ABSTRACT
Exosomes are a class of nanovesicles formed and released through the late endosomal compartment and represent an important mode of intercellular communication. The ability of anticancer chemotherapy to enhance the immunogenic potential of malignant cells mainly relies on the establishment of the immunogenic cell death (ICD) and the release of damage-associated molecular patterns (DAMPs). Here, we investigated whether genotoxic stress could promote the release of exosomes from multiple myeloma (MM) cells and studied the immunomodulatory properties they exert on NK cells, a major component of the antitumor immune response playing a key role in the immunosurveillance of MM. Our findings show that melphalan, a genotoxic agent used in MM therapy, significantly induces an increased exosome release from MM cells. MM cell-derived exosomes are capable of stimulating IFNγ production, but not the cytotoxic activity of NK cells through a mechanism based on the activation of NF-κB pathway in a TLR2/HSP70-dependent manner. Interestingly, HSP70+ exosomes are primarily found in the bone marrow (BM) of MM patients suggesting that they might have a crucial immunomodulatory action in the tumor microenvironment. We also provide evidence that the CD56high NK cell subset is more responsive to exosome-induced IFNγ production mediated by TLR2 engagement. All together, these findings suggest a novel mechanism of synergism between chemotherapy and antitumor innate immune responses based on the drug-promotion of nanovesicles exposing DAMPs for innate receptors.
Abbreviations
| BM | = | bone marrow |
| DAMP | = | damage-associated molecular pattern |
| DLS | = | dynamic light scattering |
| ER | = | endoplasmic reticulum |
| HMGB1 | = | high-mobility group box 1 |
| HSP | = | heat shock protein |
| ICD | = | immunogenic cell death |
| IFN | = | interferon |
| MDSC | = | myeloid-derived suppressor cells |
| MEL | = | melphalan |
| MM | = | multiple myeloma |
| NF-kB | = | nuclear factor-kB |
| NK | = | natural killer |
| PAMP | = | pathogen-associated molecular pattern |
| PB | = | peripheral blood |
| PC | = | plasma cell |
| TLR | = | toll-like receptor |
Introduction
Developmental patterns, tissue homeostasis and immunological responses are regulated by cellular cross-talk. Traditionally, cells communicate with each other through direct cell–cell contact and/or soluble factors. However, recent work has pointed out as novel and crucial regulators of cellular cross-talk, the exosomes, endosome-derived nanovesicles, which are released in the extracellular environment and are subsequently uptaken by homo- and/or hetero-typic cells.Citation1,2 In this context, a fast developing field is that focusing on how exosomes released by cancer cells in the tumor microenvironment are uptaken by different immune cell types, thereby modulating antitumor immune response and affecting tumor progression.Citation3,4
Natural killer (NK) cells are innate immune effectors that play a pivotal role in tumor immunosurveillance by exerting direct cytotoxic effects as well as producing a variety of cytokines. NK cell activation is tightly regulated by a fine balance between activating and inhibitory signals. Recognition of altered self on tumor cells triggers non-MHC class-I-restricted activating receptors.Citation5 In addition, NK cells can directly recognize several molecular patterns of ligands (i.e., pathogen-associated molecular patterns, PAMPs and damage-associated molecular patterns, DAMPs) through receptors belonging to the toll-like receptor family (TLRs).Citation6 Human NK cells can be subdivided into various subsets based on the relative expression of CD16 and CD56. In particular, the CD56low CD16high population is predominant in peripheral blood (PB), whereas tissue resident NK cells are primarily CD56high. The latter are more efficient producers of cytokines endowed with immunoregulatory properties, but they can also become cytotoxic upon appropriate activation and have been implicated to different disease states including cancer.Citation7
Multiple myeloma (MM) is a clonal B-cell malignancy characterized by the expansion of plasma cells (PCs) in the bone marrow (BM). NK cells have long been considered essential in the control of MM progression, because they undergo expansion and activation during the early stages of disease and have the ability to recognize and kill MM cells.Citation8 This notion has spurred great interest in developing strategies aimed at stimulating NK cell-mediated functions for MM immunotherapy.Citation9-12
The ability of anticancer chemotherapy and radiotherapy to enhance the immunogenic potential of malignant cells mainly relies on the establishment of immunogenic cell death (ICD) and release of DAMPs, including heat shock proteins (HSPs), high-mobility group box 1 proteins (HMGB1), ATP and the endoplasmic reticulum (ER) chaperone calreticulin.Citation13,14 In this context, both HSP70 and HMGB1 appear to exert an immunostimulatory activity by promoting DC maturation through TLR4, which, in turn, leads to the induction of antigen-specific T cell-mediated antitumor immune responses.Citation15,16 In this regard, several studies highlight the importance of TLRs in the modulation of innate immune response in anticancer immunity.Citation17,18 More recently, low doses of chemotherapeutic drugs have been shown to induce immunogenic senescence and stimulate NK cell-mediated recognition and clearance of drug-treated tumor cells via the upregulation of NKG2D and DNAM-1 activating ligands on the surface of cancer cells.Citation19-22
An outstanding question concerns the role of tumor-derived exosomes (Tex) on the modulation of NK cell-mediated functions. The available evidence points to the molecular composition of the exosome cargo as a major determinant of the immunoregulatory activity of Tex on NK cells pointing to a prevailing, although not exclusive, inhibitory role.Citation23 Accordingly, exosomes carrying TGF-β and/or ligands for the activating receptor NKG2D have been described to negatively affect NK cell functions.Citation24-30 In contrast, exosomes displaying high levels of HSP70 were reported to enhance NK cell migration and cytotoxicity through a still unknown mechanism.Citation31,32 Stimulation of NK cell activity by Tex was also shown to be dependent on NKp30 ligand BAT3 expressed on the exosome surface.Citation33,34
We endeavored to address whether exosomes released by MM cells exposed to sublethal doses of melphalan (MEL), a drug currently used in the clinical management of MM patients, affect NK cell effector functions. Herein, we show that MM-derived exosomes are capable of stimulating the production of IFNγ by NK cells through a mechanism based on the activation of the NF-kB pathway in a TLR-2/HSP70-dependent manner. We also provide evidence that the CD56high subset is more responsive to exosome stimulation. Our findings suggest a novel mechanism whereby a chemotherapeutic drug can enhance antitumor innate immune responses via the release of nanovesicles carrying DAMPs/HSP70 capable of triggering the activation of innate receptor/TLR2 expressed by NK cells.
Results
Structural and biochemical characterization of exosomes derived from multiple myeloma cells in steady-state conditions and upon MEL treatment
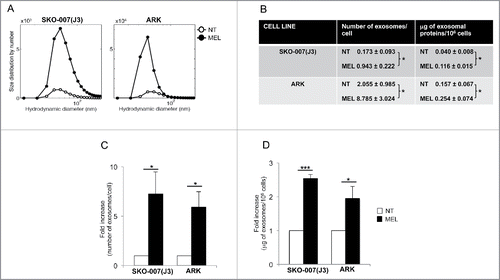
To isolate and characterize exosome release by MM cells, we used an established exosome isolation protocol that exploits their differential sedimentation properties.Citation35 Exosomes were isolated from the conditioned media of ARK and SKO-007(J3) MM cell lines either under steady-state conditions or upon treatment with sublethal doses of MEL. Dynamic light scattering (DLS) methodology was used to evaluate exosome size distribution and quantify their number (Figs. S1A–D). Our results show that exosomal preparations from both cell lines contain vesicles whose diameter ranges from 60 to 130 nm, thus matching the reported size of typical exosomes. In addition, drug treatment increased the amount of released exosomes without affecting their size distribution (). Remarkably, the higher number of exosomes released by drug-treated cells yielded an increased amount of total proteins recovered from the exosomal samples (), implying constant loading of cargo into exosomes. Interestingly, although at the steady-state ARK cells secreted more nanovesicles when compared with SKO-007(J3) cells, the relative increase of exosome production in response to MEL treatment was similar in the two MM cell lines ( and ).
Figure 1. Melphalan-treated MM cells release a higher amount of exosomes. (A) MM cells were cultured for 48 h in the presence or not of MEL, and then exosomes were isolated from the conditioned media. Size distribution of SKO-007(J3)- and ARK-derived exosomes was analyzed through DLS. A representative experiment out of three is shown. (B) The number of exosomes/cell was calculated through the DLS (details can be found in Fig. S1). In parallel, the amount of total proteins recovered from exosomal preparations derived from 106 cells was evaluated by BPA. The mean values of three (for SKO-007(J3)) or four (for ARK) independent experiments ± SEM is shown. (C) Data are expressed as fold increase of the number of particles/cell obtained from MEL-treated MM cells divided by the number of particles/cell of the untreated group. The mean of three (for SKO-007(J3)) or four (for ARK) independent experiments ± SEM is shown. (D) Data are expressed as fold increase of the μg of exosomal proteins/106 cells values obtained from MEL treated MM cells divided by μg/106 cells of the untreated group. The mean of nine (for SKO-007(J3)) or six (for ARK) independent experiments ± SEM is shown.
We confirmed through ultrastructural analysis that our MM-purified exosome preparations contained nano-sized cup-shaped vesicles (). In addition, immunogold labeling showed the expression of specific exosome markers such as CD81 and Tsg101 on the outer layer of MM vesicles (). In accordance with the results obtained by DLS, the ultrastructural analysis did not reveal any relevant morphological difference among exosomes purified from control and drug-treated cells (Fig. S2). In addition, we readily detected canonical exosome markers such as Tsg101, CD63, MHC I and HSP70 by Western blot analysis. Importantly, calreticulin, which is exclusively associated to ER, was not detected in the exosome preparations (). Again, a similar marker profile was found in exosomes derived from control and MEL-treated cells. CD63 expression on the surface of exosomes was also evaluated by immunofluorescence and flow cytometric (FACS) analysis ().
Figure 2. Characterization of MM cell-derived exosomes. (A) Electron microscope analysis of exosomes morphology and size. A representative picture of SKO-007(J3)-derived exosomes is shown. (B) Immune-gold labeling for Tsg101 and CD81 of SKO-007(J3)-derived exosomes. (C) Western blot analysis was performed on lysates derived from exosome fractions or from cell pellet, using anti-Hsp70, anti-MHC I, anti-Tsg101, anti-CD63 and anti-calreticulin antibodies. (D) Visualization of CD63-conjugated beads coated with exosomes by immunofluorescence and FACS analysis (upper panel). CD63 expression on the surface of exosomes was assessed after overnight incubation of exosomes with beads, exosomes were stained with anti-CD63 mAb (thin histograms) or control isotypic Ig (filled histograms) (bottom panel).
