptide vaccination using natural presented peptides in OvCa patients has been previously demonstrated in a clinical phase I trial. Citation22 We further improved this approach by analyzing the HLA ligandome of primary tumor material from OvCa patients and identifying peptides presented on all analyzed OvCa samples, enabling to target a broad spectrum of different ovarian tumors.
Immunohistochemical stainings revealed that strong HDAC1 overexpression was associated with decreased OS of OvCa patients, supporting previous studies Citation23,24 by analyzing a clinical series of histologically defined OvCa samples employed here. In accorandance with previous studies, we demonstrated that high numbers of CD3+ T cells (except for regulatory T cells) were indicative of improved survival in OvCa; Citation3,4,25 however, the effect of TILs was analyzed separately depending on their localization–tumor stroma vs. tumor epithelium. We confirmed the positive effect of intraepithelial CD3+T cells but additionally identified a prognostic relevance of stromal CD8+ T cells.
Using these datasets, we investigated whether there is a link between the expression of HDAC1 and the beneficial effects of high TIL numbers. We could demonstrate that the prognostic effect of HDAC1 was no longer evident in tumors with high numbers of CD3+ TILs. At the same time, HDAC overexpression was absent in the majority of cases with high TIL counts, not only providing evidence for a role of specific TAAs in shaping antitumor response, but also confirming that HDAC1 is a suitable target for the development of a multi-peptide vaccine.
Patients and methods
Patients and tumor samples
For HLA ligandome analysis, six fresh tumor samples and six benign ovarian samples were collected under informed consent during tumor debulking surgery or oophorectomy/salpingo-oophorectomy (with pathologically confirmed non-malignant status) at the University Women's Hospital in Tübingen and were snap-frozen ( ). The study cohort used for TIL quantification and candidate protein expression by immunohistochemistry included 136 consecutive patients diagnosed and treated with high-grade serous OvCa FIGO stage II–IV at the University Women's Hospital in Tübingen between 2000 and 2008. All patients had undergone surgical resection according to their clinical stage and had received, if indicated, platinum-containing chemotherapy based on current German guidelines (www.ago-online.de). Follow-up data was obtained from the patient registry of the Comprehensive Cancer Center Tübingen. The clinical follow-up ranged from 1 to 120 mo. All studies were approved by the local ethics committee.
Peptides and tetramers
Synthetic peptides RMLPHAPGV (HDAC1371-379), YTTDRVMTV (HDAC1188-196), LPHAPGVQM (HDAC1373-381), EYSKQMQRF (HDAC186-94), GILGFVFTL (M1_I34A15866) and YLLPAIVHI (DDX5148-156) (gene names according to uniprot entry names) were produced by standard 9-fluorenylmethyloxycarbonyl/tert-butyl strategy using an automated peptide synthesizer 433A (Applied Biosystems, Foster City, CA, USA). Purity was assessed by reversed phase HPLC (e2695; Waters, Eschborn, Germany) and identity affirmed by mass spectrometry. Lyophilized peptides were dissolved at 10 mg/mL in DMSO (WAK Chemie, WAK-DMSO) and further diluted in bidestilled H2O.
Biotinylated HLA-peptide complexes were manufactured as described previously Citation26 and tetramerized using PE-conjugated streptavidin (Invitrogen Life Technologies, S866) at a 4:1 molar ratio.
Blood samples
PBMCs were isolated by standard Ficoll Hypaque separation (Merck Millipore, L6115) from whole blood samples of healthy donors. Blood samples were kindly provided by the Institute for Clinical and Experimental Transfusion Medicine at the University Hospital of Tübingen after obtaining written informed consent.
