ABSTRACT
Adult T-cell leukemia (ATL) is an aggressive, chemotherapy-resistant CD4+CD25+ leukemia caused by HTLV-1 infection, which usually develops in a minority of patients several decades after infection. IFN + AZT combination therapy has shown clinical benefit in ATL, although its mechanism of action remains unclear. We have previously shown that an IFN-responsive FAS promoter polymorphism in a STAT1 binding site (rs1800682) is associated to ATL susceptibility and survival. Recently, CD4 T stem cell memory (TSCM) Fashi cells have been identified as the hierarchical cellular apex of ATL, but a possible link between FAS, apoptosis, proliferation and IFN response in ATL has not been studied.
In this study, we found significant ex vivo antiproliferative, antiviral and immunomodulatory effects of IFN-α treatment in short-term culture of primary mononuclear cells from ATL patients (n = 25). Bayesian Network analysis allowed us to integrate ex vivo IFN-α response with clinical, genetic and immunological data from ATL patients, thereby revealing a central role for FAS -670 polymorphism and apoptosis in the coordinated mechanism of action of IFN-α. FAS genotype-dependence of IFN-induced apoptosis was experimentally validated in an independent cohort of healthy controls (n = 20). The same FAS -670 polymorphism also determined CD4 TSCM levels in a genome-wide twin study (p = 7 × 10−11, n = 460), confirming a genetic link between apoptosis and TSCM levels. Transcriptomic analysis and cell type deconvolution confirmed the FAS genotype/TSCM link and IFN-α-induced downregulation of CD4 TSCM-specific genes in ATL patient cells.
In conclusion, ex vivo IFN-α treatment exerts a pleiotropic effect on primary ATL cells, with a genetic IFN/STAT1/Fas axis determining apoptosis vs. proliferation and underscoring the CD4 TSCM model of ATL leukemogenesis.
Introduction
Adult T-cell leukemia (ATL) is characterized by circulating CD4+CD25+ T-cellsCitation1 and is etiologically linked to infection with Human T-cell Leukemia Virus 1 (HTLV-1), the first isolated human pathogenic retrovirus.Citation2 The estimated lifetime risk is about 5% in infected individuals,Citation3 with a long incubation period after HTLV-1 infection, therefore infection early in life is fundamental in the development of ATL.Citation4 ATL is classified according to clinical and laboratory criteria as smoldering, chronic, lymphoma, or acute subtypes.Citation5 The median survival time for acute and lymphoma (aggressive) subtypes is less than one year, whereas patients with chronic and smoldering (indolent) subtypes survive longer.Citation6 In addition, an atypical aggressive, primary cutaneous tumoral form (PCT) of ATL with a shorter survival has been described.Citation7 The exceptional oncogenicity of HTLV-1, as compared to other viruses or infectious agents,Citation8 or even to its closest relative HTLV-2,Citation9, is most likely due to its ability to deregulate several host cell signaling pathways. In sharp contrast to HIV, plasma viral load for HTLV-1 is mostly undetectable and proviral replication is driven by spontaneous lymphoproliferation (reviewed inCitation3), which implies that ATL might be explained mainly by host factors such as immune response and host genetics.
Regarding host immune response, interleukin 2 (IL-2) promotes proliferation of IL-2 receptor-positive (IL-2R+/CD25+) T-cells. Membrane-bound CD25 is present on all activated T-cells as well as malignant T-cells infected by HTLV-1. Serum levels of IL-2R/CD25 positively correlate with circulating ATL cells (CD4+CD25+) and disease severity.Citation10 In addition, systemic IL-6 levels correlate with aggressive clinical forms and short survival.Citation11
Regarding host genetics, FAS (TNFRSF6/Apo-1/CD95) and TP53 polymorphisms have been associated to ATL disease progression.Citation12,Citation13 In addition, FAS mutations have been demonstrated in ATLCitation14,Citation15 as well as in other leukemias.Citation16 The FAS -670 A/G polymorphism is situated in a STAT1 binding site, for which the A allele has a higher binding affinity.Citation17,Citation18 We have previously shown that the A allele corresponds to increased IFN-induced Fas mRNA, and is significantly over-represented in ATL patients, as compared to healthy controls or HTLV-1-infected asymptomatic individuals.Citation12 In addition, ATL patients with the AA genotype were significantly more likely to develop aggressive (acute/lymphoma) clinical forms.Citation12 Interestingly, a specific subset of Fashi cells has recently been implicated in ATL leukemogenesis, as CD4 T stem cell memory (TSCM) cells were identified as its hierarchical cellular apex.Citation19
A combination of interferon-α (IFN-α) and zidovudine (AZT) has been found effective in pioneer ATL trials, and was confirmed by meta-analysis for non-lymphoma subtypes.Citation20-22 In spite of this clinical benefit of IFN-α + AZT combination therapy in ATL, its mechanisms of action remain elusive. IFN-α has anti-proliferative, pro-apoptotic, antiviral and immunomodulatory activity in many human cells, but limited in vitro, ex vivo or in vivo data are available in ATL.Citation23 HTLV-1 infected and/or ATL-derived cell lines are refractory to IFN-α-induced cell death in the absence or presence of AZTCitation23–25 and to IFN-β in vitro.Citation26 However, we have recently demonstrated a superior antiproliferative and pro-apoptotic activity of IFN-β vs. IFN-α in primary ATL patient cells.Citation27 Moreover, a wealth of studies has demonstrated both pro- and anti-apoptotic activity of IFN-α/β in vitro and in vivo,Citation28–33 resulting in an “IFN apoptotic paradox” on the effect of type I IFN (IFN-α/β) on programmed cell death. On the other hand, the clinical benefit of type I IFN has been convincingly demonstrated in several other leukemias, such as hairy-cell leukemia, chronic myeloid leukemia as well as myeloproliferative neoplasms, including essential thrombocytosis, polycythemia vera, and myelofibrosis.Citation34–36
We therefore engaged in a systematic study of IFN-α response and its possible link to Fas levels, apoptosis, proliferation, viral protein expression and immune activation in ATL.
Methods
Patient recruitment
This study was approved by the Ethics Review Board of HUPES, according to the Declaration of Helsinki principles. All study participants signed informed consent. In this prospective study, consecutive patients were recruited between 2001 and 2005 at “Hospital Universitário Professor Edgar Santos” (HUPES, Salvador-Bahia), according to previously described inclusion and exclusion criteria.Citation7 Samples were obtained from 25 patients with clinically definite ATL (Shimoyama criteriaCitation5), with serology, inverted PCR and/or flow cytometry as described.Citation12,Citation37,Citation38 All ATL patients were HTLV-1 seropositive and HIV/HTLV-2 seronegative. Standardized patient treatment was in agreement to a published international consensus.Citation6 Smoldering ATL forms were left untreated but under “watchful waiting”, until possible disease progression, whereas acute/chronic leukemic patients received IFN-α+AZT combination therapy and lymphoma patients received chemotherapy. Samples were obtained before treatment and at least five-year follow-up was available for each patient. Clinical, demographic and flow cytometry data of ATL patients are summarized in Table I. Twenty healthy controls from the same endemic area (seronegative for HTLV-1/HTLV-2/HIV) were recruited in parallel.
Ex vivo IFN-α stimulation
PBMCs from patients and healthy controls were purified from heparinized venous blood by Ficoll-Hypaque gradient (Sigma-Aldrich). Cells were plated in 24-well tissue culture plates (Costar, Corning Incorporated) at 4 × 106 cells/ml in RPMI1640 medium with 2mM L-glutamine, gentamycin (50 μg/ml) and 10% heat-inactivated fetal calf serum (Gibco) and incubated at 37°C, 5% CO2, for 48h in the presence or absence of IFN-α2A (1,000 U/ml, Blausiegel Ltda., SP-Brazil). Cells were immediately processed for bioassays (apoptosis, proliferation), flow cytometry or RNA extraction.
Flow cytometry
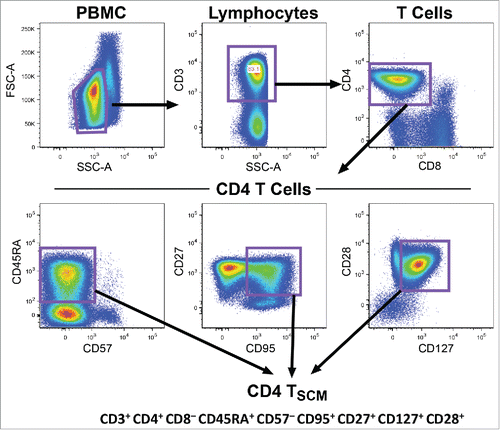
PBMC from Brazilian patients and controls were resuspended at 200,000 cells/50 μl (1% BSA + 0.1% NaN3 in PBS) and incubated for 30 min on ice with antibodies (CD3/CD4/CD25/CD95) and corresponding isotype controls (BD Biosciences). Samples were acquired using FACSort (BD Biosciences) and analyzed using CellQuest software. CD4 TSCM phenotyping (CD3+CD4+CD8−CD45RA+CD57−CD95+CD27+CD127+CD28+) and quantification was performed in the UK Twin cohort,Citation39,Citation40 using the gating strategy outlined in .