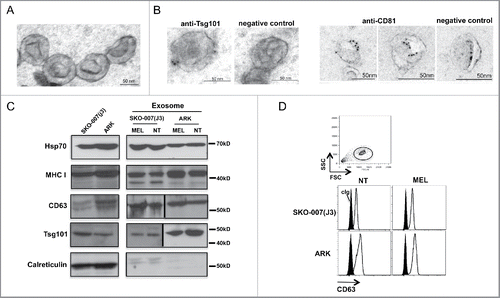
All together, these data indicate that MM cells secrete exosomes and MEL treatment increases their release without affecting their size or marker content.
MM-derived exosomes are uptaken by NK cells and stimulate CD69 expression
To investigate the effects of MM-derived exosomes on NK cell-mediated functions, we first explored whether these nanovesicles could be uptaken by NK cells. Exosomes labeled with the fluorescent PKH26 dye were incubated for different length of time with highly purified NK cells with or without IL-15 supplementation. As shown in , both resting and IL-15-activated NK cells were labeled by PKH6, with a peak of fluorescence being detected at 3 h. To directly demonstrate that exosomes were internalized and not just bound to the cell surface, exosome–NK cell conjugates were treated with trypsin before FACS analysis. We found that trypsin treatment reduced the percentage of fluorescent cells marginally, thus supporting the notion that the majority of PKH26-labeled exosomes were distributed intracellularly (). In addition, when NK cells were incubated with an excess of unlabeled exosomes, we could not detect PKH26+ cells, strongly suggesting that NK cells were labeled by PKH26 as a consequence of exosome uptake (). Fluorescence microscopy imaging of NK cells incubated for 3 h with PKH26-labeled exosomes further confirmed exosome internalization (). We verified that CD56high and CD56low NK cell subsets had a similar capability to uptake exosomes (). Next, we assessed the mechanism by which exosomes are internalized into human primary NK cells. The uptake of exosomes was strongly impaired when cells were assayed either in presence of EDTA () or at 4°C (not shown), suggesting that exosome uptake is an energy-dependent process reliant on tensile forces generated by the actin cytoskeleton. In addition, exosome uptake was markedly reduced by nystatin and dynasore, which implies its dependence on endocytic route/s requiring caveolae/raft integrity and dynamin functionCitation36 (). EIPA caused a minor reduction of exosome uptake, pointing to macropynocytosis as an additional, albeit marginally relevant, route of exosome internalization in NK cells.
Figure 3. MM cell-derived exosomes are uptaken by primary NK cells and induce CD69 expression. (A) SKO-007(J3)-derived exosomes were labeled with the red fluorescent dye PKH26. Primary peripheral blood human NK cells were incubated for different times with 20 μg/mL of PKH26-labeled exosomes with or without IL-15 (50 ng/mL). The fluorescence of internalized exosomes was evaluated by immunofluorescence and FACS analysis and measured as the percentage of PKH26+ cells. One representative experiment is shown. (B) NK cells were cultured for 3 h in the presence of PKH26-labeled exosomes and IL-15 as described in panel (A), or with a combination of PKH26-labeled exosomes and trypsin, or PKH26-labeled and unlabeled exosomes at 1:3 ratio. The mean of three independent experiments ± SEM is shown. (C) NK cells were incubated for 3 h with PKH26-labeled exosomes (20 µg/mL), washed and plated on poly-L-lysine-coated multichamber glass plates and fixed. Images were acquired using an ApoTome Observer Z.1 microscope with a 60x/1.4 NA Plan-Neofluar oil immersion objective. Upper panels: Representative images of single cells are shown as maximum intensity projection (3 Z sections with 0.2 μm spacing). Lower panels: Differential interference contrast overlay the fluorescence images were also shown. Bar represents 5 µm. (D) NK cells were incubated for 3 h with PKH26-labeled exosomes (20 µg/mL), anti-CD56APC was added the last 20 min. Cells were washed and the fluorescence of internalized exosomes was evaluated by immunofluorescence and FACS analysis and measured as the percentage of PKH26+ cells by gating on CD56low and CD56high subsets. The mean of four experiments is shown. (E) NK cells were pre-treated for 1 h with EDTA (2 mM), EIPA (50 μM), dynasore (50 μM), nystatin (40 μg/mL) and then incubated for 3 h with PKH26-labeled exosomes (20 µg/mL), the fluorescence of internalized exosomes was evaluated by immunofluorescence and FACS analysis and measured as the percentage of PKH26+ cells. Data expressed as the percentage of uptake were referred to the untreated cells considered as 100%. Values reported represent the mean of three independent experiments ± SEM. (F) NK cells were incubated for 48 h with different amounts of SKO-007(J3)-derived exosomes as indicated. CD69 expression was evaluated by immunofluorescence and FACS analysis. Numbers in each histogram represent the percentage of CD69+ cells. A representative experiment is shown. (G) NK cells were incubated with 20 μg/mL of SKO-007(J3) and ARK–derived exosomes as described in panel (F). Where indicated, exosomes were prepared from MEL-treated MM cells (exo MEL). Data are represented as the mean values of the percentage of CD69+ NK cells of four (for SKO-007(J3)-derived exosomes) or five (for ARK-derived exosomes) independent experiments ± SEM.
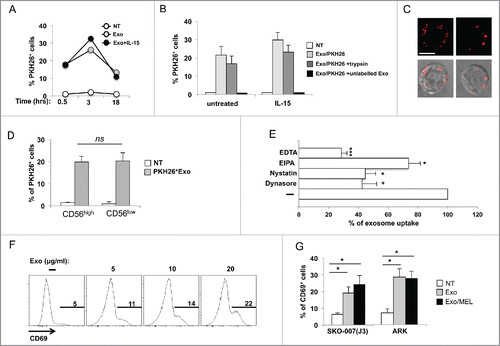
To analyze whether exosomes could affect NK cell activation, we incubated primary PB NK cells with different amounts of exosomes and evaluated expression of the activation marker CD69. Interestingly, CD69 was induced on exosome-treated NK cells in a dose-dependent manner (). Similar effects were observed using exosomes released from ARK or SKO-007(J3) MM cells, with no significant differences between exosomes derived from untreated or MEL-treated MM cells ().
MM-derived exosomes stimulate IFNγ production through a mechanism mediated by nuclear factor (NF)-kB signaling pathway
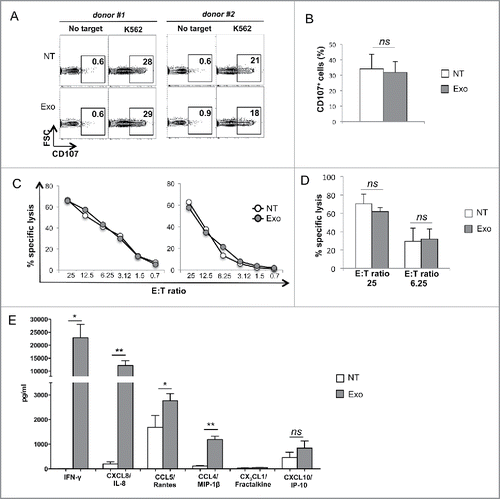
Next, we investigated whether MM cell-derived exosomes could affect NK cell-mediated effector functions. To this aim, NK cell degranulation, cytotoxicity and cytokine production were measured following incubation with exosomes. We found that treatment of NK cells with exosomes affected neither degranulation (as determined by CD107a surface expression) ( and ) nor cytotoxic activity ( and ) toward co-cultured K562 cells. In contrast, a significant increase in the production of IFNγ and some chemokines, including CXCL8/IL-8, CCL5/Rantes and CCL4/MIP-1β was observed upon exosome treatment ().
Figure 4. MM cell-derived exosomes do not affect NK cell degranulation and cytotoxicity but enhance cytokine and chemokine production. (A) NK cells were incubated for 48 h with 20 μg/mL of SKO-007(J3)-derived exosomes. The degranulation assay was performed against the sensitive target K562, using the CD107a lysosomal marker. The percentage of CD107+ NK cells is indicated in each panel. Two representative donors are shown. (B) The mean values of five independent experiments are shown. (C) NK cells were incubated in the presence of exosomes as in panel (A). The cytotoxic activity of untreated or exosome-treated NK cells was assayed against K562 target cells. Two representative experiments are shown. (D) The mean values of three independent experiments at the indicated E:T ratio are shown. (E) NK cells were incubated in the presence of exosomes as in panel (A). Cells were harvested and supernatants were collected and the amount of different cytokines as indicated, was measured by performing a Luminex assay. The mean values of three independent experiments ± SEM are shown. Medium containing exosome alone has undetectable levels of cytokine/chemokines (data not shown).
We then examine the molecular mechanism/s through which the MM-derived exosomes could induce IFNγ production by NK cells. We verified that exosome-mediated IFNγ production by NK cells was associated to increased expression of IFNG mRNA (); notably, neither exosomes derived from non-malignant cells (such as primary fibroblasts or PBMCs) nor synthetic liposomes exerted a stimulatory effect (Fig. S3).
Figure 5. MM cell-derived exosomes stimulate IFNγ production through a mechanism mediated by NF-kB pathway. (A) NK cells were incubated for 48 h with 20 μg/mL of SKO-007(J3)-derived exosomes. Real-time PCR analysis of IFNγ mRNA. Data, expressed as fold change units, were normalized with β-actin and referred to the untreated cells considered as calibrator. Values reported represent the mean of six independent experiments ± SEM. (B) NK cells were incubated with 20 μg/mL of SKO-007(J3)-derived exosomes as described in A. Western blot analysis was performed on total cell lysates using p65, phospho-p65 (p-p65) and β-actin Abs. Numbers beneath each line represent quantification of p-p65 and p65 by densitometry normalized with β-actin. (C) NK cells were pretreated for 1 h with the NF-kB inhibitor, SN50 (15 µM), and then incubated with 20 μg/mL of SKO-007(J3)-derived exosomes for 48 h. Real-time PCR analysis of IFNγ mRNA was performed as described in panel (A). The mean of three independent experiments is shown. (D) Nuclear extracts were prepared from NK cells untreated or treated with MM-derived exosomes, and analyzed by EMSA. The nuclear extract derived from NK cells treated with MM exosomes was used for competition with unlabelled probes as indicated in the right panel. (E) NK cells were cultured with 20 μg/mL of SKO-007(J3) cells-derived exosomes in the presence of IL-15 (50 ng/mL). After 24 h, BFA (5 µg/mL) was added and left for additional 24 h. Intracellular IFNγ expression was evaluated by immunofluorescence and FACS analysis. Numbers represent the percentage of IFNγ+ NK cells. One representative experiment is shown. (F) Data were represented as mean values of the percentage of IFNγ+ cells of seven independent experiments ± SEM.