Cell lines
HLA-A*02− OvCa cell lines OvCar29 (HTB 161) and SKOV3 (HTB 77) were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). HLA-A*02+ OvCa cell line GG was established from malignant ascites in our laboratory Citation27 and OAW42 (85073102) was obtained from the European Collection of Cell Cultures (ECACC, Salisbury, UK). The natural killer cell-sensitive and HLA-negative leukemia cell line K562 was obtained from ECACC (89121407). The CD40L expressing mouse fibroblast cell line t-CD40L-NIH-3T3 was a kind gift of Dr Schultze from the Dana Farber Cancer Institute (Boston, USA). All OvCa cell lines were cultured in Dulbecco's Modified Eagle Medium (DMEM; Gibco life technologies, 41965-062) containing 10% FCS (Biochrome, S0115), 100 U/mL Penicillin and 100 µg/mL Streptomycin (Pen/Strep; Sigma-Aldrich, P4333). The leukemia cell line K562 was cultivated in Iscove's Modified Dulbecco's Medium (IMDM; Lonza, 12-722F) containing 10% FCS and 100 U/mL Pen/Strep. T-CD40L-NIH-3T3 was cultured in DMEM containing 10% FCS, 100 U/mL Pen/Strep and 0.4% G418 (Invitrogen, 10131-027). Culture conditions were 37°C and 7.5% CO2 in humidified incubators. All OvCa cell lines were authenticated by PRC-based high-resolution HLA-typing. HLA-A*02 as well as HLA class I expression were checked by flow cytometry prior to the experiments using BB7.2 (anti-HLA-A*02) and W6/32 (anti-HLA class I) antibodies. Murine t-CD40L-NIH-3T3 cells were not authenticated by the author but CD40L expression was controlled by flow cytometry.
TIL preparation
OvCa samples were freshly collected under informed consent during tumor debulking surgery at the University Women's Hospital in Tübingen. After cutting tissues into small pieces < 2 mm3, they were digested using an enzymatic dissociation solution containing 400 U/mL Collagenase Type IV and 5 U/mL Dispase (both Gibco life technologies, 17104-019 and 17105-041) in DMEM with 10% FCS and incubated for 3 h at 37°C while gentle agitation. Remaining tissue fragments were removed with a 100 µm cell strainer (BD Bioscience, Heidelberg, Germany). Erythrocytes were lysed using ammonium chloride lysis buffer and single cell suspensions were washed twice with PBS prior to freezing.
Induction of peptide-specific CTLs with autologous APC
Freshly isolated PBMCs from healthy blood donors were incubated for 2 h in culture flasks (Nunc, Roskilde, Denmark) to separate monocytes and peripheral blood lymphocytes (PBL) by plastic-adherence. Maturation of DCs from monocytes was performed as described previously. Citation28 In brief, monocytes were incubated in XVivo 15 medium (Lonza, BE04-418Q), supplemented with 1000 U/mL IL-4 and 1000 U/mL GM-CSF, and activated after 7 d with 10 ng/mL TNFα, 10 ng/mL IL-1β, 1000 U/mL IL6, 1 µg/mL PGE2 and 1000 U/mL GM-CSF for 24 h (R&D systems, 204-IL, 215-GM, 210-TA, 201-LB, 206-IL; except PGE2, Calbiochem, CAS 363-24-6). T cells were isolated from the PBL fraction using a CD3+ magnetic cell separation (MACS) kit (Miltenyi Biotec, 130-050,101) and remaining PBLs were cocultured with the murine CD40L transfected NIH-3T3 cell line and 2 ng/mL IL-4 to generate activated B cells. Citation29
After isolation, 2 × 106 T cells per well were seeded into 24-well plates (Costar/Corning, New York, USA) in T-cell medium containing IMDM with 10% human autologous heat-inactivated plasma, 100 U/mL Pen/Strep and 50 µmol/L β-mercaptoethanol (Carl Roth, 4227). T-cell stimulation was performed using autologous, irradiated, peptide-pulsed DCs (1:10 ratio) and 10 ng/mL IL-7 (R&D systems, 207-IL). Stimulation with DCs was performed in three subsequent intervals with a stimulation interval of 7 d. Subsequently, T cells were stimulated at least two more times with autologous, irradiated, peptide-pulsed B cells (1:4 ratio) with an interval of 10 d. To support expansion of antigen-specific T cells, 10 ng/mL IL-2 (R&D systems, 202-IL) was added during and 2 d after peptide-stimulation, beginning with the third stimulation.
Peptide-loading of APCs was performed with 10 µg/mL peptide for 2 h in XVivo 15 medium. Culturing conditions were 37°C and 7.5% CO2 in a humidified incubator.