Figure 1. Gating strategy for Tscm cells. CD4 TSCM phenotyping and quantification in the UK Twin cohortCitation39 was performed by consecutive gating on CD3+CD4+CD8−, naïve (CD45RA+) and CD57−CD95+CD27+CD127+CD28+ lymphocytes.
FAS genotyping of Brazilian ATL patients and healthy controls
Genotyping for FAS -670 polymorphism by PCR-restriction fragment length polymorphism was performed as described.Citation12 Patient genotype distribution () is in agreement with our previous findings,Citation12 with the AA genotype being overrepresented in ATL patients (37%), as compared to healthy controls (10% AA, 30% GA, 60% GG, n = 20).
Table 1. Clinical, demographic and molecular features of ATL patients.
Combined GWAS and mass flow cytometry study of UK Twin cohort
This study was approved by the NIAID (NIH) IRB and London-Westminster NHS Research Ethics Committee; all participants provided informed consent. The discovery stage comprised 497 female participants from the UK Adult Twin Register, TwinsUK, with full genotyping data on 460 subjects. The TwinsUK cohort is described in detail in.Citation39 GWAS and immunophenotyping are described in detail in.Citation40
Microarray analysis
Total RNA from PBMCs was extracted according to manufacturer's protocol (RNeasy kit QIAgen, Venlo, Netherlands) from ATL patients (n = 8, with and without IFN-α treatment). Whole genome microarray was performed at the VIB Nucleomics Facility (Leuven, Belgium), using the Human Gene 1.0 ST Array with the WT PLUS reagent kit (Affymetrix, Santa Clara, CA) according to manufacturer's instructions. Data were analyzed as previously describedCitation27 and available at the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE85487. Expression data from GSE23321 was used to recapitulate the differentiation gradient demonstrated for CD8 TN, TSCM, TCM and TEM cell populationsCitation41 through Principal Component Analysis, using the published 278 TSCM-specific genes determined by Gattinoni et al.Citation41
Measurement of cytokine production
Cytokine levels in 48h culture supernatants (kept frozen at −80°C immediately after harvesting) were measured using Cytometric Bead Array Human Th1/Th2 Cytokine (BD Biosciences), following the manufacturer's instructions.
Quantification of HTLV-1 p19 expression
Viral p19 protein levelsCitation24,Citation33 in culture supernatants were measured using the HTLV-I/II p19 antigen ELISA (ZeptoMetrix), following the manufacturer's instructions.
Proliferation assays
PBMCs from ATL patients proliferate spontaneously upon culturing, while in healthy controls, proliferation was induced by anti-CD3 antibody stimulation. Ex vivo IFN- α stimulation (120h) was as above, except for plating in 96-well U-bottom plates (200 μl/well at 1 × 106 cells/ml). Lymphoproliferation was quantified by [3H]-thymidine incorporation after a 12–16h pulse (1μCi/well), using gas phase scintillation (Direct Beta Counter Matrix9600, PerkinElmer Life Sciences). Results are expressed as the mean counts per minute in triplicate cultures.
Apoptosis assays
Apoptosis was measured by microscopic quantification of nuclear fragmentation in at least 100 cells (duplicate Hoechst33432 or hematoxylin/eosin staining), as well as flow cytometry (annexin V staining), as above.
Bayesian network learning
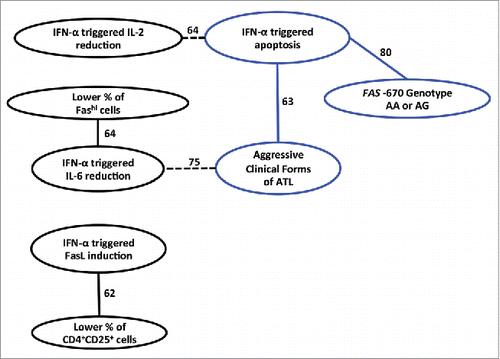
A Bayesian network (BN) is a probabilistic graphical model that describes statistical conditional dependencies between multiple variables. Although originally developed for large pharmacogenetic data sets, Bayesian network learning can discover robust interactions between variables in small datasets.Citation42,Citation43 Dependencies are visualized in a directed acyclic graph and form the qualitative component of the BN. In this graph, each node corresponds to an attribute, and a direct arc between nodes represents a direct influence. Mathematically, a Bayesian network provides a refractoring of the Joint Probability Distribution (JPD) of the data, using Bayes’ rules. As a BN simplifies the JPD, it provides an effective model that summarizes statistical properties of the data. In this way, the best Bayesian network is searched that explains a maximum of the observed associations in the data using a minimum number of direct influences. Bayesian network learning was performed using B-course software adapted by Deforche et al.Citation44 Herein, we learned Bayesian networks from observation of continuous attributes discretized as quartiles (age, proportion of CD4+, CD4+CD25+, and Fas+ cells). Gender was discretized as Male/Female, clinical forms as smoldering, chronic, acute and lymphoma/PCT. FAS -670 genotypes were discretized as GG/GA/AA. All IFN-α responses (proliferation/apoptosis/p19/IL-2/IL-6/FasL, ) were discretized as positive, negative or neutral. The confidence interval of neutral effect (1±0.19) was determined based on 80% power to detect a difference between positive and negative effect for the variable available for the lowest number of patients (i.e. 12 patients for apoptosis). In this non-linear model, the arcs (dependency) were scored based on the stability of the conditional dependency, assessed with a nonparametric bootstrap (100x replicates).Citation45 All arcs with bootstrap over 60% were considered and depicted in the consensus network.
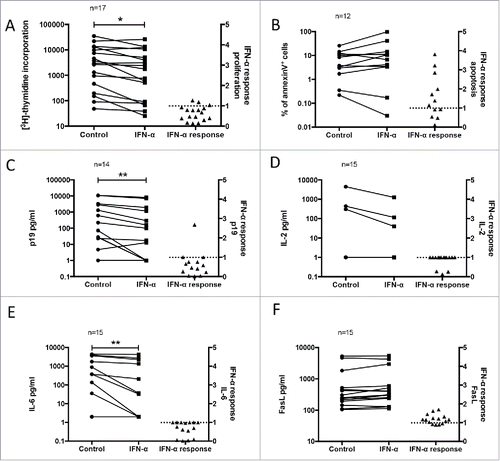
Figure 2. Pleiotropic ex vivo effect of IFN-α in PBMC of ATL patients. PBMC from ATL patients were treated ex vivo with IFN-α (1000 U/ml) and assessed for (A) proliferation (3H-thymidine incorporation), (B) apoptosis (% of annexin V+ cells), (C) virus expression (p19 pg/ml) and (D, E, F) immunomodulation (IL-2, IL-6 and FasL expression, respectively), as described in Methods. A scatter dot plot of within-patient paired observations connected by lines of control non-treated and ex vivo IFN-α-treated PBMC (Wilcoxon matched pairs test, *p < 0.05, **p < 0.01), and their IFN-α response (IFN-α treated/control value) are indicated in each graph. No IFN-α response is indicated by a dotted line.
Statistical analysis
Parametric and non-parametric tests were used according to Kolmogorov-Smirnov test for normality: Pearson/Spearman correlation, t-test/Mann-Whitney/Wilcoxon tests, all two-tailed (GraphPad Prism 5.0 software), differences were considered significant at p<0.05. To maximize data mining output, outliers (Tukey's test) were included for all univariate and multivariate analyses (–) and were excluded only for post-hoc testing of the models (Supplementary Fig. S1-S2).