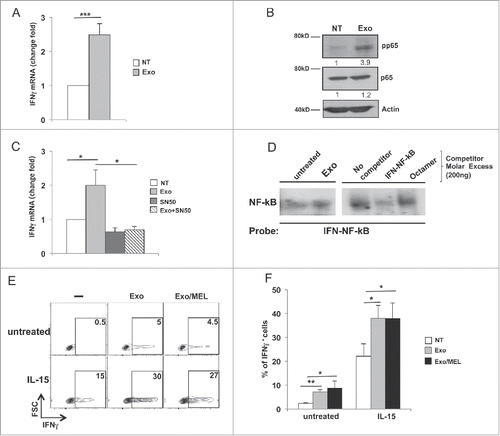
To investigate possible signaling pathways involved in exosome-induced INFγ production, we focused our attention on NF-kB, since this transcription factor was shown to be involved in the transcription of several cytokines, including IFNγ.Citation37 Our results show that exosomes caused an increase of p65 phosphorylation, a major activating component of NF-kB signaling, leaving p65 total levels unchanged (). To further support the possible involvement of NF-kB in the exosome-induced IFNγ production, NK cells were pre-treated with SN50, a cell permeable peptide that inhibits translocation of the NF-kB active complex into the nucleus, and then incubated with exosomes. As shown in , SN50 treatment inhibited exosome-induced IFNγ production. Importantly, this compound did not affect exosome uptake (data not shown). Moreover, as shown in the EMSA assay, MM-derived exosomes enhanced specific binding to a IFNγ/NF-kB site previously identified within the promoter region of this geneCitation37 confirming the involvement of this pathway in the upregulation of IFNγ expression (). Interestingly, the combined stimulation of NK cells with IL-15 and exosomes, further enhanced IL-15 induced IFNγ production, with no differences between exosomes-derived from untreated or MEL-treated MM cells ( and ). Finally, we explored the possible effect of conditioned-media obtained from untreated or exosome-treated NK cells, on MM cell proliferation and survival. The rate of cell proliferation (Figs. S4A and B) and apoptosis (Fig. S4C) was comparable in SKO-007(J3) cells cultured with conditioned-media obtained from either control or exosome-treated NK cells.
These data demonstrate that MM-derived exosomes can stimulate IFNγ production but not the degranulation or cytotoxicity of NK cells, and this event requires the activation of NF-kB signaling pathway.
MM-derived exosomes stimulate IFNγ production in a toll-like receptor 2 (TLR2) dependent manner
It has been shown that exosomes from different cellular sources have the capability to trigger immune cell functions through a mechanism requiring receptors belonging to the TLR family, namely TLR7, 8, 1 and 2.Citation38-41 Since NF-kB activation is a major signaling event downstream to TLRs, we asked whether exosome-induced NF-kB activation could be mediated by one or more TLRs. To this aim, we used a panel of 293-derived reporter cell lines engineered to co-express a specific TLR along with an NF-kB-driven luciferase reporter. Interestingly, exposure to exosomes induced luciferase activity only in the reporter cell line expressing TLR2, whereas no effect was observed in cells expressing TLR3, TLR4, TLR7, TLR8 and TLR9 (Fig. S5A) or in cells expressing the NF-κB-driven luciferase reporter gene alone (Fig. S5B). These data suggest that MM cell-derived exosomes can activate NF-kB signaling by engaging TLR2.
Since we defined an important role for TLR2, the effect of a specific agonist on NK cell activation was promptly examined. Thus, highly purified PB NK cells were treated with different doses of the TLR2 agonist, Pam3CSK4 or exosomes, and IFNγ production was evaluated by immunofluorescence and FACS analysis on both CD56high and CD56low NK cells. Interestingly, Pam3CSK4 treatment strongly stimulated IFNγ expression in the majority of CD56high NK cells, whereas just a small percentage of CD56low NK cells responded to TLR2 agonist. Remarkably, also MM-derived exosome-induced IFNγ production was prevalent in CD56high NK cells ( and ). To further prove that the CD56high NK cell subset was more responsive to both exosomes and Pam3CSK4, CD56high and CD56low NK cell subsets were FACS sorted based on CD56 expression levels (), and IFNγ mRNA expression was evaluated upon exosome or Pam3CSK4 treatment. As shown in , in response to exosome or TLR2 agonist stimulation, upregulation of IFNγ mRNA expression was observed only in the CD56high NK cell subset. Of note, the above differences in responsiveness to TLR2 agonist and exosomes were not attributable to different levels of cell surface TLR2 expressed by the two NK cell subsets ().
Figure 6. MM exosomes stimulate IFNγ production in a TLR2-dependent manner. (A) NK cells were incubated with increasing doses of Pam3CSK4 or SKO-007(J3) cell-derived exosomes, as indicated. After 24 h, BFA (5 µg/mL) was added and left for additional 24 h. Intracellular IFNγ expression was evaluated by immunofluorescence and FACS analysis. The gating strategy used consists in separating CD56high cells from CD56low NK cells. Numbers indicate the percentage of IFNγ+ cells. A representative experiment is shown. (B) NK cells were incubated with exosomes (20 µg/mL) as described in panel (A). The mean values of four independent experiments ± SEM is shown. (C) NK cells were FACs sorted based on CD56 expression levels, and incubated for 48 h with SKO-007(J3)-derived exosomes (20 µg/mL) or Pam3CSK4 (1 μM). Real-time PCR analysis of IFNγ mRNA was performed and the data, expressed as fold change units, were normalized with β-actin and referred to the untreated cells considered as calibrator. The mean values of five independent experiments ± SEM are shown. (D) Cell surface expression of TLR2 was evaluated on CD56+CD3− total NK cells, on CD56highCD3− and CD56lowCD3− NK cell subsets of total PBMC derived from 10 different healthy donors. Values represent the mean fluorescence intensity (MFI) of TLR2 substracted from the MFI value of the isotype control Ig. (E) CD56high NK cells were pretreated for 20 min with anti-TLR2 neutralizing antibody (1μg/106 cells), and then incubated with both TLR2 agonist and exosomes as described in panel (B). Real-time PCR analysis of IFNγ mRNA was performed and the data, expressed as fold change units, were normalized with β-actin and referred to the untreated cells considered as calibrator. Results relative to two representative donors are shown.
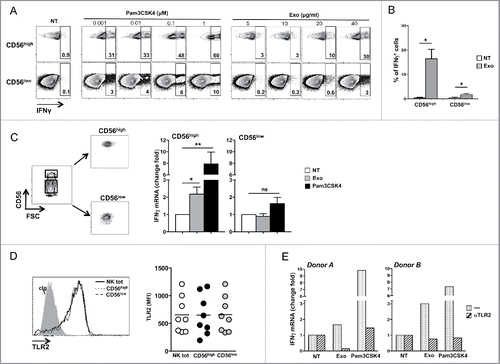
To further investigate the contribution of TLR2 in the exosome-mediated IFNγ production by CD56high NK cells, an anti-TLR2 neutralizing antibody was added to FACS-sorted CD56high NK cells before exosome stimulation. This resulted in a blockade of both Pam3CSK4 and exosome-mediated IFNγ production by CD56high NK cells from two different healthy donors ().
Collectively, our results indicate that (a) MM-cell derived exosomes stimulate IFNγ production by human primary NK cells through the engagement of TLR2 and (b) the CD56high subset is prevalently responsible for IFNγ production by exosome-targeted NK cells.
HSP70 is associated with MM-derived exosomes and contributes to IFNγ production
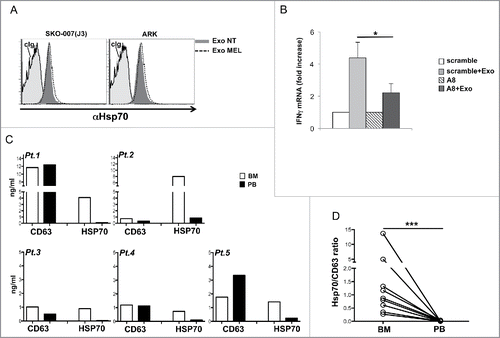
A variety of DAMPs have been classified as TLR2 ligands including Versican, HMGB1 and HSPs.Citation13 Having detected that lysates prepared from MM-derived exosomes displayed strong anti-HSP70 immunoreactivity (), we investigated HSP70 localization on the outer surface of the exosome membrane. To this aim, the cmHsp70.1 mAb, previously shown to recognize cell surface HSP70,Citation42 was used to stain exosome-bead conjugates. Our findings show that exosomes derived from both ARK and SKO-007(J3) MM cells displayed surface HSP70 expression. Similar levels were detected in exosomes derived from untreated or MEL-treated cells (), despite the fact that MEL treatment upregulated cell surface HSP70 expression in both MM cell lines (Fig. S6).
Figure 7. HSP70 is associated to MM cell-derived exosomes and contribute to IFNγ production. (A) HSP70 expression on the surface of exosomes was assessed after overnight incubation of exosomes with latex-beads. Exosomes derived from untreated or MEL-treated MM cells were stained with anti-HSP70 mAb or control isotypic Ig. One out of three experiments is shown. (B) Purified NK cells were incubated for 24 h with 20 µg/mL of SKO-007(J3)-derived exosomes in the presence of A8 aptamer or with a scramble peptide both used at 30 μM. Real-time PCR analysis of IFNγ mRNA was performed and the data, expressed as fold change units, were normalized with β-actin and referred to the untreated cells or scramble/treated cells considered as calibrators. The mean values of three independent experiments ± SEM are shown. (C–D) Exosomes were extracted from the plasma of PB or BM-derived from MM patients (Pt), lysed in RIPA buffer and ELISA for HSP70 or CD63 was performed. In panel (C), absolute values of both CD63 and HSP70 are shown for five patients. In panel (D), values of HSP70 were normalized with CD63 and used to compare the relative amount of BM HSP70 vs. PB HSP70. Results relative to nine patients are shown.