Induction of peptide-specific CTLs with artificial APC (aAPC)
CD8+ T cells were isolated from PBMCs of healthy donors using a MACS kit (Miltenyi Biotec, 130-045-201). 1 × 106 T cells per well were cultured in round bottom 96-well plates (Costar/Corning) and stimulated with 1 × 106 aAPCs per well and 5 ng/mL IL-12 (PromoCell, C-62212) three times with a 7 d stimulation interval. 40 U/mL IL-2 were added 2 d after each stimulation. Citation30
For generation of aAPCs, 5.6-µm-diameter streptavidin-coated polystyrene beads (Bangs Laboratories, Fishers, IN, USA) were resuspended at 2 × 106 particles per mL. Beads were incubated with 10 nM biotinylated pMHC complexes and 10 nM stimulating anti-CD28 antibody (hybridoma 9.3, kind gift of Prof. Gundram Jung, University of Tübingen) for 30 min at RT.
Short-time stimulation of T cells and IFNγ-ELISpot assays
24 h after thawing, PBMC were stimulated with 1 µg/mL candidate peptide or negative control peptide. IL-2 was added on day 2, 5 and 7 with a final concentration of 20 U/mL. After 12 d, PBMCs were harvested and IFNγ-ELISpot was performed by seeding PBMCs at a concentration of 500,000 cells/well in T-cell medium onto nitrocellulose plates (Immunospot M200, Merck Millipore, MSHAN4B50). Thus, T cells were stimulated for 24 h with 1 µg/mL of peptide or phytohaemagglutinin (PHA; Sigma Life Science, L1668). Secretion of IFNγ was detected using an ELISpot kit (Mabtech, 3420-2A) according to the manufacturer's recommendation and the cancer immunotherapy monitoring panel. Citation31 Spot number was counted by an ImmunoSpot Series 6 ELISPOT Reader (CTL, Bonn, Germany). Peptide responses were considered positive if the mean number of spots per well was at least ten and more than three times the mean number of spots of the negative control.
Flow cytometric analysis of antigen-specific T cells
All flow cytometric measurements were performed on a FACSCanto II (BD) and evaluated using FlowJo version 9.2 (Tree Star, Ashland, OR, USA). For HLA tetramer analysis, T cells were washed in FACS buffer containing 2% FCS, 2 mM EDTA (Carl Roth, 8040) and 0.02% sodium azide (Merck Millipore, 8223350100) in PBS, followed by a staining for dead cells using Live/Dead fixable Aqua dead cell stain kit (Invitrogen, L34957). Afterwards, cells were washed and incubated with 2.5 µg/mL of the respective tetramer in tetramer solution (FACS buffer with 50% FCS) for 30 min at RT. Finally, cell surface molecules were stained with CD8-PerCP (Biolegend, 301030) for 20 min at 4°C.
Intracellular cytokine staining was performed by incubating in vitro primed T cells with 10 µg/mL candidate or control peptide, Golgi-Stop-Solution (BD Bioscience, 554724) and anti-CD107a-FITC antibody (BD Bioscience, 555800) for 6 h. Afterwards, cells were stained for dead cells using Live/Dead fixable Aqua dead cell stain kit followed by staining with CD8-PerCP for 20 min. Intracellular staining was performed after 30 min permeabilization with Cytoperm/Cytofix solution (BD Bioscience, 554722) using the following antibodies: IFNγ-PE-Cy7, IL-2-APC, TNFα-PacificBlue and MIP-1β-PE (all BD Bioscience, 557844, 554567 and 550078; except TNFα, Biolegend, 502920).
Cytotoxicity assay
Cytolytic activity of T cells was assessed in a standard 51chromium release assay as described previously Citation32 and detailed in the Supplementary Information.
Isolation of HLA presented peptides
HLA presented peptides were obtained by immunoaffinity chromatography of HLA molecules using a protocol developed for solid tissue analysis, published recently. Citation33 A detailed protocol can be found in the Supplementary Information.