Results
Ex vivo antiproliferative, pro-apoptotic, antiviral and immunomodulatory effect of IFN-α in PBMC of ATL patients
Freshly isolated primary cells (PBMC) of ATL patients were treated ex vivo with or without IFN-α for 48h and possible pro-apoptotic, anti-proliferative, antiviral and/or immunomodulatory activity was quantified (). To take into account strong inter-patient variation, data were normalized and expressed as IFN-α response (IFN-α value/control value). IFN-α significantly decreased ex vivo lymphoproliferation by a median of 41% (p = 0.027) (). In contrast, the pro-apoptotic effect of IFN-α (median 43% increase) did not reach statistical significance (p = 0.11), but a large inter-patient variability was observed (). As an antiviral readout,Citation24,Citation33 IFN-α significantly decreased viral p19 protein levels in cell culture supernatants (median 60% decrease, p = 0.0015) (). The immunomodulatory effect of IFN-α was examined by measuring Th1/Th2 cytokines in supernatants. No significant differences were observed for TNF-α, IFN-γ, IL-2, IL-4 and IL-10 production (p > 0.05 for all, data not shown). However, a median 74% decrease in IL-2 production was observed after IFN-α treatment in all 3 IL-2 producing patients (). Furthermore, IFN-α treatment significantly reduced IL-6 levels (median 99% decrease, p = 0.0039) (). Fas ligand (FasL) levels were not significantly increased after IFN-α treatment (median 24% increase, p = 0.055) (). To investigate possible interdependencies between the different biological measures, correlation between all six measures of IFN-α response was assessed. In these pairwise comparisons, we found that IFN-α-induced apoptotic and FasL response were positively correlated (r = 0.75, p = 0.013). Similarly, IFN-α-induced antiproliferative and antiviral activities were also positively correlated (r = 0.77, p = 0.003) (Supplementary Fig. S1A-B). Together, these results argue for a pleiotropic effect of IFN-α in primary ATL cells. Therefore, we decided to model the interdependencies between molecular, cellular and clinical data using a data mining approach.
IFN-α response is dependent on clinical and molecular variables: A Bayesian Network approach
Clinical, cellular and molecular attributes were included and discretized as detailed in Methods. Among all 13 attributes, Bayesian network learning discovered several robust interactions: 6 arcs had a bootstrap support over 60% (). The most connected variable was IFN-α-triggered apoptosis, directly linked to FAS -670 genotype, aggressive ATL, and IFN-α-triggered IL-2 reduction. This confirmed that IFN-α triggered apoptosis does play a role in the overall mechanism of action of IFN- α, but that it is heavily dependent on other variables, explaining why it was not significant in univariate analysis (). This Bayesian network suggests that IFN-α-triggered apoptosis is strongly dependent on FAS -670 genotype and especially so in aggressive clinical forms of ATL (blue lines in ), confirming our earlier findings.Citation12 In addition, IFN-α-triggered IL-6 reduction depends on the proportion of Fas+ cells, while IFN-α-induced FasL levels depend on the proportion of CD4+CD25+ (comprising ATL) cells, consistent with the reported importance of IL-6 and Fas in ATL pathogenesis.Citation11-15
Figure 3. IFN-α response is dependent on clinical and molecular variables: A Bayesian Network approach. Annotated Bayesian Network including the following 13 clinical, molecular and cellular attributes: Clinical forms, Age, Gender, FAS -670 genotype, CD4+ %, CD4+CD25+ %, Fashi % (see ) and all six IFN-α response measures (proliferation, apoptosis, p19, IL-2, IL-6 and FasL, see ). Variables that are strongly interdependent are shown by arcs, solid arcs when the association is positive, dashed arcs when negative. The stability of the dependency was assessed with a non-parametric bootstrap (100x replicates). All arcs with bootstrap over 60% are depicted in the network. Apoptosis triggered by IFN-α is strongly dependent on the presence of the FAS -670 A allele, linked to aggressive clinical forms of ATL (blue circles and arcs), and inversely linked to IFN-α induced reduction of IL-2 levels. Likewise, with regard to reduction of IL-6, IFN-α response is more pronounced when there are fewer Fas+ cells and when ATL is less aggressive; while IFN-α induced FasL induction is more pronounced when there are fewer CD4+CD25+ cells (comprising ATL cells).
Pro-apoptotic effect of IFN-α is FAS -670 genotype-dependent in both ATL patients and healthy controls
To confirm the dependencies seen in the BN, we examined IFN-α response in function of FAS -670 genotype. Indeed, the presence of the FAS -670 A allele was highly associated with IFN-α-inducible apoptosis (p = 0.0069) () but not antiproliferative or antiviral IFN-α effect (data not shown). No significant difference in IFN-α response was observed between homozygous AA and heterozygous GA individuals, indicating that one functional allele A is sufficient to determine the pro-apoptotic phenotype.
Figure 4. FAS -670 genotype-dependent effects of IFN-α induced ex vivo apoptosis in PBMCs from ATL patients and healthy donors. PBMCs from ATL patients and healthy donors were treated ex vivo with IFN-α (1000U/ml) (see methods). Apoptosis (A and B, % annexinV+ cells) and FasL levels in supernatants (C and D) were stratified according to FAS -670 genotype (GG vs. GA+AA). (A) IFN-α-induced apoptosis (IFN-α value/control value) in PBMCs of ATL patients depends on FAS -670 genotype (unpaired t test, **p = 0.0069). (B) IFN-α- induced apoptosis in PBMCs of healthy donors depends on FAS -670 genotype (Wilcoxon signed rank test, *p = 0.023). (C) IFN-α does not increase FasL levels in supernatants of PBMCs of ATL patients, independent of FAS -670 genotype. (D) IFN-α significantly increases FasL levels in supernatants of PBMCs of healthy donors (Wilcoxon signed rank test, ***p = 0.0005, **p = 0.0078) but IFN-α response (IFN-α value/control value) does not differ according to FAS -670 genotype.
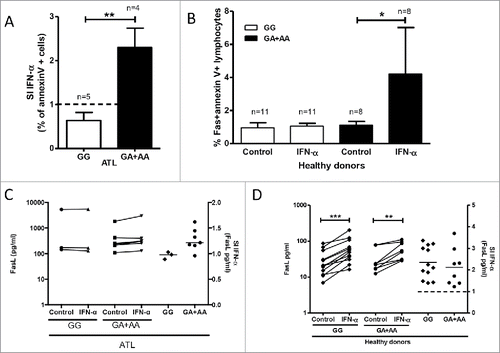
Due to its unusually high penetrance (80% of phenotypic difference explained by genotype), FAS -670 genotype-dependent apoptosis upon IFN-α stimulation was tested in an additional cohort of twenty healthy donors from the same endemic area. Lymphocytes from healthy donors were mostly Faslo (16.8±2.0% Fas+, n = 20). Therefore, Fas-expressing cells were gated separately for apoptosis quantification by annexinV staining. A strikingly similar genotype-phenotype effect was observed in healthy donors (), where IFN-α again selectively induced ex vivo apoptosis in AA and GA donors (p = 0.023, n = 8), but not in GG donors (p = 0.23, n = 12), confirming high penetrance (90%) of the phenotype.
As shown in , there is no significant IFN-α-induced increase in FasL levels in ATL, and in it is shown that this remains so, independent of FAS -670 genotype. In healthy donors, FasL levels can be induced by IFN-α, again independent of FAS -670 genotype (). Of note, compared to healthy controls, FasL levels in ATL patients are constitutively high, at levels that seem not to be further inducible. Taken together, these data suggest a pivotal role of the Fas receptor, rather than its ligand in IFN-α-induced apoptosis in both ATL patients and in healthy donors.
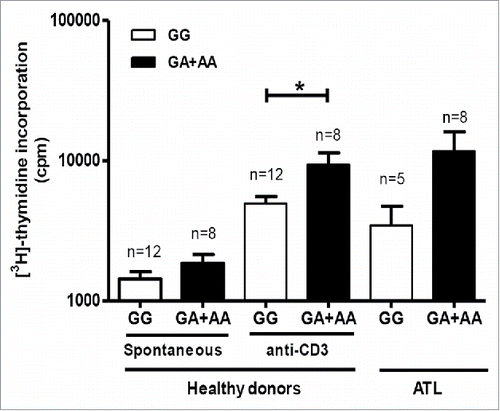
FAS -670 genotype determines TCR-triggered proliferation in PBMC of healthy donors
To investigate the possible effect of FAS -670 genotype on lymphoproliferation, PBMC from genotyped healthy donors were treated with anti-CD3 antibody to induce TCR-triggered polyclonal proliferation. Since this is also known to result in activation-induced, Fas-mediated cell death,Citation46–48 we expected a decrease in proliferation for the “pro-apoptotic” GA/AA genotypes. However, the presence of FAS -670 GA/AA genotypes resulted in significantly higher anti-CD3-stimulated proliferation (p = 0.049), as compared to GG homozygotes (), suggesting Fas-mediated proliferation may be independent of Fas-mediated apoptosis. Interestingly, a 3.4 fold increase in spontaneous lymphoproliferation was also observed in ATL patients with a functional FAS -670 A allele, although not statistically significant (p = 0.11) ().
Figure 5. FAS -670 genotype-dependent proliferation in PBMC from healthy donors and ATL patients. Spontaneous proliferation of PBMCs from healthy donors and ATL patients and anti-CD3 induced proliferation for healthy donors were stratified according to FAS -670 genotype (GG vs. GA + AA) (Mann-Whitney test, *p < 0.05).