Recent evidence show that the C-terminal domain of surface-exposed HSP70 can be bound by the A8 peptide aptamer, thereby preventing HSP70/TLR2 association and the TLR2-dependent activation of myeloid-derived suppressor cells (MDSCs) by tumor-derived exosomes.Citation43
The addition of the A8 peptide to the culture medium partially suppressed the induction of IFNγ expression by MM cell-derived exosomes, whereas a scrambled peptide had no effect (), indicating that HSP70 expressed on the outer membrane of exosomes could contribute to TLR2 triggering in NK cells.
Overexpression of HSP70 in cancer cells has been associated to tumor progression and the majority of HSP70 is mainly associated with lipid vesicles secreted by tumor cells displaying HSP70 on their surface.Citation44,45 We thus investigated HSP70 expression on exosomes purified from the PB and BM of MM patients (Figs. S7A and B). Interestingly, although the levels of CD63 were comparable in the two pools of exosome preparations, HSP70 was mainly present on BM-derived exosomes ( and ). Next, we examined whether primary malignant PCs could effectively contribute to the HSP70+ exosome pool in the BM, and our results show that highly purified PCs isolated from the BM of two different MM patients had the capability to produce HSP70+ exosomes in short-term (48 h) cultures (Fig. S7C). All together these data indicate that HSP70 is expressed on the surface of MM exosomes and contributes to exosome-induced IFNγ production by NK cells. Moreover, exosomes expressing HSP70 are mainly found in the BM tumor microenvironment.
Discussion
Exosomes play a relevant role in intercellular communication, thanks to their ability to convey many proteins, lipids, miRNAs and mRNA onto target cells.Citation2,4 Several studies describe the involvement of exosomes in the modulation of both innate and adaptive immune response through different mechanisms.Citation1,3,46 As for NK cells, the immunomodulatory role of exosomes appears to be multi-faceted and dependent on the identity of both the exosomes-associated cargo end exosome releasing cells.Citation23,47 In this context, the inhibitory action of Tex on NK cell functions has been mostly attributed to TGF-β and/or NKG2D ligands associated to these vesicles.Citation24-30
Focusing on MM as a clinically and biologically relevant model of tumor-NK cell interactions, we report herein that (i) melphalan, a drug currently used in anti-MM chemotherapy, enhances the release of exosomes from MM cells; (ii) MM-derived exosomes are capable of inducing INFγ production by NK cells through a mechanism that entails activation of NF-kB downstream to TLR2; (iii) HSP70 displayed on the outer surface of MM-derived exosomes is a major trigger of TLR2 activation in NK cells. Hence, our study describes a novel paradigm of drug-induced modifications in MM cells capable of affecting tumor development through the regulation of NK cell activity.
We initially observed that MM cell-derived exosomes had the typical size and morphology as assessed by electron microscopy and DLS together with the expression analysis of some characteristic markers as described previously.Citation48,49 Our findings also show that both resting or IL-15 activated primary NK cells internalize, and not only bind MM cell-derived exosomes with a rapid kinetics. Interestingly, we found that NK cell exosome uptake is an active process that mainly depends on endocytic route/s requiring dynamin and caveolae/raft endocytosis () with TLR2 laying no apparent role (data not shown).
We provide novel evidence that the MM-derived exosomes induced IFNγ production on CD56high NK cells is mediated by TLR2 engagement. The different response to TLR2 triggering among CD56high and CD56low subsets was not attributable to differences in either the levels of cell surface TLR2 expression or to a different capability to uptake exosomes. In line with these results, it was previously shown that IFNγ production in response to PSK2, one of the known TLR2 agonists, was mainly associated to the NK CD56high subset.Citation50 Thus, it is likely that the higher responsiveness of the CD56high cell subset to exosome treatment mainly relies on its intrinsic ability to produce cytokines.
Besides IFNγ production, MM exosomes also induced the expression of the CD69 activation marker and the production of several cytokines whereas they did not affect NK cell degranulation and cytotoxicity. Our data are in agreement with previous work showing that TLR engagement on NK cells can stimulate cytokine production without affecting cytotoxicity.Citation51-53 Similar to our results, exposure of NK cells to TLR2 agonists was found to allow the induction of CD69 expression as well as the production of IFNγ.Citation54,55 Of interest, unlike other studies reporting that different types of exosomes have the capability to trigger immune cell functions through different receptors belonging to TLR family, including TLR7, 8, 1 and 2,Citation38-41 only TLR2 is involved in the MM exosome-induced NK cell activation.
One of the main pathway triggered by TLRs is NF-kB, a transcription factor implicated in activation of several cytokine genes, including IFNG.Citation37 We found that exosome-induced IFNγ production was dependent on NF-kB signaling. Consistent with these findings, a recent study has proposed a model of TLR2-mediated NF-kB activation and consequent cytokine production in human macrophages in response to breast cancer cell-derived exosomes.Citation38
TLR2 can recognize different types of DAMPs including HSPs.Citation56,57 In general, these molecular chaperones are usually expressed intracellularly where they support the folding and the transport of a great variety of proteins. In contrast, membrane-bound and extracellular-located HSPs act as potent danger signals.Citation58 Several pieces of evidence demonstrate that extracellular-located HSPs can be associated to extracellular vesicles.Citation31,32,43,59 Our results show that MM exosomes expressed high levels of HSP70 on the surface, and we also demonstrate that this molecule contributed to exosome-induced IFNγ production by NK cells.
Notably, the presence of HSP70+ exosomes primarily in the BM of MM patients allows us to suggest that these vesicles play a pivotal role in the tumor microenvironment by affecting immune cell functions. In this regard, our preliminary data indicate that CD56high NKcells are present in the BM of MM patients suggesting that this subset might promptly respond to HSP70/exosomes-mediated stimulation (Zingoni et al. data not shown). In addition, other TLR2+ immune cells (i.e., MDSC, DC, etc.) resident in the MM BM microenvironment could also be affected by the exosomes. In this regard, Chalmin and colleagues demonstrated that Hsp72 located on the tumor exosome surface was able to engage TLR2 expressed on MDSCs.Citation39 Moreover, MM exosomes overexpressing membrane-bound Hsp70 have the capability to induce DC maturation and stimulate a type 1 CD4+ T and CD8+ T-cell responses along with the induction of NK cell-mediated immunity in mice.Citation60 Thus, MM cell-derived exosomes in vivo could affect tumor survival and progression, thanks to their ability to regulate different subsets of immune cells localized in the BM microenvironment and responsive to HSP70.
Of interest, we also provide evidence that low doses of the genotoxic agent MEL stimulate an increased release of exosomes from MM cells. In terms of modulatory effects on NK cells, no differences were observed between exosomes derived from untreated and MEL-treated cells. Moreover, since we found a dose-dependent correlation between the exosome amount and NK cell activation, chemotherapy could play a crucial role in the induction of a stronger NK cell-mediated immune response consequent to the enhancement of exosome release. In this regard, TLR2 agonist treatment was proposed as adjuvant in cancer immunotherapy,Citation61 because it caused strong activation of NK cells in the context of ADCC induced by the therapeutic antibody Trastuzumab.Citation50 Our results are in line with recent evidence that exosome release in tumor cells was augmented in response to different drugs, including etoposide,Citation32 cisplatin and 5-fluorouracil.Citation43
In summary, our findings add new information on the immunomodulatory action of MM-derived exosomes during the course of chemotherapeutic interventions. We propose a novel mechanism whereby chemotherapeutic drugs act in synergy with antitumor innate immune response based by enhancing the release of nanovesicles exposing DAMPs, such as HSP70. We envisage that a better characterization of exosome molecular phenotype and immunomodulatory properties will provide new insights into their immune-regulatory role during chemotherapy and, possibly, into their use as prognostic biomarkers.
Materials and methods
Antibodies and reagents
The following antibodies were from BD Biosciences (San Jose, CA): anti-CD3/APC-H7 (clone SK7), anti-CD3/APC (clone SK7), anti-CD56/PE (clone NCAM16.2), anti-IFNγ/APC (clone B27), anti-CD38/APC (clone HIT2), anti-CD138/FITC (clone MI15), anti-BrdU/FITC (clone B44). Anti-CD63 (H-193), anti-Hsp70/Hsc70 (W27), anti-CD81 (H-121), NF-kB p65 (C-20), anti-Tsg101 (M-19) were purchased from Santa Cruz Biotechnology (CA). Phospho-NF-kB p65 (Ser 536) (clone 7F1) was from Cell Signaling (Danvers, MA). Anti-calreticulin was from Thermo Fisher Scientific (Rockford, USA). Anti-MHC I (clone HC10) was kindly provided from Dr P. Giacomini, Regina Elena Cancer Institute, Rome, Italy. Anti-human Hsp70/FITC (clone cmHsp70.1) was provided from Prof. Multhoff G,Citation35 Department of Radiation Oncology, Technische Universität München, Munich, Germany. The following antibodies were from BioLegend (San Diego, CA): control mouse IgG1 (clone MOPC-21), anti-human CD69/APC (clone FN50), anti-human CD63/PE (clone H5C6). F(ab)2 fragments of APC conjugated goat-anti-mouse (GAM-APC or GAM-PE) IgG were from Jackson Immunoresearch Laboratories (West Grove, PA). Anti-β-actin (clone AC-74, IgG2a) was from Sigma-Aldrich (St Louis, MO); anti-hTLR2-IgA neutralizing antiboby (clone B4H2) was from InvivoGen (San Diego, CA); anti-human CD282 (TLR2) APC (clone TL2.1) and Annexin V apoptosis detection kit were from eBioscience (California, USA). The NF-kB SN50 inhibitor peptide was from Calbiochem (Los Angeles, USA). Other reagents used were as follows: bovine serum albumin (BSA), brefeldin A (BFA), saponin, paraformaldehyde (PFA), propidium iodide (PI), trypan blue, dimethylformamide (DMF), puromycin, Melphalan, dynasore, nystatin, 5-(N-ethyl-N-isopropyl) amiloride (EIPA), all from Sigma-Aldrich (St Louis, MO). In addition, hygromycin, blasticidin, R848, Pam3CSK4, ODN 2216, Poly (I:C), LPS were from InvivoGen (San Diego, CA). Recombinant Human IL-15 was purchased from Peprotech (Rocky Hill, NJ). The peptide aptamer A8 (SPWPRPTY)Citation62 and the scramble (TPWYPSRP) were synthesized and highly purified by BioFab Research (Italy, Rome).