Nanoflow LC-MS/MS
Mass spectrometry was performed on an LTQ OrbitrapXL mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) equipped with a nanoelectron spray ion source and coupled to an Ultimate 3000 RSLC Nano UHPLC System (Dionex, Sunnyvale, CA, USA). After injection, samples were loaded with 3% of solvent B (20% H2O, 80% acetonitrile and 0.04% formic acid) on a 2 cm PepMap 100 C18 Nanotrap column (Dionex) at a flowrate of 4 μL/min for 10 min. Peptides were then separated on a 50 cm PepMap C18 column with a particle size of 2 μm (Dionex) running at 45°C with a gradient ranging from 3 to 30% solvent B within 140 min and a flow rate of 300 nL/min.
Data-dependent acquisition was enabled for analysis using a top five method (the five most-intense ions with a positive charge between 2 and 4 observed during survey scan were selected for fragmentation during each scan cycle). Survey scans were performed in the orbitrap at a resolution of 60k with a scan range of 450–650 m/z. Peptides were fragmented using collision induced dissociation (normalized collision energy 35%, activation time 30 ms, isolation width 1.3 m/z) with resulting fragment ions (MS/MS scans) analyzed in the linear ion trap. Dynamic exclusion was enabled for all runs (exclusion size list 500). Duration of exclusion was dependent on peptide concentration varying between 1 and 30 s with initial runs usually performed with 1 s.
Data analysis
Analysis of MS Data was carried out using Proteome Discoverer 1.3 (Thermo Scientific). Peak lists were searched against Swissprot database (date: July. 2012, taxonomy filter: Homo sapiens) using Mascot software version 2.2.04. Mass tolerance used for processing were 5 parts per million (ppm) for precursor ions and 0.5 Da for product ions. Percolator tool was used to evaluate peptide confidence based on q-value with a strict target FDR of 0.01 (high confidence) and a relaxed target FDR of 0.05 (medium con-fidence).
Additional post-processing filters were a peptide length between 8 and 12 amino acids and a search engine rank of 1. All fragment spectra derived from HDAC peptides were additionally confirmed by positive matching of collected fragment spectra to fragment spectra of respective synthetic peptides acquired using the same setup.
HLA annotation of identified HLA ligands
Amino acid sequences of identified HLA ligands were scored using the online databases SYFPEITHI 1.0 (www.syfpeithi.de) and NetMHC 3.0 (www.cbs.dtu.dk/services/NetMHC-3.0). SYFPEITHI scores were normalized to the different maximum scores of individual prediction matrices. NetMHC scores are shown as logscore (maximal logscore = 1). The five highest scored HLA restrictions of both databases were compared and matched against the donors' HLA types.
Construction of tissue microarrays (TMA)
Consecutive paraffin embedded samples of high-grade serous OvCas were retrieved from the archives of the Institute of Pathology. Cases were included in the study after confirmation of histological subtype and grading according to published criteria. Citation34 The construction of TMAs and preparation of the slices for immunohistochemistry was performed as published previously Citation35 and described in the Supplementary Information.
Immunohistochemistry
The following primary antibodies and dilutions were used for immunohistochemistry: CD3 (1:100, rat monoclonal SP7, DCS Innovative Diagnostics, CI597R06), CD8+ (1:200, mouse monoclonal C8/144B, M710301-2), HDAC1 (ready to use, rabbit polyclonal, ABCAM, ab15316) and pan-cytokeratin (1:250, mouse monoclonal AE1/AE3, DAKO, M3515). The tissue sections were pre-treated with EDTA-buffer solution (pH 8.6; Cell Conditioning Solution, Roche, 950-124) at 95°C for 36 min. Immunohistochemical stainings were performed on an automated immunostainer according to the manufacturer's instructions (Ventana Medical Systems Inc.., Tucson, AZ, USA) using the iView DAB detection kit (Roche, 760-091) for single stain protocols. For double staining of HDAC1 and pan-cytokeratin, Ultraview universal alkaline phosphatase red detection kit (Roche, 760-501) was used in a second step.
Immunoscoring
Lymphocyte subsets were visualized by immunohistochemistry and quantification of TILs was carried out by first assessing the average number of immunostained cells per high power field (HPF = 400×) by counting at least two HPF for each core. In a second step, the average number of lymphocytes per HPF for the left and right triple core set was calculated, and for all the cores together. This bilateral average count was used for further calculations. The fibrovascular tumor stroma (CD3S and CD8S), and the intraepithelial compartment of the tumor (CD3E and CD8E) were evaluated separately.