CD4+CD25+ levels correlate positively with IFN-α induced apoptosis
In order to appreciate whether a different T-cell population might be proliferating vs. undergoing apoptosis, CD4+ and CD4+CD25+ levels were correlated with IFN-α-proliferative/apoptotic response in both healthy donors and ATL patients. A significant positive correlation between CD4+CD25+ levels was found for IFN-α-induced apoptosis, but not proliferation, in both healthy donors (p = 0.044) and ATL patients (p = 0.0037) (Suppl. Fig. 2). Since we showed above that FAS genotype is significantly associated to ex vivo IFN-α-induced apoptosis and TCR-triggered proliferation, this suggests a differential effect on different T-cell types, which alerted us to the possible involvement of the recently described Fashi memory stem cell subset in our ex vivo ATL model.
FAS -670 genotype determines systemic CD4 TSCM levels in a large twin study
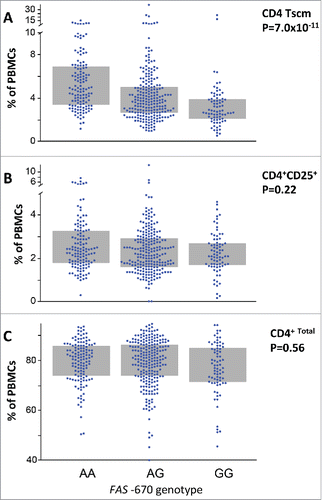
A recent report has identified CD4 TSCM cells as the hierarchical apex of ATL.Citation19 Due to the prospective design of our ATL cohort, patient cells were no longer available to test a possible association between FAS -670 genotype and CD4 TSCM. To investigate this possible association in healthy donors, a large cohort would be required to achieve sufficient statistical power. Therefore, we tested this hypothesis in the unique twin cohort recently analyzed by simultaneous GWAS and mass cytometry.Citation40 As shown in , FAS -670 genotype was significantly associated with circulating CD4 TSCM levels (p = 7 × 10−11, n = 460). Individuals with the functional (i.e. IFN-responsive) AA genotype exhibited a two-fold increase in circulating CD4 TSCM levels, as compared to the non-functional GG. However, FAS -670 genotype was not associated to levels of either activated (CD4+CD25+ cells, ) or total CD4 cells (), highlighting its cell-type specificity.
Figure 6. FAS -670 genotype determines systemic CD4 TSCM levels in a large twin study. (A) FAS -670 genotype is significantly associated with circulating CD4 TSCM levels (p = 7 × 10−11, n = 460). Individuals with the functional (i.e. IFN-responsive) AA genotype exhibited a two-fold increase in circulating CD4 TSCM levels (5% of total PBMC), as compared to the non-functional GG genotype (2.5% of total PBMC). However, FAS -670 genotype was not associated to either activated CD4 + CD25 + cells (B), or total CD4 cells (C). Twin study and GWAS statistics using GenABEL software package as described.Citation39,Citation40
Transcriptomic analysis and cell type deconvolution confirms IFN/FAS/TSCM link
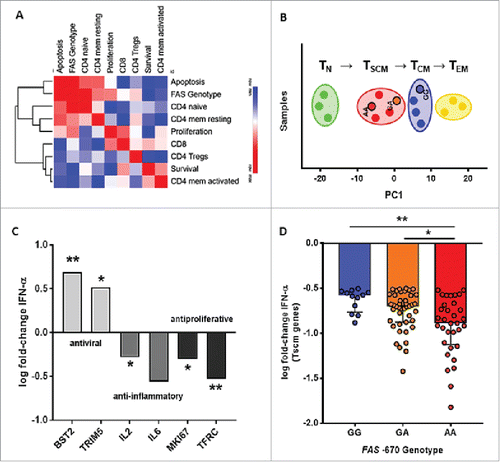
Since the TSCM cellular phenotype was unknown at the time of patient recruitment for this study, our flow cytometry analysis was limited to broader, overlapping phenotypes (CD4/CD8/CD25/CD95). However, using archived RNA samples from a subset of 8 ATL patients with paired IFN-α-treated samples, we were able to perform transcriptome profiling by microarray and cell type deconvolution using CIBERSORT.Citation49 Thus, using validated cell type-specific gene sets, the relative size of total CD8 cells and CD4 subsets (naïve, memory resting, memory activated and Treg) was predicted in silico and correlated to patient survival, apoptosis and proliferation. As shown in , unsupervised hierarchical clustering revealed the strong link between FAS genotype, IFN-induced apoptosis, CD4 naïve cells and CD4 resting memory cells. This is in agreement with Gattinoni et al., with TSCM displaying an intermediate transcriptome profile between naïve and central memory T cells.Citation41 Therefore, we used 278 TSCM-specific genes identified by Gattinoni et al.Citation41 (Suppl. Table I) to perform principal component analysis of purified naive T cells, memory stem T cells, central memory T cells and effector memory T cells (n = 3 each, fromCitation41). and primary cells of three distinct ATL patients from this study cohort with GG, GA or AA genotypes (one each). As shown in , the first principal component clearly recapitulates the described differentiation gradient of TN→TSCM→TCM→TEM. Clustering (k-means) allowed to distinguish all four memory subsets, and to discriminate between functional (AA, GA) and non-functional (GG) FAS genotypes in ATL patients. Strikingly, only AA and GA patient samples clustered with TSCM cells, whereas the GG patient sample did not, replicating the intricate link between FAS -670 genotype and TSCM phenotype we identified in the large twin cohort (). Clustering results were robust to the inclusion of our patient samples (predominantly CD4 subpopulations, by flow cytometry and CIBERSORT) with the published data used to define TSCM-specific genes in CD8 subpopulations (Supplementary Figure 3). suggesting the “stemness” of CD4 and CD8 cells might be conserved at the transcriptional level. To test the hypothesis of IFN-induced apoptosis of TSCM cells, we quantified the effect of IFN-α upon overall vs. TSCM-specific gene expression in the ATL transcriptome. When analyzing all patients together, irrespective of genotypes, IFN-α signficantly decreased transcript levels of four out of six Naive/TSCM signature genesCitation41 (data not shown), including transcription regulators LEF1 (p = 0.005, n = 8), CERS6 (p = 0.036, n = 8) and TAF4B (p = 0.004, n = 8), whereas no significant effect was observed for T-cell effector or memory signature genes (e.g. Eomes, T-bet, PRDM1, data not shown). Furthermore, we were able to confirm the antiviral, anti-inflammatory and antiproliferative effects of IFN-α () at the transcriptional level (), as evidenced by pronounced upregulation of antiretroviral effectors (BST2/TRIM5) and downregulation of cytokines (IL2/IL6) and proliferation markers Ki67 and CD71 (encoded by MKI67/TFRC genes), in agreement with our previous results.Citation27 When considering FAS genotype, we found IFN-induced downregulation of TSCM-specific genes was dependent on the number of functional A alleles (0-1-2 for GG-GA-AA genotypes, ANOVA with post-test for linear trend, p = 0.001). As shown in , IFN-α caused the strongest downregulation of TSCM genes in AA genotype (>GA, p = 0.014; >GG, p = 0.0011), argueing in favour of Fas-dependent TSCM cell death. Of note, we also confirmed our previous observationCitation12 of a significant negative correlation between the number of functional FAS -670 A alleles and survival (r = -0.72, p = 0.030, n = 8), indicating this small subset is representative of the larger cohort.
