ABSTRACT
Whether PD-L1 expression is associated with survival outcomes in gastric cancer (GC) is controversial. The inhibition of the PD-1/PD-L1 pathway is effective against genomically unstable tumors. Hypothesizing that also the clinical significance of PD-L1 might be dependent on the activation of molecular circuits ensuring genomic stability, we evaluated PD-L1 expression in tissue samples from 72 advanced GC patients treated with first-line chemotherapy. Samples were already characterized for DNA damage repair (DDR) component expression (pATM, pChk1, pWee1, γ-H2AX and pRPA2) along with mutations in DDR-linked genes (TP53 and ARID1A). Overall, PD-L1 expression was not associated with progression-free survival (PFS) and overall survival (OS), independently on whether we considered its expression in tumor cells (PD-L1-TCs) or in the immune infiltrate (PD-L1-TILs). In subgroup analysis, positive PD-L1-TC immunostaining was associated with better PFS in patients whose tumors did not carry DDR activation (multivariate Cox: HR 0.34, 95%CI: 0.15–0.76, p = 0.008). This subset (DDRoff) was characterized by negative pATM expression or the presence of ARID1A mutations. Conversely, the relationship between PD-L1-TC expression and PFS was lost in a molecular scenario denoting DDR activation (DDRon), as defined by concomitant pATM expression and ARID1A wild-type form. Surprisingly, while PD-L1-TC expression was associated with better OS in the DDRoff subset (multivariate Cox: HR 0.41, 95% CI: 0.17–0.96, p = 0.039), in the DDRon subgroup we observed an opposite impact on OS (multivariate Cox: HR 2.56, 95%CI: 1.06–6.16, p = 0.036). Thus, PD-L1-TC expression may impact survival outcomes in GC on the basis of the activation/inactivation of genome-safeguarding pathways.
Introduction
Antibodies directed against the programmed cell death protein-1 (PD-1)/programmed cell death ligand-1 (PD-L1) pathway demonstrated significant efficacy in a variety of malignancies.Citation1-Citation7 Expression of PD-L1, assessed by immunohistochemical assays, is widely used in clinical trials in the attempt of delineating the patient population with greater likelihood of response to PD-1-blocking approaches.Citation8,Citation9 More recently, genetic biomarkers gained popularity, as accumulating evidence conveyed the message that abnormalities leading to log-fold increases in tumor mutational burden predict responsiveness to immune checkpoint blockade.Citation9-Citation11 Consistently, pembrolizumab was found to be active across a range of solid tumors harboring mismatch-repair (MMR) deficiency (microsatellite instability, MSI),Citation10,Citation11 a condition stemming from epigenetic inactivation or germline mutations in the MMR machinery that gives rise to replication infidelity, a hypermutated phenotype and formation of neoantigens.Citation12-Citation16 At the same time, a wave of studies strived to investigate the prognostic significance of PD-L1 expression, independently of the administration of PD-1-targeting agents.Citation17 In gastric cancer (GC), efforts toward understanding whether PD-L1 affects survival outcomes yielded mixed and in some instances opposite findings, as emerging when evaluating results from individual studies included in three recent meta-analyses.Citation18-Citation20 Moreover, available evidence did not point to the advanced setting.
Taking into account the inconsistencies emerging from studies evaluating the relationship between PD-L1 expression and survival outcomes in GC, and considering that the efficacy of immune checkpoint inhibitors is tied to genomic instability, it is plausible to hypothesize that PD-L1 is not a standalone prognostic marker, but it should rather be framed into a broader molecular context mirroring the activation/inactivation status of genome-protecting pathways.