For HDAC1 expression, nuclear intensity was graded from 0 to 3, multiplied by a score from 1 to 4 for the percentage of tumor cells (0–10%/1; 10–50%/2; 50–80%/3; 80–100%/4) Citation36. Scores 9–12 were considered strongly positive.
136 of the initial 154 patients could be successfully evaluated.
Statistical analysis
To analyze survival data, a Kaplan–Meier analysis was performed using SSPS statistical software (Version 22, IBM Corp., Armonk, NY, USA). p values of less than 0.05 were considered significant.
Disclosure of potential conflicts of interest
Hans-Georg Rammensee declares to be the shareholder of immatics biotechnologies, Tübingen, and CureVac GmbH, Tübingen. The other authors disclose no potential conflicts of interest.
1065369_supplemental_files.zip
Download Zip (966.6 KB)Acknowledgments
We thank Lynne Yakes for expert proof reading. The authors also thank Patricia Hrstić, Nicole Zuschke, Katharina Graf, Beate Pömmerl, Karen Greif and Christine Beschorner for their technical support.
Funding
This work was supported by the DFG (Deutsche Forschungsgemeinschaft, Collaborative Research Center 685), the German Cancer Consortium (DKTK) and the Ludwig Hiermaier Stiftung für Angewandte Krebsforschung.
References
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin 2010; 60:277-300; PMID:20610543; http://dx.doi.org/10.3322/caac.20073
- Vaughan S, Coward JI, Bast RC, Jr., Berchuck A, Berek JS, Brenton JD, Coukos G, Crum CC, Drapkin R, Etemadmoghadam D et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer 2011; 11:719-25; PMID:21941283; http://dx.doi.org/10.1038/nrc3144
- Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, Jungbluth AA, Frosina D, Gnjatic S, Ambrosone C et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci U S A 2005; 102:18538-43; PMID:16344461; http://dx.doi.org/10.1073/pnas.0509182102
- Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, Makrigiannakis A, Gray H, Schlienger K, Liebman MN et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med 2003; 348:203-13; PMID:12529460; http://dx.doi.org/10.1056/NEJMoa020177
- Hwang WT, Adams SF, Tahirovic E, Hagemann IS, Coukos G. Prognostic significance of tumor-infiltrating T cells in ovarian cancer: a meta-analysis. Gynecol Oncol 2012; 124:192-8; PMID:22040834; http://dx.doi.org/10.1016/j.ygyno.2011.09.039
- Galon J, Pages F, Marincola FM, Angell HK, Thurin M, Lugli A, Zlobec I, Berger A, Bifulco C, Botti G et al. Cancer classification using the Immunoscore: a worldwide task force. J Transl Med 2012; 10:205; PMID:23034130; http://dx.doi.org/10.1186/1479-5876-10-205
- Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010; 363:711-23; PMID:20525992; http://dx.doi.org/10.1056/NEJMoa1003466
- Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014; 515:563-7; PMID:25428504; http://dx.doi.org/10.1038/nature14011
- Topalian SL, Weiner GJ, Pardoll DM. Cancer immunotherapy comes of age. J Clin Oncol 2011; 29:4828-36; PMID:22042955; http://dx.doi.org/10.1200/JCO.2011.38.0899
- Diefenbach CS, Gnjatic S, Sabbatini P, Aghajanian C, Hensley ML, Spriggs DR, Iasonos A, Lee H, Dupont B, Pezzulli S et al. Safety and immunogenicity study of NY-ESO-1b peptide and montanide ISA-51 vaccination of patients with epithelial ovarian cancer in high-risk first remission. Clin Cancer Res 2008; 14:2740-8; PMID:18451240; http://dx.doi.org/10.1158/1078-0432.CCR-07-4619