Figure 7. Transcriptomic analysis and cell type deconvolution confirms IFN/FAS/TSCM link in ATL primary cells. (A) Transcriptome profiling by microarray and cell type deconvolution (CIBERSORT) was used to quantify total CD8 cells and CD4 subsets (naïve, memory resting, memory activated and Treg). Data were correlated to patient survival, apoptosis and proliferation, followed by unsupervised hierarchical clustering. (B) For Principal Component (PC) analysis, we used microarray signal intensity data of 278 selected TSCM genes of purified naive (TN cells) (n = 3), stem cell memory (TSCM cells) (n = 3), central memory (TCM cells) (n = 3) and effector memory T cells (TEM cells) (n = 3) from the Gattinoni study,Citation41 and primary cells of three distinct ATL patients with FAS -670 GG, GA or AA genotypes (one each) from this study cohort. Clusters were defined by k-means algorithm. (C) Confirmation of antiviral, anti-inflammatory and antiproliferative effect of IFN-α in ATL patients (n = 8) at the transcriptional level, by upregulation of antiretroviral effector molecules (BST2/TRIM5) and downregulation of cytokines (IL2/IL6) and proliferation markers (MKI67/TFRC). (D) Among 278 TSCM-specific genes (Suppl. Table I), downregulation by IFN-α treatment was calculated as log fold-change (with a cut-off at -0.5) in ATL patients with selected genotypes (AA-GA-GG, one each). TSCM-specific genes were most strongly downregulated in AA genotype, vs both GA and GG genotypes (Kruskall-Wallis with FDR correction, p = 0.014 and p = 0.0011, respectively). *p < 0.05, **p < 0.01
Proposed model for a coordinated mechanism of action of IFN-α in ATL
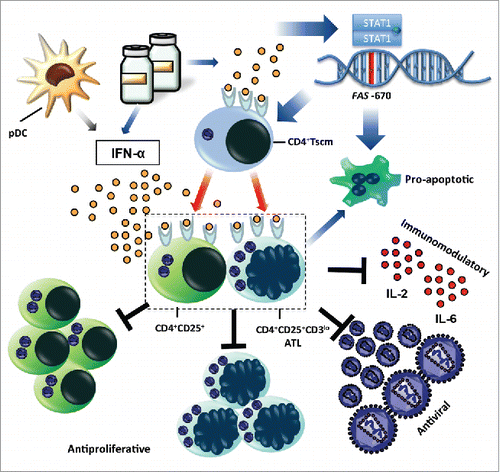
Endogenous IFN-α has been shown to be produced by plasmacytoid dendritic cells (pDC) upon stimulation with free virions,Citation50 which can be assumed to occur in vivo during ATL pathogenesis and/or disease progression.Citation51 Furthermore, exogenous IFN-α treatment exerts pleiotropic effects as demonstrated in this study. A simplified model () illustrates how endogenous or exogenous IFN-α might exert its coordinated effects in vivo. Our data strongly suggest that IFN-α-induced apoptosis, intrinsically linked to the presence of the FAS -670 A allele, is driving the antiviral and antiproliferative activity of IFN-α. The FAS -670 A allele is also linked to increased CD4 TSCM cells (), resulting in an ATL leukemic phenotype characterized as CD4+CD25+CD3lo and Fashi.(Citation52,Citation53 and this study) In addition, IFN-α results in a decrease of both IL-2 (growth factor for CD4+CD25+ cells) and IL-6, high levels of which have been previously associated with decreased ATL survival.Citation11 Furthermore, a direct activity of IFN-α on CD4+CD25+ cells can be assumed (Suppl. ), which might be independent of FAS genotype (). Apoptosis of CD4+CD25+ cells, the main reservoir of infected cells,Citation54 likely results in decreased viral protein levels, as well as decreased CD4+CD25+ proliferation. Due to the high significance of the FAS/CD4 TSCM connection, beyond the genome-wide association threshold (), it is tempting to speculate that the model proposed here would be applicable and of clinical relevance in other leukemias, as well as other viral pathologies treated with IFN-α.
Figure 8. Proposed model for coordinated mechanism of action of IFN-α in ATL. Endogenous (from plasmacytoid dendritic cells – pDC) or exogenous IFN-α (ex vivo or in vivo treatment) elicits pro-apoptotic, antiproliferative, antiviral and immunomodulatory effects on CD4+CD25+ cells, the major HTLV-1-infected reservoir, as well as on leukemic CD4+CD25+CD3loFas+ cells. Red arrows indicate the link to FAS -670 polymorphism and TSCM cells (this study,Citation12,Citation19).
Discussion
Conflicting data exist on the cellular and molecular mechanisms of action of type I IFN (IFN-α and IFN-β) in several major human diseasesCitation23-36 including ATL. This HTLV-1-associated adult T-cell leukemia responds at varying degrees to IFN-α + AZT combination therapy.Citation20-22 In this study, we show that ex vivo IFN-α treatment exerts a pleiotropic and strongly heterogeneous response in ATL primary cells. IFN-α significantly decreased proliferation, viral protein levels and IL-6 levels for a majority of patients, while it induced apoptosis and reduced IL-2 levels for a subset of patients (). These results are in contrast with our results in another HTLV-1-associated disease (HTLV-1-associated myelopathy/Tropical spastic paraparesis), where we found no significant effect of ex vivo IFN-α upon proliferation, cell death or IL-6 production.Citation25 In addition, a recently published systems biology approach revealed an IFN-inducible gene signature in HAM/TSP patients, surprisingly paralleled by limited antiviral activity.Citation55 Therefore, we set out to model the interdependencies between ex vivo molecular and cellular IFN-α-response and the in vivo clinical and immunological data in ATL. In this study, we reveal a high-penetrance, IFN-dependent, pro-apoptotic phenotype for the functional A allele of the FAS -670 A/G polymorphism in both ATL patients and healthy controls. Strikingly, the association of this A allele with IFN-induced apoptosis underscores the recently established “CD4 TSCM apex” model for ATL,Citation19 since we also found a strong link between the A allele and higher in vivo circulating CD4 TSCM levels in a large twin study ().
Since ATL is a relatively rare disease, even in highly endemic areas such as Salvador-Bahia, Brasil,Citation56 only 25 patients with clinically definite ATL could be recruited during a five-year period, followed by a five-year clinical follow-up, which precluded confirmation of our data in a second cohort of ATL patients. However, we were able to confirm the IFN-dependent link between CD4 TSCM cells, Fas genotype and apoptosis in a subset of ATL patients by transciptomic analysis (). In addition, the dependence of the IFN-α pro-apoptotic effect on the presence of the FAS -670 A allele was validated in healthy donors from the same endemic area. The remarkably high penetrance (80% in ATL, 90% in normal donors) of the FAS polymorphism suggests a decisive effect of the receptor in Fas/FasL signaling, at least upon IFN-α stimulation. Although FasL polymorphisms have also been significantly associated with an in vitro pro-apoptotic phenotype in cervical cancer,Citation57 three lines of evidence support our receptor-mediated hypothesis for the genotype-phenotype association of IFN-induced apoptosis in this study. First, FASL -844 polymorphism was not associated to in vitro apoptosis in both ATL patients and healthy controls (Silva-Santos et al., unpublished). Second, IFN-α strongly increased FasL levels in 9 out of 13 ATL patients () and in all healthy controls tested (, p < 0.001), resulting in excess ligand and hence indicating receptor signaling as the limiting step. Third, a recent GWAS twin study linking 80.000 immune phenotypes to SNPs identified polymorphisms in FAS, but not FASL, as significantly associated with circulating T-cell subset levels.Citation40 Following up on this study, we show that the FAS -670 polymorphism is significantly associated to circulating CD4 TSCM, but not CD4+CD25+ or total CD4 levels ().
Surprisingly, functional GA/AA genotypes displayed higher ex vivo proliferation upon TCR stimulation (), which somehow contrasts with the pro-apoptotic genotype/phenotype, but is consistent with a proliferative TSCM phenotype in vitro.Citation41 Non-apoptotic Fas signaling has indeed been demonstrated to stimulate cellular proliferation in primary CD4 cells,Citation58 but also phenotypic differentiation of naïve CD8 T cells into memory subsets, through an Akt-driven mechanism.Citation59 Similar to these observations, we have recently identified a Fashi lymphoproliferative phenotype in another HTLV-1-associated pathology, the neuroinflammatory disease HAM/TSP60. Likewise, it has been shown that most ATL cells display a Fashi phenotype,Citation61 whereas most T-cell leukemias display decreased Fas expression and/or mutations in Fas/FasL pro-apoptotic signaling.Citation15 Increased Fas expression might induce survival of (pre-)leukemic cells, through increased proliferation of CD4+Fas+ T-cell clones upon antigenic stimulation and eventually escape apoptosis,Citation62 through increased c-FLIP or similar pathways.Citation63,Citation64 In fact, Fas was previously identified as a proto-oncogene, capable of inducing tumorogenesis in a different type of cancer.Citation65
Our results indicate that this Fas-mediated dichotomy of apoptosis vs. proliferation, previously described in vitro in CD4+ cells,Citation58 might be implicated in ATL pathogenesis, by tipping the balance between leukemic CD4+CD25+CD3lo(Fashi) and anti-leukemic CD8+ cells ( and ). Conversely, high levels of leukemic and/or Treg cells with a CD4+CD25+ phenotype were positively correlated with IFN-α-induced apoptosis (Suppl. ). Elimination of both cell types will most probably result in clinical benefit in ATL patients either directly or indirectly, since Tregs have been shown to inhibit the ex vivo CTL response in ATL patients,Citation66 and negatively correlate to patient survival in our analysis (). Taken together, the functionality of the FAS -670 polymorphism resolves the IFN-α apoptotic paradox and reveals this polymorphism as a candidate pharmacogenetic marker for several other IFN-α-treated leukemias and cancers.Citation67 Thus, our model does not contradict the previous findings of absent cytotoxic or antiproliferative activity of IFNsCitation23,Citation26 but highlights the role of FAS genotype in determining cell fate: proliferation vs. apoptosis.Citation58–65,Citation67
Moreover, patient-derived individual T-cell clones represent only a small percentage of the whole T-cell population, whereas the majority has been eliminated, either through “death by neglect”, activation-induced cell death or direct CD8+ T-cell-mediated killing.Citation46-48,Citation61-68 Therefore, our ex vivo results are not in contrast, but complimentary to previous in vitro results in patient-derived T-cell clones.Citation26,Citation64,Citation68 In addition, the use of total PBMCs also allowed us to measure and integrate the impact of other cellular subtypes, such as DCs,Citation50,Citation51 monocytesCitation69 and NK cells,Citation70 among others, upon apoptosis, proliferation and antiviral response. It is well described that pDC are the main source of IFN-α production in vivo.Citation52 Therefore, we believe our ex vivo model, in spite of its limitations, reflects the in vivo situation in ATL patients.