The transmission of an unaltered genetic code to the progeny is ensured by DNA damage repair (DDR) pathways.Citation21 When DNA single- and double-strand breaks (SSBs and DSBs) arise, the DDR network halts the cell cycle and coordinates DNA repair or self-elimination of irreversibly damaged cells.Citation21 Central to this process are two crosstalking signaling avenues: i) ATM-Chk2 (Ataxia-Telangiectasia Mutated-Checkpoint Kinase 2), and ii) ATR-Chk1-Wee1 (Ataxia Telangiectasia and Rad3-related Protein-Checkpoint Kinase 1-Wee1-like Protein Kinase).Citation22-Citation24 This system is often aberrantly activated in cancer cells, being a protective response against a number of extracellular and intracellular cues that threaten the genome.Citation25-Citation27 These include cytotoxic therapies, reactive oxygen species, the replicative stress generated by mutations in oncogenes that control cell proliferation, and mutations in genes that control cell-cycle checkpoints.Citation25-Citation28
On this premise, we herein investigated the connection between PD-L1 expression, DDR pathways and survival outcomes exploiting tissue samples from 72 advanced GC patients treated with first-line chemotherapy in prospective phase II trials or in routine clinical practice.Citation29-Citation32 The present patient population represents a subset of a wider cohort already characterized for the expression levels of DDR kinases (pATM, pChk1 and pWee1) and DNA damage markers, namely the DNA DSB marker phosphorylated H2A Histone Family Member X (γ-H2AX) and the single-stranded DNA/SSB marker phosphorylated replication protein A2 (pRPA2, best known as pRPA32).Citation33 Mutational status of TP53 and ARID1A, two recurrently mutated genes in GC that intersect the DDR at the protein level, was also available.Citation33 Indeed, TP53 mutations hinder correct function of the G1-S checkpoint, thus rendering cancer cells reliant on intact cell cycle control systems to deal with DNA damage.Citation27 The nexus between ARID1A and the DDR machinery was more recently described, in a model envisioning the recruitment of ARID1A to DNA DSBs via ATM/ATR signaling.Citation34 Consistently, ARID1A-deficient cells are characterized by impaired initiation of the G2-M checkpoint and reduced non-homologous end joining activity.Citation34,Citation35 Overall, the present study was designed with the following goals: i) investigating the connection between PD-L1 expression and survival outcomes in advanced GC patients treated with chemotherapy, with a special focus on progression-free survival (PFS), the most direct indicator of efficacy/inefficacy of anticancer treatments, and ii) investigating whether underlying molecular backgrounds mirroring the activation of genome-protecting molecular circuits impact the clinical significance of PD-L1 expression.
Results
Baseline characteristics of the patients and PD-L1 expression pattern
Baseline characteristics of the 72 patients included in the present analysis are summarized in . In this subset of the original cohort, 40 (55.6%) patients received three-drug regimens, taxane-containing chemotherapy was administered to 38 (52.8%) patients, and 36 (50%) patients were treated in prospective phase II trials. PD-L1 expression in tumor cells (PD-L1-TCs) was detected in 41 samples (57%), whereas 30 samples (41.5%) displayed PD-L1 expression in the immune infiltrate (TILs, PD-L1-TILs). Associations between PD-L1 and baseline characteristics of the patients are provided in Supplementary Table 1, whereas associations between PD-L1 and DDR biomarkers are detailed in Supplementary Table 2. Immunohistochemical staining of four representative cases is presented in Supplementary Fig. 1.
Table 1. Baseline characteristics of gastric cancer patients included in this study (N = 72).