- Disis ML, Gooley TA, Rinn K, Davis D, Piepkorn M, Cheever MA, Knutson KL, Schiffman KGeneration of T-cell immunity to the HER-2/neu protein after active immunization with HER-2/neu peptide-based vaccines. J Clin Oncol 2002; 20:2624-32; PMID:12039923; http://dx.doi.org/10.1200/JCO.2002.06.171
- Weinzierl AO, Lemmel C, Schoor O, Müller M, Krüger T, Wernet D, Hennenlotter J, Stenzl A, Klingel K, Rammensee HG et al. Distorted relation between mRNA copy number and corresponding major histocompatibility complex ligand density on the cell surface. Mol Cell Proteomics 2007; 6:102-13; PMID:17074750; http://dx.doi.org/10.1074/mcp.M600310-MCP200
- Walter S, Weinschenk T, Stenzl A, Zdrojowy R, Pluzanska A, Szczylik C, Staehler M, Brugger W, Dietrich PY, Mendrzyk R et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat Med 2012; 18:1254-61; PMID:22842478; http://dx.doi.org/10.1038/nm.2883
- Seidman JD, Cho KR, Ronnett BM, Kurman RJ. Surface epithelial tumors of the ovary. In: Kurman RJ, Ellenson LH, Ronnett BM, eds. Blaustein's Pathology of the Female Genital Tract. New York: Springer-Verlag, 2011:679-784
- Hayashi A, Horiuchi A, Kikuchi N, Hayashi T, Fuseya C, Suzuki A, Konishi I, Shiozawa T. Type-specific roles of histone deacetylase (HDAC) overexpression in ovarian carcinoma: HDAC1 enhances cell proliferation and HDAC3 stimulates cell migration with downregulation of E-cadherin. Int J Cancer 2010; 127:1332-46; PMID:20049841; http://dx.doi.org/10.1002/ijc.25151
- Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene 2007; 26:5420-32; PMID:17694083; http://dx.doi.org/10.1038/sj.onc.1210610
- Lampen MH, Hassan C, Sluijter M, Geluk A, Dijkman K, Tjon JM, de Ru AH, van der Burg SH, van Veelen PA, van Hall T. Alternative peptide repertoire of HLA-E reveals a binding motif that is strikingly similar to HLA-A2. Mol Immunol 2013; 53:126-31; PMID:22898188; http://dx.doi.org/10.1016/j.molimm.2012.07.009
- Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 2006; 107:4781-9; PMID:16467198; http://dx.doi.org/10.1182/blood-2005-12-4818
- Yuan J, Gnjatic S, Li H, Powel S, Gallardo HF, Ritter E, Ku GY, Jungbluth AA, Segal NH, Rasalan TS et al. CTLA-4 blockade enhances polyfunctional NY-ESO-1 specific T cell responses in metastatic melanoma patients with clinical benefit. Proc Natl Acad Sci U S A 2008; 105:20410-5; PMID:19074257; http://dx.doi.org/10.1073/pnas.0810114105
- Rahma OE, Ashtar E, Czystowska M, Szajnik ME, Wieckowski E, Bernstein S, Herrin VE, Shams MA, Steinberg SM, Merino M et al. A gynecologic oncology group phase II trial of two p53 peptide vaccine approaches: subcutaneous injection and intravenous pulsed dendritic cells in high recurrence risk ovarian cancer patients. Cancer Immunol Immunother 2012; 61:373-84; PMID:21927947; http://dx.doi.org/10.1007/s00262-011-1100-9
- Odunsi K, Jungbluth AA, Stockert E, Qian F, Gnjatic S, Tammela J, Intengan M, Beck A, Keitz B, Santiago D et al. NY-ESO-1 and LAGE-1 cancer-testis antigens are potential targets for immunotherapy in epithelial ovarian cancer. Cancer Res 2003; 63:6076-83; PMID:14522938
- Morse MA, Secord AA, Blackwell K, Hobeika AC, Sinnathamby G, Osada T, Hafner J, Philip M, Clay TM, Lyerly HK et al. MHC class I-presented tumor antigens identified in ovarian cancer by immunoproteomic analysis are targets for T-cell responses against breast and ovarian cancer. Clin Cancer Res 2011; 17:3408-19; PMID:21300761; http://dx.doi.org/10.1158/1078-0432.CCR-10-2614