In conclusion, we show that a functional FAS polymorphism resolves the current “IFN apoptotic paradox”, by determining ex vivo apoptosis in both ATL patients and healthy controls with high penetrance. The same polymorphism also determines lymphoproliferation and CD4 TSCM levels in healthy controls. In line with the CD4 TSCM ATL origin, our data help explain both in vivo IFN+AZT responsiveness and chemotherapy resistance in ATL patients. FAS -670 polymorphism thus represents a promising pharmacogenetic marker in the clinical follow-up of several major human diseases currently or previously treated by type I IFN, such as other leukemias, melanoma, HCV infection and multiple sclerosis.
Authorship and conflict of interest statements
RK and JVW designed research; RK, GSS, DD, SMM, ACS and JVW performed research; TD, KT, and AMV contributed vital new analytical tools; AB and LF provided patient samples; MM and MR provided genetic and flow cytometry data from the twin cohort; RK and JVW analyzed data and wrote the paper. None of the authors has any conflict of interest to declare.
IndependentRunPCA.pdf
Download PDF (30.5 KB)SUPPLEMENTARY_DATA.docx
Download MS Word (14.6 KB)2017ONCOIMM0008R1-s04.xlsx
Download MS Excel (52.3 KB)2017ONCOIMM0008R1-s03.tif
Download TIFF Image (911.1 KB)2017ONCOIMM0008R1-s02.tif
Download TIFF Image (926.3 KB)Acknowledgments
This work was supported by: Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), “Science without Borders” Program, PRONEX, FWO (Grant: ZKC1280-00-W10 and G0D6817N), Alban Program (European Union Program of High Level Scholarships for Latin America, scholarship no. E06D103200BR), VLAIO (TD scholarship) and KU Leuven (IRO and “Vaast Leysen Leerstoel voor Infectieziekten in Ontwikkelingslanden”). KT is funded as a Postdoctoral Researcher by the Research Foundation – Flanders (FWO).
Additional information
Funding
References
- Uchiyama T, Yodoi J, Sagawa K, Takatsuki K, Uchino H. Adult T-cell leukemia: clinical and hematologic features of 16 cases. Blood. 1977;50(3):481–92. PMID:301762.
- Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci United States Am. 1980;77(12):7415–9. doi: 10.1073/pnas.77.12.7415.
- Verdonck K, González E, Van Dooren S, Vandamme AM, Vanham G, Gotuzzo E. Human T-lymphotropic virus 1: recent knowledge about an ancient infection. Lancet Infect Dis. 2007;7(4):266–81.doi: 10.1016/S1473-3099(07)70081-6 PMID:17376384.
- Cleghorn FR, Manns A, Falk R, Hartge P, Hanchard B, Jack N, Williams E, Jaffe E, White F, Bartholomew C, et al. Effect of human T-lymphotropic virus type I infection on non-Hodgkin's lymphoma incidence. J Natl Cancer Inst. 1995;87(13):1009–14. doi:10.1093/jnci/87.13.1009. PMID:7629870.
- Shimoyama M. Diagnostic criteria and classification of clinical subtypes of adult T-cell leukaemia-lymphoma. A report from the Lymphoma Study Group (1984-87). Br J Haematol. 1991;79(3):428–37. doi:10.1111/j.1365-2141.1991.tb08051.x. PMID:1751370.
- Tsukasaki K, Hermine O, Bazarbachi A, Ratner L, Ramos JC, Harrington W, Jr, O'Mahony D, Janik JE, Bittencourt AL, Taylor GP, et al. Definition, prognostic factors, treatment, and response criteria of adult T-cell leukemia-lymphoma: a proposal from an international consensus meeting. J Clin Oncol. 2009;27(3):453–9. doi:10.1200/JCO.2008.18.2428. PMID:19064971.
- Bittencourt AL, da Graças Vieira M, Brites CR, Farre L, Barbosa HS. Adult T-cell leukemia/lymphoma in Bahia, Brazil: analysis of prognostic factors in a group of 70 patients. Am J Clin Pathol. 2007;128(5):875–82. doi:10.1309/2YGD1P0QCVCWBLDX. PMID:17951212.
- Tagaya Y, Gallo RC. The Exceptional Oncogenicity of HTLV-1. Front Microbiol. 2017;8:1425. doi:10.3389/fmicb.2017.01425. PMID:28824561.
- Ciminale V, Rende F, Bertazzoni U, Romanelli MG. HTLV-1 and HTLV-2: highly similar viruses with distinct oncogenic properties. Front Microbiol. 2014;5:398. doi:10.3389/fmicb.2014.00398. PMID:25120538.
- Yasuda N, Lai PK, Ip SH, Kung PC, Hinuma Y, Matsuoka M, Hattori T, Takatsuki K, Purtilo D. Soluble interleukin 2 receptors in sera of Japanese patients with adult T cell leukemia mark activity of disease. Blood. 1988;71(4):1021–6. PMID:2895675.
- Yamamura M, Yamada Y, Momita S, Kamihira S, Tomonaga M. Circulating interleukin-6 levels are elevated in adult T-cell leukaemia/lymphoma patients and correlate with adverse clinical features and survival. Br. J. Haematol. 1998;100(1):129–34. doi:10.1046/j.1365-2141.1998.00538.x. PMID:9450801.
- Farre L, Bittencourt AL, Silva-Santos G, Almeida A, Silva AC, Decanine D, Soares GM, Alcantara LC, Jr, Van Dooren S, Galvão-Castro B, et al. Fas 670 promoter polymorphism is associated to susceptibility, clinical presentation, and survival in adult T cell leukemia. J Leukoc Biol. 2008;83(1):220–2. doi:10.1189/jlb.0407198. PMID:17962369.
- Takeuchi S, Matsushita M, Tsukasaki K, Takeuchi N, Tomonaga M, Komatsu N, Ikezoe T, Uehara Y, Koeffler HP. P53 codon 72 polymorphism is associated with disease progression in adult T-cell leukaemia/lymphoma. Br J Haematol. 2005;131(4):552–3. doi:10.1111/j.1365-2141.2005.05798.x. PMID:16281948.
- Tamiya S, Etoh K, Suzushima H, Takatsuki K, Matsuoka M. Mutation of CD95 (Fas/Apo-1) gene in adult T-cell leukemia cells. Blood. 1998;91(10):3935–42. PMID:9573032.
- Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga J, Totoki Y, Chiba K, Sato-Otsubo A, Nagae G, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015;47(11):1304–15. doi:10.1038/ng.3415. PMID:26437031.
- Beltinger C, Kurz E, Böhler T, Schrappe M, Ludwig WD, Debatin KM. CD95 (APO-1/Fas) mutations in childhood T-lineage acute lymphoblastic leukemia. Blood. 1998;91(10):3943–51. PMID:9573033.
- Huang QR, Morris D, Manolios N. Identification and characterization of polymorphisms in the promoter region of the human Apo-1/Fas (CD95) gene. Mol Immunol. 1997;34(8-9):577–82. doi:10.1016/S0161-5890(97)00081-3. PMID:9393960.
- Kanemitsu S, Ihara K, Saifddin A, Otsuka T, Takeuchi T, Nagayama J, Kuwano M, Hara T. A functional polymorphism in fas (CD95/APO-1) gene promoter associated with systemic lupus erythematosus. J. Rheumatol. 2002;29(6):1183–8. PMID:12064832.
- Nagai Y, Kawahara M, Hishizawa M, Shimazu Y, Sugino N, Fujii S, Kadowaki N, Takaori-Kondo A. T memory stem cells are the hierarchical apex of adult T-cell leukemia. Blood. 2015;125(23):3527–35. doi:10.1182/blood-2014-10-607465. PMID:25847015.