Cells and culture conditions
The human MM cell lines SKO-007(J3) and ARK, were provided by P. Trivedi (“Sapienza” University of Rome). The cell lines were maintained at 37°C and 5% CO2 in RPMI 1640 (Life Technologies, Gaithersburg, MD) supplemented with 10% FBS. Primary human foreskin fibroblasts (HFFs) were purchased from the American Type Culture Collection and were grown in DMEM supplemented with 10% FBS. All cell lines were mycoplasma free (EZ-PCR Mycoplasma test kit; Biological Industries).
Patients
Clinical samples from patients with MM were managed at the Division of Hematology (“Sapienza” University of Rome). Informed consent, in accordance with the Declaration of Helsinki, was obtained from all patients, and approval was obtained from the Ethics Committee of “Sapienza” University of Rome. The BM aspirates were processed, as described previously.Citation19,22 In some experiments, myeloma cells were purified using anti-CD138 magnetic beads (Miltenyi Biotec, Auburn, CA).Citation19,22 More than 95% of the purified cells expressed CD138 and CD38.
Exosome purification
Exosome-free medium was obtained as follows: FBS was centrifuged at 100,000 g for 3 h in a Beckman ultracentrifuge (Beckman Coulter, Brea, CA) to remove microvesicles-like exosomes. RPMI 1640 was supplemented with 10% of FBS-exosome free and antibiotics. ARK and SKO-007(J3) cell lines were cultured at 0.8–1×106 cells/mL in exosome-free medium for 48 h. In some experiments, cells were treated with a sublethal dose of Melphalan (MEL) determined by the MTT assay and was 3μM for ARK and 5μM for SKO-007(J3). Exosome purification protocol consists of different sequential centrifugations as previously reported.Citation35 In some experiments, HFFs were grown for 5 d up to reach confluence and the conditioned medium was used to isolate exosomes. PBMCs were plated at 3×106 cells/mL for 48 h and the conditioned medium was used to isolate exosomes. Briefly, cells were harvested by centrifugation at 300 g for 10 min and supernatants were collected. Cell-free supernatants were then centrifuged at 2,000 g for 20 min followed by centrifugation at 10,000 g for 30 min to remove cells and debris. Supernatants were filtered using a 0.22 μm filter and centrifuged at 100,000 g for 70 min at 4°C in a Beckman ultracentrifuge to pellet exosomes. The resulting pellet was washed in a large volume of cold PBS and again centrifuged at 100,000 g for 70 min at 4°C. Finally, exosomes were resuspended in PBS for further analyses and functional studies.
MM patients plasma was collected from BM aspirates and from PB by centrifugation at 400 g for 10 min, stored at −80°C and further used for exosome extraction. One mL of MM patient plasma was centrifuged at 2,000 g for 30 min, supernatants were collected and 0.2 volumes of the Total Exosome Isolation reagent (Invitrogen) were added. The samples were incubated for 30 min at 4°C followed by centrifugation at 10,000 g for 10 min at RT. The pellet was resuspended in PBS, filtered using a 0.22 μm filter and washed two times by centrifugation at 100,000 g for 70 min at 4°C. Finally, exosomes were resuspended in PBS.
Size experiments through DLS
DLS experiments were performed to measure exosome size and concentration. All the measurements were made at 25°C on a Zetasizer Nano ZS90 spectrometer (Malvern, UK) equipped with a 5 mW HeNe laser (wavelength λ = 632.8 nm) and a non-invasive back-scattering optical setup (NIBS). For each sample, the detected intensity was processed by a digital logarithmic correlator, which computes a normalized intensity autocorrelation functions. Then, the distribution of the diffusion coefficient D was obtained by using the CONTIN method.Citation63 D was converted into an effective hydrodynamic diameter DH through the Stokes–Einstein equation: DH = KBT/(3πηD), where KBT is the system's thermal energy and η represents the solvent viscosity. Solvent-resistant micro cuvettes (ZEN0040, Malvern, Herrenberg, Germany) have been used for experiments with a sample volume of 40 μL. The count rates obtained were then corrected for the attenuator used. Results are given as mean ± standard deviation of five replicates. Exosome size distribution and concentration were calculated by a recently proposed DLS-based non-invasive tool. More details can be found in the supplementary methodology and in Fig. S1.Citation64,65
Ultrastructural analysis and immunoelectron microscopy
Transmission electron microscopy (TEM) of MM cells-isolated exosomes was performed as follow. Briefly, exosomes were fixed in 2% PFA and adsorbed on formvar-carbon-coated copper grids. The grids were then incubated in 1% glutaraldehyde for 5 min, washed with deionized water eight times, and then negatively stained with 2% uranyl oxalate (pH 7) for 5 min and methyl cellulose/uranyl for 10 min at 4°C. Excess methyl cellulose/uranyl was blotted off, and the grids were air-dried and observed with a TEM (Philips Morgagni268D) at an accelerating voltage of 80 kV. Digital images were taken with Mega View imaging software. For immunoelectron microscopy, exosomes were fixed in 2% PFA and adsorbed on formvar-carbon-coated nickel grids. The grids were then incubated with primary antibodies anti-TSG101 and anti-CD81 overnight. After several washes with PBS the grids were then incubated with specific secondary antibodies conjugated with colloidal gold (Sigma Aldrich, Milan, Italy). The grids were then washed in PBS, fixed in 1% glutaraldehyde, washed with deionized water, and then negatively stained with 2% uranyl oxalate (pH 7) and methyl cellulose/uranyl as described previously. The grids were air-dried and observed with a TEM (Philips Morgagni268D) at an accelerating voltage of 80 kV. Digital images were taken with Mega View imaging software.
Liposome preparation
The zwitterionic lipid 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and cholesterol (Chol) were purchased from Avanti Polar Lipids (AL, USA) and used without further purification. DMPC:Chol (4:1 molar ratio) liposomes were routinely prepared. In brief, a lipid mixture was dissolved in chloroform and then let to evaporate under vacuum for 24 h. The obtained lipid film was hydrated with PBS (final lipid concentration = 1 mg/mL). The obtained liposome solutions were extruded 20 times through a 0.1 μm polycarbonate carbonate filter with the Avanti Mini-Extruder (Avanti Polar Lipids, AL, USA).
Flow cytometry analysis of exosomes
FACS analysis of CD63+ exosomes was performed by positive magnetic selection using CD63+ dynabeads (Invitrogen). About 5–10 μg of exosomes were incubated with 20 µL of magnetic beads for 18–22 h at 4°C with gentle tilting. The bead-bound exosomes were labeled with cIg/PE or anti-CD63/PE (BioLegend). In other experiments, Aldehyde/Sulfate Latex Beads (Thermo Scientific) were incubated 18-22 h with about 2–4 μg of exosomes. The bead-bound exosomes were incubated with 100 mM of glycine for 30 min at RT, then washed three times and labeled with cIg/FITC (BioLegend) or anti-Hsp70/FITC (clone cmHsp70.1). Samples were acquired using a FACSCanto (BD Biosciences, San Jose, CA) and data analysis was performed using the FlowJo program.
RNA isolation, RT-PCR and real-time PCR
Total RNA from human primary purified NK cells was extracted using Total RNA Mini Kit (Geneaid, New Taipei City, Taiwan). Total RNA (100 ng–1 μg) was used for cDNA first-strand synthesis using oligo-dT (Promega, Madison, WI) in a 25 μL reaction volume. Real-time PCR was performed using the ABI Prism 7900 Sequence Detection system (Applied Biosystems, Foster City, CA). cDNAs were amplified in triplicate with primers for IFNγ (Hs00989291_m1), and human β-actin (Hs99999903_m1), all conjugated with fluorochrome FAM (Applied Biosystems). The cycling conditions were: 50°C for 10 min, followed by 40 cycles of 95°C for 30 sec, and 60°C for 2 min. Data were analyzed using the Sequence Detector v1.7 analysis software (Applied Biosystems). The level of gene expression was measured using threshold cycle (Ct). The Ct was obtained by subtracting the Ct value of the gene of interest from the housekeeping gene (β-actin) Ct value. In the current study, we used Ct of the untreated sample as the calibrator. The fold change was calculated according to equation 2−ΔΔCt, where ΔΔCt was the difference between Ct of the sample and the Ct of the calibrator (according to the formula, the value of the calibrator in each run is 1).
SDS-PAGE and Western blot analysis
For Western blot analysis, SKO-007(J3) or ARK cells or exosomal preparations were lysed in 1X RIPA lysis buffer (1% NP-40, 0.1% SDS 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.5% Sodium Deoxycholate, 1 mM EDTA) plus complete protease inhibitor mixture and phosphatase inhibitors as described previously.Citation66 Protein concentration was determined with the Bio-Rad Protein Assay (BPA). 40 to 50 μg of cell or exosome extract was run on 8% or 10% denaturing sodium dodecyl sulfate (SDS)-polyacrylamide gels. Proteins were then electro-blotted onto nitrocellulose membranes (Schleicher & Schuell, Keene, NJ) and blocked in 5% milk in TBST buffer for 1 h. Immunoreactive bands were visualized on the nitrocellulose membranes, using Horseradish Peroxidase (HRP)-coupled goat anti-rabbit or goat anti-mouse Igs and the enhanced chemiluminescence kit (ECL) detection system (GE Healthcare Amersham), following the manufacturer's instructions.