- Khabele D, Son DS, Parl AK, Goldberg GL, Augenlicht LH, Mariadason JM, Rice VM. Drug-induced inactivation or gene silencing of class I histone deacetylases suppresses ovarian cancer cell growth: implications for therapy. Cancer Biol Ther 2007; 6:795-801; PMID:17387270; http://dx.doi.org/10.4161/cbt.6.5.4007
- Jin KL, Pak JH, Park JY, Choi WH, Lee JY, Kim JH, Nam JH. Expression profile of histone deacetylases 1, 2 and 3 in ovarian cancer tissues. J Gynecol Oncol 2008; 19:185-90; PMID:19471575; http://dx.doi.org/10.3802/jgo.2008.19.3.185
- Tomsova M, Melichar B, Sedlakova I, Steiner I. Prognostic significance of CD3+ tumor-infiltrating lymphocytes in ovarian carcinoma. Gynecol Oncol 2008; 108:415-20; PMID:18037158; http://dx.doi.org/10.1016/j.ygyno.2007.10.016
- Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science 1996; 274:94-6; PMID:8810254; http://dx.doi.org/10.1126/science.274.5284.94
- Kayser S, Watermann I, Rentzsch C, Weinschenk T, Wallwiener D, Gückel B. Tumor-associated antigen profiling in breast and ovarian cancer: mRNA, protein or T cell recognition? J Cancer Res Clin Oncol 2003; 129:397-409; PMID:12836015; http://dx.doi.org/10.1007/s00432-003-0445-7
- Inzkirweli N, Gückel B, Sohn C, Wallwiener D, Bastert G, Lindner M. Antigen loading of dendritic cells with apoptotic tumor cell-preparations is superior to that using necrotic cells or tumor lysates. Anticancer Res 2007; 27:2121-9; PMID:17695495
- Schultze JL, Michalak S, Seamon MJ, Dranoff G, Jung K, Daley J, Delgado JC, Gribben JG, Nadler LM. CD40-activated human B cells: an alternative source of highly efficient antigen presenting cells to generate autologous antigen-specific T cells for adoptive immunotherapy. J Clin Invest 1997; 100:2757-65; PMID:9389740; http://dx.doi.org/10.1172/JCI119822
- Walter S, Herrgen L, Schoor O, Jung G, Wernet D, Bühring HJ, Rammensee HG, Stevanovic S. Cutting edge: predetermined avidity of human CD8 T cells expanded on calibrated MHC/anti-CD28-coated microspheres. J Immunol 2003; 171:4974-8; PMID:14607891; http://dx.doi.org/10.4049/jimmunol.171.10.4974
- Britten CM, Gouttefangeas C, Welters MJ, Pawelec G, Koch S, Ottensmeier C, Mander A, Walter S, Paschen A, Muller-Berghaus J et al. The CIMT-monitoring panel: a two-step approach to harmonize the enumeration of antigen-specific CD8+ T lymphocytes by structural and functional assays. Cancer Immunol Immunother 2008; 57:289-302; PMID:17721783; http://dx.doi.org/10.1007/s00262-007-0378-0
- Brunner KT, Mauel J, Cerottini JC, Chapuis B. Quantitative assay of the lytic action of immune lymphoid cells on 51-Cr-labelled allogeneic target cells in vitro; inhibition by isoantibody and by drugs. Immunology 1968; 14:181-96; PMID:4966657
- Kowalewski DJ, Stevanovic S. Biochemical large-scale identification of MHC class I ligands. Methods Mol Biol 2013; 960:145-57; PMID:23329485; http://dx.doi.org/10.1007/978-1-62703-218-6_12
- Kurman R. Blaustein's pathology of the female genital tract. New York Springer Verlag 2002; Fifth edition:791-904
- Pham DL, Scheble V, Bareiss P, Fischer A, Beschorner C, Adam A, Bachmann C, Neubauer H, Boesmueller H, Kanz L et al. SOX2 expression and prognostic significance in ovarian carcinoma. Int J Gynecol Pathol 2013; 32:358-67; PMID:23722508; http://dx.doi.org/10.1097/PGP.0b013e31826a642b
- Remmele W, Schicketanz KH. Immunohistochemical determination of estrogen and progesterone receptor content in human breast cancer. Computer-assisted image analysis (QIC score) vs. subjective grading (IRS). Pathol Res Pract 1993; 189:862-6; PMID:8302707; http://dx.doi.org/10.1016/S0344-0338(11)81095-2