- Gill PS, Harrington W, Kaplan MH, Ribeiro RC, Bennett JM, Liebman HA, Bernstein-Singer M, Espina BM, Cabral L, Allen S, et al. Treatment of adult T-cell leukemia-lymphoma with a combination of interferon alfa and zidovudine. N Engl J Med. 1995;332(26):1744–8. doi:10.1056/NEJM199506293322603. PMID:7760890.
- Hermine O, Bouscary D, Gessain A, Turlure P, Leblond V, Franck N, Buzyn-Veil A, Rio B, Macintyre E, Dreyfus F, et al. Brief report: treatment of adult T-cell leukemia-lymphoma with zidovudine and interferon alfa. N Engl J Med. 1995;332(26):1749–51. doi:10.1056/NEJM199506293322604. PMID:7760891.
- Bazarbachi A, Plumelle Y, Carlos Ramos J, Tortevoye P, Otrock Z, Taylor G, Gessain A, Harrington W, Panelatti G, Hermine O. Meta-analysis on the use of zidovudine and interferon-alfa in adult T-cell leukemia/lymphoma showing improved survival in the leukemic subtypes. J Clin Oncol. 2010;28(27):4177–83. doi:10.1200/JCO.2010.28.0669. PMID:20585095.
- Bazarbachi A, Nasr R, El-Sabban ME, Mahé A, Mahieux R, Gessain A, Darwiche N, Dbaibo G, Kersual J, Zermati Y, et al. Evidence against a direct cytotoxic effect of alpha interferon and zidovudine in HTLV-I associated adult T cell leukemia/lymphoma. Leukemia. 2000;14(4):716–21. doi:10.1038/sj.leu.2401742. PMID:10764160.
- Moens B, Pannecouque C, López G, Talledo M, Gotuzzo E, Khouri R, Bittencourt A, Farré L, Galvão-Castro B, Vandamme AM, et al. Simultaneous RNA quantification of human and retroviral genomes reveals intact interferon signaling in HTLV-1-infected CD4+ T cell lines. Virol J. 2012;9:171. doi:10.1186/1743-422X-9-171. PMID:22917064.
- Moens B, Decanine D, Menezes SM, Khouri R, Silva-Santos G, Lopez G, Alvarez C, Talledo M, Gotuzzo E, de Almeida Kruschewsky R, et al. Ascorbic acid has superior ex vivo antiproliferative, cell death-inducing and immunomodulatory effects over IFN-α in HTLV-1-associated myelopathy. PLoS Negl Trop Dis. 2012;6(7):e1729. doi:10.1371/journal.pntd.0001729. PMID:22848768.
- Smith D, Buckle GJ, Hafler DA, Frank DA, Höllsberg P. HTLV-I-infected T cells evade the antiproliferative action of IFN-beta. Virology. 1999;257(2):314–21. doi:10.1006/viro.1999.9679. PMID:10329542.
- Dierckx T, Khouri R, Menezes SM, Decanine D, Farre L, Bittencourt A, Vandamme AM, Van Weyenbergh J. IFN-β induces greater antiproliferative and proapoptotic effects and increased p53 signaling compared with IFN-α in PBMCs of Adult T-cell Leukemia/Lymphoma patients. Blood Cancer J. 2017;7(1):e519.25. doi:10.1038/bcj.2016.126..
- Kaser A, Nagata S, Tilg H. Interferon alpha augments activation-induced T cell death by upregulation of Fas (CD95/APO-1) and Fas ligand expression. Cytokine. 1999;11(10):736–43. doi:10.1006/cyto.1998.0484. PMID:10525311.
- Van Weyenbergh J, Wietzerbin J, Rouillard D, Barral-Netto M, Liblau R. Treatment of multiple sclerosis patients with interferon-beta primes monocyte-derived macrophages for apoptotic cell death. J Leukoc Biol. 2001;70(5):745–8. PMID:11698494.
- Gniadek P, Aktas O, Wandinger K-P, Bellmann-Strobl J, Wengert O, Weber A, von Wussow P, Obert HJ, Zipp F. Systemic IFN-beta treatment induces apoptosis of peripheral immune cells in MS patients. J Neuroimmunol. 2003;137(1-2):187–96. doi:10.1016/S0165-5728(03)00074-2. PMID:12667663.
- Zipp F, Beyer M, Gelderblom H, Wernet D, Zschenderlein R, Weller M. No induction of apoptosis by IFN-beta in human antigen-specific T cells. Neurology. 2000;54(2):485–7. doi:10.1212/WNL.54.2.485. PMID:10668719.
- Dondi E, Roué G, Yuste VJ, Susin SA, Pellegrini S. A dual role of IFN-alpha in the balance between proliferation and death of human CD4+ T lymphocytes during primary response. J Immunol. 2004;173(6):3740–7. doi:10.4049/jimmunol.173.6.3740. PMID:15356120.
- Feng X, Vander HN, Ratner L. Alpha interferon inhibits human T-cell leukemia virus type 1 assembly by preventing Gag interaction with rafts. J Virol. 2003;77(24):13389–95. doi:10.1128/JVI.77.24.13389-13395.2003. PMID:14645593.
- Kiladjian J-J, Giraudier S, Cassinat B. Interferon-alpha for the therapy of myeloproliferative neoplasms: targeting the malignant clone. Leukemia. 2016;30(4):776–81. doi:10.1038/leu.2015.326. PMID:26601783.
- Mullally A, Bruedigam C, Poveromo L, Heidel FH, Purdon A, Vu T, Austin R, Heckl D, Breyfogle LJ, Kuhn CP, et al. Depletion of Jak2V617F myeloproliferative neoplasm-propagating stem cells by interferon-α in a murine model of polycythemia vera. Blood. 2013;121(18):3692–702. doi:10.1182/blood-2012-05-432989. PMID:23487027.
- Talpaz M, Mercer J, Hehlmann R. The interferon-alpha revival in CML. Ann. Hematol. 2015;94 Suppl 2:S195–207. doi:10.1007/s00277-015-2326-y. PMID:25814086.
- Bittencourt AL, Barbosa HS, Requião C, da Silva AC, Vandamme AM, Van Weyenbergh J, Farré L. Adult T-cell leukemia/lymphoma with a mixed CD4+ and CD8+ phenotype and indolent course. J Clin Oncol. 2007;25(17):2480–2. doi:10.1200/JCO.2007.11.3043. PMID:17557960.
- Farre L, de Oliveira M, de FP, Primo J, Vandamme AM, Van Weyenbergh J, Bittencourt AL. Early sequential development of infective dermatitis, human T cell lymphotropic virus type 1-associated myelopathy, and adult T cell leukemia/lymphoma. Clin Infect Dis. 2008;46(3):440–2. doi:10.1086/524695. PMID:18173359.
- Moayyeri A, Hammond CJ, Hart DJ, Spector TD. Effects of age on genetic influence on bone loss over 17 years in women: the Healthy Ageing Twin Study (HATS). J Bone Miner Res. 2012;27(10):2170–8. doi:10.1002/jbmr.1659. PMID:22589082.
- Roederer M, Quaye L, Mangino M, Beddall MH, Mahnke Y, Chattopadhyay P, Tosi I, Napolitano L, Terranova Barberio M, Menni C, et al. The genetic architecture of the human immune system: A bioresource for autoimmunity and disease pathogenesis. Cell. 2015;161(2):387–403. doi:10.1016/j.cell.2015.02.046. PMID:25772697.
- Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, Almeida JR, Gostick E, Yu Z, Carpenito C, et al. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17(10):1290–7. doi:10.1038/nm.2446. PMID:21926977.
- Kouri V, Khouri R, Alemán Y, Abrahantes Y, Vercauteren J, Pineda-Peña AC, Theys K, Megens S, Moutschen M, Pfeifer N, et al. CRF19_cpx is an Evolutionary fit HIV-1 Variant Strongly Associated With Rapid Progression to AIDS in Cuba. EBioMedicine. 2015;2(3):244–54. doi:10.1016/j.ebiom.2015.01.015. PMID:26137563.
- Snoek J, Casado C, Colombo S, et al. A Bayesian network approach to study host and viral genetic correlates of HIV-1 disease progression. Retrovirology. 2011;8(Suppl 2):P70. doi:10.1186/1742-4690-8-S2-P70.
- Deforche K, Silander T, Camacho R, Grossman Z, Soares MA, Van Laethem K, Kantor R, Moreau Y, Vandamme AM. Analysis of HIV-1 pol sequences using Bayesian Networks: implications for drug resistance. Bioinformatics. 2006;22(24):2975–9. doi:10.1093/bioinformatics/btl508. PMID:17021157.