Electrophoretic mobility-shift assay (EMSA)
Nuclear proteins were prepared as described previously.Citation67 Protein concentration of extracts was determined by the BPA method (Pierce, Rockford, IL). The nuclear proteins (4 μg) were incubated with radiolabeled DNA probes in a 20 μL reaction mixture containing 20 mM Tris (pH 7.5), 60 mM KCl, 2 mM EDTA, 0.5 mM dithiothreitol (DTT), 1 μg of poly(dI-dC) and 4% ficoll. Where indicated, a molar excess of double-strand oligomer was added as a cold competitor and the mixture was incubated at room temperature for 10 min before adding the DNA probe. Nucleoprotein complexes were resolved as described previously.Citation67 Oligonucleotides were purchased by BioFab (Rome, Italy). Complementary strands were annealed and end-labeled as described.Citation67 Approximately 3×104 cpm of labeled DNA was used in a standard electrophoretic mobility-shift assay reaction. The following double-strand oligomers were used as specific labeled probes or cold competitors (sense strand): Octamer-(h-Histone H2b), 5′-agctcttcaccttatttgcataagcgat-3′ and the oligo containing an NF-kB binding site C3–3P (−278 to −268) of the IFNG promoter 5′-gggaggtacaaaaaaatttccagtccttga-3′.Citation37
Human NK cell isolation
Highly purified primary NK cells were obtained from human peripheral blood mononuclear cells (PBMCs) by negative selection using magnetic beads (Miltenyi Biotec). NK cell purity was more than 96% CD56+CD3− as assessed by immunofluorescence and flow cytometry analysis (data not shown). In some experiments, NK cells were sorted based on CD56 expression levels for high and low NK cells using a BD FACSAria III (BD Biosciences) equipped with FACSDiva software (BD Biosciences version 6.1.3) as shown in .
Intracellular cytokine production, degranulation assay, 51Cr-release assay and cell proliferation
For intracellular cytokine detection, NK cells were seeded at 2–3×106/mL in exosome-free medium, and incubated with different doses of MM cell-derived exosomes (1–20 µg/mL) for 24 h. BFA was added at 5 µg/mL and left for additional 24 h. In some experiments, NK cells were treated with recombinant human IL-15 (50 ng/mL) and sometimes, before exosomes exposure, NK cells were pre-incubated with the NF-kB peptide inhibitor SN50 (15 μM) for 1 h at 37°C. Degranulation assay was performed as described previously.Citation22 The 51Cr-release assay was used to measure cytotoxic activity against K562 target cells, as described previously.Citation68 Cell proliferation of SKO-007(J3) cells was evaluated as follow: BrdU (20 μM) was added to the culture for 7 h, cells were harvested, fixed in a solution containing 30% methanol, 4% PFA. After washing with PBS, cells were incubated at −20°C for at least 18 h. Cells were permealized with a solution containing 1% Tween-20 and treated with DNAse (0.250 μg/mL) for 15 min at RT. After washing, cells were stained with anti-BrdU/FITC or with a control Ig-FITC.
Immunofluorescence and FACS analysis
For intracellular cytokine detection, human primary NK cells were first labeled with anti-CD56/PE and then PBS washed and fixed in 1% PFA for 20 min at room temperature (RT). After washing, cells were incubated with PBS containing 2% BSA for 15 min at RT and then permeabilized with PBS containing 0.5% saponin and 1% FBS for 20 min at RT. After washing, cells were labeled with anti-IFNγ/APC. In other experiments, NK cells were stained with anti-CD69/APC or TLR2/APC together with antiCD56/PE and CD3/APCH7. All the samples were acquired using a FACSCanto (BD Biosciences, San Jose, CA) and data analysis was performed using the FlowJo program.
Exosome uptake
100 μg of exosomes diluted in PBS were incubated with the red fluorescent dye PKH26 (Sigma-Aldrich). Exosomes were washed twice with PBS by centrifugation at 100,000 g for 1 h. PKH26-labeled exosomes were diluted with PBS and used for uptake experiments. Primary NK cells were plated on poly-L-lysine-coated multichamber glass plates in complete medium and incubated with PKH26-labeled exosomes (20 μg/mL) for different times. Medium was removed and the cell monolayer was gently washed with PBS, and fixed with 4% PFA. Coverslips were mounted using SlowFade Gold reagent (Life Technologies) and acquired at room temperature using an ApoTome Observer Z.1 microscope with an Axiocam MR equipped with AxioVision Version 4.6.3 software for image acquisition as described previously.Citation69 In some experiments, PKH26-labeled exosomes were incubated with highly purified NK cells for different times. Cells were collected, washed twice with PBS and then samples were acquired using a FACSCanto (BD Biosciences, San Jose, CA). Data analysis was performed using the FlowJo program.
ELISA/Luminex
Detection of Hsp70 or CD63 in total exosomes lysates was performed using specific ELISA kits from R&D Systems (DY1663–2) and BioSource (MBS763944), respectively. Plates were developed using a peroxidase substrate system (R&D Systems), and then read with the Victor3 multilabel plate reader (Model # 1420–033, Perkin Elmer, Santa Clara, CA) capable of measuring absorbance in 96-well plates using dual wavelength of 450–540 nm. Results were expressed as picograms per milliliter (pg/mL) and referred to a standard curve obtained by plotting the mean absorbance for each standard on the y-axis against the concentration on the x-axis and drawing a best-fit curve through the points on the graph. Detection of cytokines and chemokines (i.e., IFNγ, CXCL8/IL-8, CCL5/Rantes, CCL4/MIP1β, CX3CL1/fractalkine, CXCL10/IP-10) in the same supernatants was performed with a Milliplex MAP Human Cytokine/Chemokine Magnetic Bead Panel—Immunology Multiplex Assay according to the manufacturer's instructions (Millipore). Plates were read with Bio-Plex MAGPIX Multiplex Reader (BIO-RAD).
NF-κB luciferase reporter assay
TLR-specific activation assays were performed using human embryonic kidney 293 (HEK293) cells expressing luciferase under control of the NF-κB promoter and stably transfected with either TLR4, MD2, and CD14 (TLR4-HEK293), TLR2 (TLR2-HEK293), TLR3 (TLR3-HEK293), TLR7 (TLR7-HEK293), TLR8 (TLR8-HEK293) and TLR9 (TLR9-HEK293). HEK293-transfected cells were maintained in DMEM supplemented with 4.5 g/L glucose and 10% FBS, 1% penicillin/streptomycin solution (Invitrogen), and specific antibiotics for the different cell lines were added. All the HEK293-transfected cells were kindly provided by Dr U. D'Oro (GlaxoSmithKline Vaccines, Siena). For the NF-κB luciferase assay, 30,000 cells/well were seeded in 100 μL of complete DMEM without antibiotics in 96-well plates and incubated for 18 h at 37°C. Different concentrations of exosomes (1–20 μg/mL) were added and left for different times (2–18 h). As positive control for each transfectant, specific TLR agonists were used as follow: TRL2 (Pam3CSK4, 1 μM), TLR3 (Poly(I:C), 1 μg/mL), TLR4 (LPS, 10 μg/mL), TLR7 and TLR8 (R848, 10 μM), TLR9 (CpG, 50 μM). After incubation, supernatants were aspired from each well, cells were washed with PBS and then were lysed for 15 min at room temperature using 100 μL/well of 1:5 diluted “passive lysis buffer” (Promega, Madison WI). Protein concentration was evaluated by BPA. 3 μg of total proteins for each sample were diluted in 50 μL of PBS and 50 μL of luciferase assay substrate (Promega, Madison WI) was added. Emitted light was immediately quantified using a luminometer GloMax-Multi Detection System (Promega, Madison WI).
Statistics
Error bars represent SD or where indicated SEM. Statistical analysis was performed with the Student paired test; *p < 0.05, **p < 0.01, ***p < 0.001.
Disclosure of potential conflicts of interest
The authors declare no competing financial interests. UD is employee of the GSK group of companies and is listed as an inventor on patents owned by the GSK group of companies.
KONI_A_1279372_supplementary_data.zip
Download Zip (2.5 MB)Acknowledgments
The expert technical assistance of Bernardina Milana is gratefully acknowledged. We thank Dr Oreste Segatto for his careful and critical reading of the manuscript.
Funding
This work was supported by Italian Association for Cancer Research (AIRC Investigator Grant and AIRC 5×1000). E.V. is supported by a fellowship from AIRC.
References
- Bobrie A, Thery C. Exosomes and communication between tumours and the immune system: are all exosomes equal? Biochem Soc Trans 2013; 41:263-7; PMID:23356294; http://dx.doi.org/10.1042/BST20120245
- Tkach M, Thery C. Communication by extracellular vesicles: Where we are and where we need to go. Cell 2016; 164:1226-32; PMID:26967288; http://dx.doi.org/10.1016/j.cell.2016.01.043
- Liu Y, Gu Y, Cao X. The exosomes in tumor immunity. Oncoimmunology 2015; 4:e1027472; PMID:26405598; http://dx.doi.org/10.1080/2162402X.2015.1027472
- Pitt JM, Kroemer G, Zitvogel L. Extracellular vesicles: masters of intercellular communication and potential clinical interventions. J Clin Invest 2016; 126:1139-43; PMID:27035805; http://dx.doi.org/10.1172/JCI87316
- Lanier LL. NK cell recognition. Ann Rev Immunol 2005; 23:225-74; PMID:15771571; http://dx.doi.org/10.1146/annurev.immunol.23.021704.115526
- Adib-Conquy M, Scott-Algara D, Cavaillon JM, Souza-Fonseca-Guimaraes F. TLR-mediated activation of NK cells and their role in bacterial/viral immune responses in mammals. Immunol Cell Biol 2014; 92:256-62; PMID:24366517; http://dx.doi.org/10.1038/icb.2013.99
- Michel T, Poli A, Cuapio A, Briquemont B, Iserentant G, Ollert M, Zimmer J. Human CD56bright NK cells: An update. J Immunol 2016; 196:2923-31; PMID:26994304; http://dx.doi.org/10.4049/jimmunol.1502570
- Godfrey J, Benson DM, Jr. The role of natural killer cells in immunity against multiple myeloma. Leuk Lymphoma 2012; 53:1666-76; PMID:22423650; http://dx.doi.org/10.3109/10428194.2012.676175
- Guillerey C, Nakamura K, Vuckovic S, Hill GR, Smyth MJ. Immune responses in multiple myeloma: role of the natural immune surveillance and potential of immunotherapies. Cell Mol Life Sci 2016; 73:1569-89; PMID:26801219; http://dx.doi.org/10.1007/s00018-016-2135-z
- Fionda C, Soriani A, Zingoni A, Santoni A, Cippitelli M. NKG2D and DNAM-1 ligands: molecular targets for NK cell-mediated immunotherapeutic intervention in multiple myeloma. Biomed Res Int 2015; 2015:178698; PMID:26161387; http://dx.doi.org/10.1155/2015/178698
- Fionda C, Abruzzese MP, Zingoni A, Cecere F, Vulpis E, Peruzzi G, Soriani A, Molfetta R, Paolini R, Ricciardi MR et al. The IMiDs targets IKZF-1/3 and IRF4 as novel negative regulators of NK cell-activating ligands expression in multiple myeloma. Oncotarget 2015; 6:23609-30; PMID:26269456; http://dx.doi.org/10.18632/oncotarget.4603
- Abruzzese MP, Bilotta MT, Fionda C, Zingoni A, Soriani A, Vulpis E, Borrelli C, Zitti B, Petrucci MT, Ricciardi MR et al. Inhibition of bromodomain and extra-terminal (BET) proteins increases NKG2D ligand MICA expression and sensitivity to NK cell-mediated cytotoxicity in multiple myeloma cells: role of cMYC-IRF4-miR-125b interplay. J Hematol Oncol 2016; 9:134; PMID:27903272; http://dx.doi.org/10.1186/s13045-016-0362-2
- Garg AD, Galluzzi L, Apetoh L, Baert T, Birge RB, Bravo-San Pedro JM, Breckpot K, Brough D, Chaurio R, Cirone M et al. Molecular and translational classifications of DAMPs in immunogenic cell death. Front Immunol 2015; 6:588; PMID:26635802; http://dx.doi.org/10.3389/fimmu.2015.00588
- Kepp O, Senovilla L, Vitale I, Vacchelli E, Adjemian S, Agostinis P, Apetoh L, Aranda F, Barnaba V, Bloy N et al. Consensus guidelines for the detection of immunogenic cell death. Oncoimmunology 2014; 3:e955691; PMID:25941621; http://dx.doi.org/10.4161/21624011.2014.955691
- Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, Mignot G, Maiuri MC, Ullrich E, Saulnier P et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nature Med 2007; 13:1050-9; PMID:17704786; http://dx.doi.org/10.1038/nm1622
- Chen T, Guo J, Han C, Yang M, Cao X. Heat shock protein 70, released from heat-stressed tumor cells, initiates antitumor immunity by inducing tumor cell chemokine production and activating dendritic cells via TLR4 pathway. J Immunol 2009; 182:1449-59; PMID:19155492; http://dx.doi.org/10.4049/jimmunol.182.3.1449
- Vacchelli E, Eggermont A, Sautes-Fridman C, Galon J, Zitvogel L, Kroemer G, Galluzzi L. Trial Watch Toll-like receptor agonists for cancer therapy. Oncoimmunology 2013; 2:e22789; PMID:23482847; http://dx.doi.org/10.4161/onci.22789
- Pradere JP, Dapito DH, Schwabe RF. The Yin and Yang of Toll-like receptors in cancer. Oncogene 2014; 33:3485-95; PMID:23934186; http://dx.doi.org/10.1038/onc.2013.302
- Soriani A, Zingoni A, Cerboni C, Iannitto ML, Ricciardi MR, Di Gialleonardo V, Cippitelli M, Fionda C, Petrucci MT, Guarini A et al. ATM-ATR-dependent up-regulation of DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic agents results in enhanced NK-cell susceptibility and is associated with a senescent phenotype. Blood 2009; 113:3503-11; PMID:19098271; http://dx.doi.org/10.1182/blood-2008-08-173914
- Antonangeli F, Soriani A, Ricci BM, Ponzetta A, Benigni G, Morrone S, Bernardini G, Santoni A. Natural killer cell recognition of in vivo drug-induced senescent multiple myeloma cells. Oncoimmunology 5:e1218105; PMID:27853638; http://dx.doi.org/10.1080/2162402X.2016.1218105
- Cerboni C, Fionda C, Soriani A, Zingoni A, Doria M, Cippitelli M, Santoni A. The DNA damage response: a common pathway in the regulation of NKG2D and DNAM-1 ligand expression in normal infected, and cancer cells. Front Immunol 2014; 4:508; PMID:24432022; http://dx.doi.org/10.3389/fimmu.2013.00508
- Zingoni A, Cecere F, Vulpis E, Fionda C, Molfetta R, Soriani A, Petrucci MT, Ricciardi MR, Fuerst D, Amendola MG et al. Genotoxic stress induces senescence-associated ADAM10-dependent release of NKG2D MIC ligands in multiple myeloma cells. J Immunol 2015; 195:736-48; PMID:26071561; http://dx.doi.org/10.4049/jimmunol.1402643
- Reiners KS, Dassler J, Coch C, Pogge von Strandmann E. Role of exosomes released by dendritic cells and/or by tumor targets: regulation of NK cell plasticity. Front Immunol 2014; 5:91; PMID:24639679; http://dx.doi.org/10.3389/fimmu.2014.00091
- Clayton A, Mitchell JP, Court J, Linnane S, Mason MD, Tabi Z. Human tumor-derived exosomes down-modulate NKG2D expression. J Immunol 2008; 180:7249-58; PMID:18490724; http://dx.doi.org/10.4049/jimmunol.180.11.7249
- Ashiru O, Boutet P, Fernandez-Messina L, Aguera-Gonzalez S, Skepper JN, Vales-Gomez M, Reyburn HT. Natural killer cell cytotoxicity is suppressed by exposure to the human NKG2D ligand MICA*008 that is shed by tumor cells in exosomes. Cancer Res 2010; 70:481-9; PMID:20068167; http://dx.doi.org/10.1158/0008-5472.CAN-09-1688
- Berchem G, Noman MZ, Bosseler M, Paggetti J, Baconnais S, Le Cam E, Nanbakhsh A, Moussay E, Mami-Chouaib F, Janji B et al. Hypoxic tumor-derived microvesicles negatively regulate NK cell function by a mechanism involving TGF-beta and miR23a transfer. Oncoimmunology 2016; 5:e1062968; PMID:27141372; http://dx.doi.org/10.1080/2162402X.2015.1062968
- Labani-Motlagh A, Israelsson P, Ottander U, Lundin E, Nagaev I, Nagaeva O, Dehlin E, Baranov V, Mincheva-Nilsson L. Differential expression of ligands for NKG2D and DNAM-1 receptors by epithelial ovarian cancer-derived exosomes and its influence on NK cell cytotoxicity. Tumour Biol 2016; 37:5455-66; PMID:26563374; http://dx.doi.org/10.1007/s13277-015-4313-2
- Liu C, Yu S, Zinn K, Wang J, Zhang L, Jia Y, Kappes JC, Barnes S, Kimberly RP, Grizzle WE et al. Murine mammary carcinoma exosomes promote tumor growth by suppression of NK cell function. J Immunol 2006; 176:1375; PMID:16424164; http://dx.doi.org/10.4049/jimmunol.176.3.1375
- Szcepanski MJ, Szajinik M, Welsh A, Whiteside TL, Boyiadzis M. Blast-derived microvesicles in sera from patients with acute myeloid leukemia suppress natural killer cell function via membrane-associated transforming growth factor beta1. Haematologica 2011; 96:1302-09; PMID:21606166; http://dx.doi.org/10.3324/haematol.2010.0397433
- Clayton A, Mitchell JP, Court J, Mason MD, Tabi Z. Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res 2007; 67:7458-66; PMID:17671216; http://dx.doi.org/10.1158/0008-5472.CAN-06-3456
- Gastpar R, Gehrmann M, Bausero MA, Asea A, Gross C, Schroeder JA, Multhoff G. Heat shock protein 70 surface-positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. Cancer Res 2005; 65:5238-47; PMID:15958569; http://dx.doi.org/10.1158/0008-5472.CAN-04-3804
- Lv LH, Wan YL, Lin Y, Zhang W, Yang M, Li GL, Lin HM, Shang CZ, Chen YJ, Min J. Anticancer drugs cause release of exosomes with heat shock proteins from human hepatocellular carcinoma cells that elicit effective natural killer cell antitumor responses in vitro. J Biol Chem 2012; 287:15874-85; PMID:22396543; http://dx.doi.org/10.1074/jbc.M112.340588
- Reiners KS, Topolar D, Henke A, Simhadri VR, Kessler J, Sauer M, Bessler M, Hansen HP, Tawadros S, Herling M et al. Soluble ligands for NK cell receptors promote evasion from chronic lymphocytic leukemia cells from NK cell anti-tumor activity. Blood 2013; 121:3658-65; PMID:23509156; http://dx.doi.org/10.1182/blood-2013-01-476606
- Daβler-Plenker J, Reiners KS, van den Boorn JG, Hansen HP, Putschli B, Barnert S, Schuberth-Wagner C, Schubert R, Tüting T, Hallek M et al. RIG-I activation induces the release of extracellular vesicles with antitumor activity. Oncoimmunology 2016; 19:e1218827; PMID:27853642; http://dx.doi.org/10.1080/2162402X.2016.1219827
- Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Prot Cell Biol 2006; Chapter 3:Unit 3.22; PMID:18228490; http://dx.doi.org/10.1002/0471143030.cb0322s30
- Nabi IR, Le PU. Caveolae/raft-dependent endocytosis. J Cell Biol 2003; 161:673-77; PMID:12771123; http://dx.doi.org/10.1083/jcb.200302028
- Sica A, Dorman L, Viggiano V, Cippitelli M, Ghosh P, Rice N, Young HA. Interaction of NF-kappaB and NFAT with the interferon-gamma promoter. J Biol Chem 1997; 272:30412-20; PMID:9374532; http://dx.doi.org/10.1074/jbc.272.48.30412
- Chow A, Zhou W, Liu L, Fong MY, Champer J, Van Haute D, Chin AR, Ren X, Gugiu BG, Meng Z et al. Macrophage immunomodulation by breast cancer-derived exosomes requires Toll-like receptor 2-mediated activation of NF-kappaB. Sci Rep 2014; 4:5750; PMID:25034888; http://dx.doi.org/10.1038/srep05750
- Chalmin F, Ladoire S, Mignot G, Vincent J, Bruchard M, Remy-Martin JP, Boireau W, Rouleau A, Simon B, Lanneau D et al. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J Clin Invest 2010; 120:457-71 PMID:20093776; http://dx.doi.org/10.1172/JCI40483
- Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA 2012; 109:E2110-6; PMID:22753494; http://dx.doi.org/10.1073/pnas.1209414109
- He S, Chu J, Wu LC, Mao H, Peng Y, Alvarez-Breckenridge CA, Hughes T, Wei M, Zhang J, Yuan S et al. MicroRNAs activate natural killer cells through Toll-like receptor signaling. Blood 2013; 121:4663-71; PMID:23580661; http://dx.doi.org/10.1182/blood-2012-07-441360
- Stangl S, Gehrmann M, Riegger J, Kuhs K, Riederer I, Sievert W, Hube K, Mocikat R, Dressel R, Kremmer E et al. Targeting membrane heat-shock protein 70 (Hsp70) on tumors by cmHsp70.1 antibody. Proc Natl Acad Sci U S A 2011; 108:733-8; PMID:21187371; http://dx.doi.org/10.1073/pnas.1016065108
- Gobbo J, Marcion G, Cordonnier M, Dias AM, Pernet N, Hammann A, Richaud S, Mjahed H, Isambert N, Clausse V et al. Restoring anticancer immune response by targeting tumor-derived exosomes with a HSP70 peptide aptamer. J Natl Cancer Inst 2016; 108:djv330; PMID:26598503; http://dx.doi.org/10.1093/jnci/djv330
- Gunther S, Ostheimer C, Stangl S, Specht HM, Mozes P, Jesinghaus M, Vordermark D, Combs SE, Peltz F, Jung MP et al. Correlation of Hsp70 serum levels with gross tumor volume and composition of lymphocyte subpopulations in patients with squamous cell and adeno non-small cell lung cancer. Front Immunol 2015; 6:556; PMID:26579130; http://dx.doi.org/10.3389/fimmu.2015.00556
- Ciocca DR, Calderwood SK. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 2005; 10:86-103; PMID:16038406; http://dx.doi.org/10.1379/CSC-99r.110.1379/CSC-99r.1
- Pitt JM, Andre F, Amigorena S, Soria JC, Eggermont A, Kroemer G, Zitvogel L. Dendritic cell-derived exosomes for cancer therapy. J Clin Invest 2016; 126:1224-32; PMID:27035813; http://dx.doi.org/10.1172/JCI81137
- Zhang X, Pei Z, Chei J, Ji C, Xu J, Zhang X, Wang J. Exosomes for immunoregulation and therapeutic interventention in cancer. J Cancer 2016; 7:1081-7; PMID:27326251; http://dx.doi.org/10.7150/jca.14866
- Umezu T, Tadokoro H, Azuma K, Yoshizawa S, Ohyashiki K, Ohyashiki JH. Exosomal miR-135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor-inhibiting HIF-1. Blood 2014; 124:3748-57; PMID:25320245; http://dx.doi.org/10.1182/blood-2014-05-576116
- Purushothaman A, Bandari SK, Liu J, Mobley JA, Brown EE, Sanderson RD. Fibronectin on the Surface of Myeloma Cell-derived Exosomes Mediates Exosome-Cell Interactions. J Biol Chem 2016; 291:1652-63; PMID:26601950; http://dx.doi.org/10.1074/jbc.M115.686295
- Lu H, Yang Y, Gad E, Inatsuka C, Wenner CA, Disis ML, Standish LJ. TLR2 agonist PSK activates human NK cells and enhances the antitumor effect of HER2-targeted monoclonal antibody therapy. Clin Cancer Res 2011; 17:6742-53; PMID:21918170; http://dx.doi.org/10.1158/1078-0432.CCR-11-1142
- Deng Y, Chu J, Ren Y, Fan Z, Ji X, Mundy-Bosse B, Yuan S, Hughes T, Zhang J, Cheema B et al. The natural product phyllanthusmin C enhances IFN-gamma production by human NK cells through upregulation of TLR-mediated NF-kappaB signaling. J Immunol 2014; 193:2994-3002; PMID:25122922; http://dx.doi.org/10.4049/jimmunol.1302600
- Batoni G, Esin S, Favilli F, Pardini M, Bottai D, Maisetta G, Florio W, Campa M. Human CD56bright and CD56dim natural killer cell subsets respond differentially to direct stimulation with Mycobacterium bovis bacillus Calmette-Guerin. Scand J Immunol 2005; 62:498-506; PMID:16316416; http://dx.doi.org/10.1111/j.1365-3083.2005.01692.x
- Girart MV, Fuertes MB, Domaica CI, Rossi LE, Zwirner NW. Engagement of TLR3, TLR7, and NKG2D regulate IFN-gamma secretion but not NKG2D-mediated cytotoxicity by human NK cells stimulated with suboptimal doses of IL-12. J Immunol 2007; 179:3472-9; PMID:17804388; http://dx.doi.org/10.4049/jimmunol.179.6.3472
- Millard AL, Spirig R, Mueller NJ, Seebach JD, Rieben R. Inhibition of direct and indirect TLR-mediated activation of human NK cells by low molecular weight dextran sulfate. Mol Immunol 2010; 47:2349-58; PMID:20541808; http://dx.doi.org/10.1016/j.molimm.2010.05.284
- Esin S, Counoupas C, Aulicino A, Brancatisano FL, Maisetta G, Bottai D, Di Luca M, Florio W, Campa M, Batoni G. Interaction of Mycobacterium tuberculosis cell wall components with the human natural killer cell receptors NKp44 and Toll-like receptor 2. Scand J Immunol 2013; 77:460-9; PMID:23578092; http://dx.doi.org/10.1111/sji.12052
- Asea A, Rehli M, Kabingu E, Boch JA, Bare O, Auron PE, Stevenson MA, Calderwood SK. Novel signal transduction pathway utilized by extracellular HSP70: role of toll-like receptor (TLR) 2 and TLR4. J Biol Chem 2002; 277:15028-34; PMID:11836257; http://dx.doi.org/10.1074/jbc.M200497200
- Mathur S, Walley KR, Wang Y, Indrambarya T, Boyd JH. Extracellular heat shock protein 70 induces cardiomyocyte inflammation and contractile dysfunction via TLR2. Circ J 2011; 75:2445-52; PMID:21817814; http://dx.doi.org/10.1253/circj.CJ-11-0194
- Multhoff G. Heat shock protein 70 (Hsp70): membrane location, export and immunological relevance. Methods 2007; 43:229-37; PMID:17920520; http://dx.doi.org/10.1016/j.ymeth.2007.06.006
- Lancaster GI, Febbraio MA. Exosome-dependent trafficking of HSP70: a novel secretory pathway for cellular stress proteins. J Biol Chem 2005; 280:23349-55;PMID:15826944; http://dx.doi.org/10.1074/jbc.M502017200
- Xie Y, Bai O, Zhang H, Yuan J, Zong S, Chibbar R, Slattery K, Qureshi M, Wei Y, Deng Y et al. Membrane-bound HSP70-engineered myeloma cell-derived exosomes stimulate more efficient CD8(+) CTL- and NK-mediated antitumour immunity than exosomes released from heat-shocked tumour cells expressing cytoplasmic HSP70. J Cell Mol Med 2010; 14:2655-66; PMID:19627400; http://dx.doi.org/10.1111/j.1582-4934.2009.00851.x
- Seya T, Shime H, Takeda Y, Tatematsu M, Takashima K, Matsumoto M. Adjuvant for vaccine immunotherapy of cancer–focusing on Toll-like receptor 2 and 3 agonists for safely enhancing antitumor immunity. Cancer Sci 2015; 106:1659-68; PMID:26395101; http://dx.doi.org/10.1111/cas.12824
- Rerole AL, Gobbo J, De Thonel A, Schmitt E, Pais de Barros JP, Hammann A, Lanneau D, Fourmaux E, Demidov ON, Micheau O et al. Peptides and aptamers targeting HSP70: a novel approach for anticancer chemotherapy. Cancer Res 2011; 71:484-95; PMID:21224349; http://dx.doi.org/10.1158/0008-5472.CAN-10-1443
- Provencher SW. CONTIN: a general purpose constrained regularization program for inverting noisy linear algebraic and integral equations. Computer Physics Communications 1982; 27:229-42; http://dx.doi.org/10.1016/0010-4655(82)90174-6
- Palmieri V, Lucchetti D, Gatto I, Maiorana A, Marcantoni M, Maulucci G, Papi M, Pola R, De Spirito M, Sgambato A. Dynamic light scattering for the characterization and counting of extracellular vesicles: a powerful noninvasive tool. J Nanopart Res 2014; 16:1-8; http://dx.doi.org/10.1007/s11051-014-2583-z
- Papi M, Arcovito G, De Spirito M, Amiconi G, Bellelli A, Boumis G. Simultaneous static and dynamic light scattering approach to the characterization of the different fibrin gel structures occurring by changing chloride concentration. Appl Phys Lett 2005; 86:18391; http://dx.doi.org/10.1063/1.1915526
- Bernardini G, Kim JY, Gismondi A, Butcher EC, Santoni A. Chemoatractant induces LFA-1 associated PI 3K activity and cell migration that are dependent on Fyn signaling. FASEB J 2005; 19:1305-7; PMID:15955842; http://dx.doi.org/10.1096/fj.04-3352fje
- Fionda C, Nappi F, Piccoli M, Frati L, Santoni A, Cippitelli M. 15-deoxy-Delta12,14-prostaglandin J2 negatively regulates rankl gene expression in activated T lymphocytes: role of NF-kappaB and early growth response transcription factors. J Immunol 2007; 178:4039-50;PMID:17371958;http://dx.doi.org/10.4049/jimmunol.178.7.4039
- Zingoni A, Palmieri G, Morrone S, Carretero M, Lopez-Botet M, Santoni A. CD69-triggered ERK activation and functions are negatively regulated by CD94/NKG2A inhibitory receptor. Eur J Immunol 2000; 30:644-51; PMID:10671222; http://dx.doi.org/10.1002/1521-4141(200002)30:2%3c644::AID-IMMU644%3e3.0.CO;2-H
- Gasparrini F, Molfetta R, Quatrini L, Frati L, Santoni A, Paolini R. Syk-dependent regulation of Hrs phosphorylation and ubiquitination upon FcϵRI engagement: impact on Hrs membrane/cytosol localization. Eur J Immunol 2012; 42:2744-53; PMID:22706924; http://dx.doi.org/10.1002/eji.201142278