- Friedman N, Goldszmidt M, Wyner A. Data Analysis with Bayesian Networks: A Bootstrap Approach. CoRR. 2013;abs–1301–6695. http://dblp.org/rec/bib/journals/corr/abs-1301-6695
- Dhein J, Walczak H, Bäumler C, Debatin KM, Krammer PH. Autocrine T-cell suicide mediated by APO-1/(Fas/CD95). Nature. 1995;373(6513):438–41. doi:10.1038/373438a0. PMID:7530335.
- Brunner T, Mogil RJ, LaFace D, Yoo NJ, Mahboubi A, Echeverri F, Martin SJ, Force WR, Lynch DH, Ware CF, et al. Cell-autonomous Fas (CD95)/Fas-ligand interaction mediates activation-induced apoptosis in T-cell hybridomas. Nature. 1995;373(6513):441–4. doi:10.1038/373441a0. PMID:7530336.
- Ju ST, Panka DJ, Cui H, Ettinger R, el-Khatib M, Sherr DH, Stanger BZ, Marshak-Rothstein A. Fas(CD95)/FasL interactions required for programmed cell death after T-cell activation. Nature. 1995;373(6513):444–8. doi:10.1038/373444a0. PMID:7530337.
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–7. doi:10.1038/nmeth.3337. PMID:25822800.
- Siegal FP, Kadowaki N, Shodell M, Fitzgerald-Bocarsly PA, Shah K, Ho S, Antonenko S, Liu YJ. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284(5421):1835–7. doi:10.1126/science.284.5421.1835. PMID:10364556.
- Jones KS, Petrow-Sadowski C, Huang YK, Bertolette DC, Ruscetti FW. Cell-free HTLV-1 infects dendritic cells leading to transmission and transformation of CD4(+) T cells. Nat Med. 2008;14(4):429–36. doi:10.1038/nm1745. PMID:18376405.
- Yokote T, Akioka T, Oka S, Hara S, Kobayashi K, Nakajima H, Yamano T, Ikemoto T, Shimizu A, Tsuji M, et al. Flow cytometric immunophenotyping of adult T-cell leukemia/lymphoma using CD3 gating. Am J Clin Pathol. 2005;124(2):199–204. doi:10.1309/KEN4MXM5Y9A1GEMP. PMID:16040289.
- Tian Y, Kobayashi S, Ohno N, Isobe M, Tsuda M, Zaike Y, Watanabe N, Tani K, Tojo A, Uchimaru K. Leukemic T cells are specifically enriched in a unique CD3(dim) CD7(low) subpopulation of CD4(+) T cells in acute-type adult T-cell leukemia. Cancer Sci. 2011;102(3):569–77. doi:10.1111/j.1349-7006.2010.01833.x. PMID:21205081.
- Yamano Y, Cohen CJ, Takenouchi N, Yao K, Tomaru U, Li HC, Reiter Y, Jacobson S. Increased expression of human T lymphocyte virus type I (HTLV-I) Tax11-19 peptide-human histocompatibility leukocyte antigen A*201 complexes on CD4+ CD25+ T Cells detected by peptide-specific, major histocompatibility complex-restricted antibodies in patients with HTLV-I-associated neurologic disease. J Exp Med. 2004;199(10):1367–77. doi:10.1084/jem.20032042. PMID:15136590.
- Tattermusch S, Skinner JA, Chaussabel D, Banchereau J, Berry MP, McNab FW, O'Garra A, Taylor GP, Bangham CR. Systems biology approaches reveal a specific interferon-inducible signature in HTLV-1 associated myelopathy. PLoS Pathog. 2012;8(1):e1002480. doi:10.1371/journal.ppat.1002480. PMID:22291590.
- Dourado I, Alcantara LCJ, Barreto ML, da Gloria Teixeira M, Galvão-Castro B. HTLV-I in the general population of Salvador, Brazil: a city with African ethnic and sociodemographic characteristics. J Acquir Immune Defic Syndr. 2003;34(5):527–31. doi:10.1097/00126334-200312150-00013. PMID:14657765.
- Sun T, Zhou Y, Li H, Han X, Shi Y, Wang L, Miao X, Tan W, Zhao D, Zhang X, et al. FASL -844C polymorphism is associated with increased activation-induced T cell death and risk of cervical cancer. J. Exp. Med. 2005;202(7):967–74. doi:10.1084/jem.20050707. PMID:16186185.
- Paulsen M, Valentin S, Mathew B, Adam-Klages S, Bertsch U, Lavrik I, Krammer PH, Kabelitz D, Janssen O. Modulation of CD4+ T-cell activation by CD95 co-stimulation. Cell Death Differ. 2011;18(4):619–31. doi:10.1038/cdd.2010.134. PMID:21052094.
- Klebanoff CA, Scott CD, Leonardi AJ, Yamamoto TN, Cruz AC, Ouyang C, Ramaswamy M, Roychoudhuri R, Ji Y, Eil RL, et al. Memory T cell-driven differentiation of naive cells impairs adoptive immunotherapy. J Clin Invest. 2016;126(1):318–34. doi:10.1172/JCI81217. PMID:26657860.
- Menezes SM, Leal FE, Dierckx T, Khouri R, Decanine D, Silva-Santos G, Schnitman SV, Kruschewsky R, López G, Alvarez C, et al. A Fas(hi) Lymphoproliferative Phenotype Reveals Non-Apoptotic Fas Signaling in HTLV-1-Associated Neuroinflammation. Front Immunol. 2017;8:97. doi:10.3389/fimmu.2017.00097. PMID:28261198.
- Debatin KM, Goldman CK, Waldmann TA, Krammer PH. APO-1-induced apoptosis of leukemia cells from patients with adult T-cell leukemia. Blood. 1993;81(11):2972–7. PMID:7684622.
- Strauss G, Knape I, Melzner I, Debatin K-M. Constitutive caspase activation and impaired death-inducing signaling complex formation in CD95-resistant, long-term activated, antigen-specific T cells. J. Immunol. 2003;171(3):1172–82. doi:10.4049/jimmunol.171.3.1172. PMID:12874203.
- Krueger A, Fas SC, Giaisi M, Bleumink M, Merling A, Stumpf C, Baumann S, Holtkotte D, Bosch V, Krammer PH, et al. HTLV-1 Tax protects against CD95-mediated apoptosis by induction of the cellular FLICE-inhibitory protein (c-FLIP). Blood. 2006;107(10):3933–9. doi:10.1182/blood-2005-06-2567. PMID:16403915.
- Zane L, Sibon D, Legras C, Lachuer J, Wierinckx A, Mehlen P, Delfau-Larue MH, Gessain A, Gout O, Pinatel C, et al. Clonal expansion of HTLV-1 positive CD8+ cells relies on cIAP-2 but not on c-FLIP expression. Virology. 2010;407(2):341–51. doi:10.1016/j.virol.2010.07.023. PMID:20863547.
- Chen L, Park S-M, Tumanov AV, Hau A, Sawada K, Feig C, Turner JR, Fu YX, Romero IL, Lengyel E, et al. CD95 promotes tumour growth. Nature. 2010;465(7297):492–6. doi:10.1038/nature09075. PMID:20505730.
- Toulza F, Heaps A, Tanaka Y, Taylor GP, Bangham CRM. High frequency of CD4+FoxP3+ cells in HTLV-1 infection: inverse correlation with HTLV-1-specific CTL response. Blood. 2008;111(10):5047–53. doi:10.1182/blood-2007-10-118539. PMID:18094326.
- Sibley K, Rollinson S, Allan JM, Smith AG, Law GR, Roddam PL, Skibola CF, Smith MT, Morgan GJ. Functional FAS promoter polymorphisms are associated with increased risk of acute myeloid leukemia. Cancer Res. 2003;63(15):4327–30. PMID:12907599.
- Sibon D, Gabet A-S, Zandecki M, Pinatel C, Thête J, Delfau-Larue MH, Rabaaoui S, Gessain A, Gout O, Jacobson S, et al. HTLV-1 propels untransformed CD4 lymphocytes into the cell cycle while protecting CD8 cells from death. J Clin Invest. 2006;116(4):974–83. doi:10.1172/JCI27198. PMID:16585963.
- Enose-Akahata Y, Oh U, Grant C, Jacobson S. Retrovirally induced CTL degranulation mediated by IL-15 expression and infection of mononuclear phagocytes in patients with HTLV-I-associated neurologic disease. Blood. 2008;112(6):2400–10. doi:10.1182/blood-2008-02-138529. PMID:18509087.
- Norris PJ, Hirschkorn DF, DeVita DA, Lee TH, Murphy EL. Human T cell leukemia virus type 1 infection drives spontaneous proliferation of natural killer cells. Virulence. 2010;1(1):19–28. doi:10.4161/viru.1.1.9868. PMID:20640055.