The relationship between PD-L1 expression and progression-free survival is dependent on DDR markers
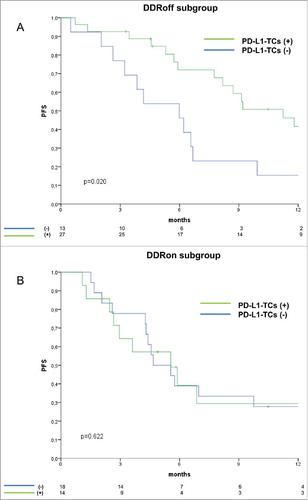
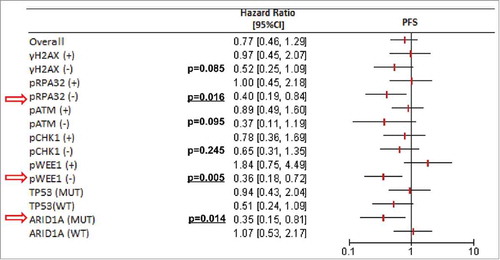
In the entire cohort, PD-L1 expression was neither significantly associated with PFS nor with OS (, panel A-B (left) for PD-L1-TCs and C-D (right) for PD-L1-TILs). Reasoning that the significance of PD-L1 might be dependent on the molecular contexts denoting inactivation/over-activation of genome-protecting mechanisms, we verified whether PD-L1 was associated with PFS in a DDR-dependent way. Thus, univariate Cox regression analyses were carried out by stratifying for DDR marker status (positive vs negative for protein biomarkers, wild-type vs mutated for ARID1A and TP53). While PD-L1-TILs immunostaining pattern was not associated with PFS even when stratifying for DDR marker status (data available upon request), patients with PD-L1-TC-positive tumors had a decreased risk of disease progression exclusively in the subgroups with DDR-negative biomarkers (). A similar pattern was documented in the ARID1A mutated background, whereas TP53 mutational status seemed to be irrelevant (). Prompted by the observation that the protective significance of PD-L1-TCs emerges in DDR-negative tumors, and considering that both ATM and ARID1A participate in the repair of DNA DSBs,Citation34 we then focused on a molecular scenario reflecting this process. On this ground, we separately analyzed two distinct molecular subgroups: i) tumors in which ARID1A wild-type co-existed with the expression of pATM (DDRon). This molecular context plausibly reflects conserved ARID1A/ATM function and then an efficient processing of DNA lesions, and ii) their negative counterparts, defined by the presence of ARID1A mutations or negative nuclear pATM expression (DDRoff). As illustrated in , PD-L1-TC expression was associated with better PFS exclusively in the DDRoff subgroup (log-rank p = 0.020), whereas its protective effect was no longer evident in the subset of patients with DDRon GC (log-rank p = 0.622). Results from uni- and multivariate Cox regression models confirmed that PD-L1-TCs expression was an independent predictor of better PFS only in the DDRoff subset (multivariate Cox: HR 0.34, 95%CI: 0.15–0.76, p = 0.008) (). These findings suggest that PD-L1-TC expression is associated with better PFS only in specific molecular contexts that may denote suboptimal processing of DNA DSBs.
Table 2. Uni- and multivariate Cox regression models for progression-free survival (PFS) performed in the DDRon group (left columns) and in the DDRoff group (right columns).
Figure 1. Kaplan-Meier survival curves of progression-free survival (PFS) and overall survival (OS) comparing PD-L1-TC-positive versus negative cases (panel A and B); and PD-L1-TIL-postive versus negative cases (panel C and D).
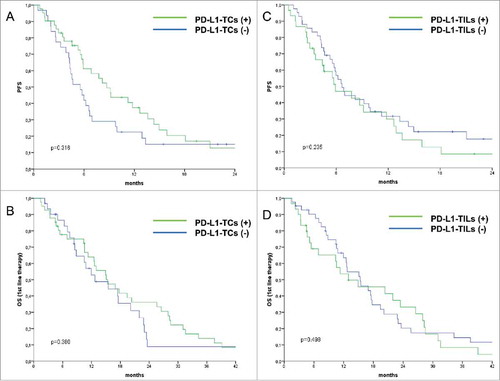
Figure 2. Forest plot illustrating the relationship between PD-L1-TCs expression and progression-free survival (univariate Cox regression analyses) by stratifying for DDR markers (positive versus negative for protein biomarkers, wild-type versus mutated for ARID1A and TP53). Statistically significant analyses are indicated with red arrows.
DDR status is an effect modifier of the relationship between PD-L1 expression and overall survival
We next investigated whether PD-L1-TC expression was associated with overall survival (OS). As expected, the multivariate Cox regression model for OS indicated that PD-L1-TC expression conferred a decreased risk of death in the DDRoff population (HR 0.41, 95%CI: 0.17–0.96, p = 0.039, ). Surprisingly, the multivariate Cox regression models carried out in the subgroup of patients with DDRon tumors showed that PD-L1-TC expression was associated with an increased risk of death (HR 2.56, 95%CI: 1.06–6.16, p = 0.036, ), even though PD-L1 expression did not test statistically significant at the univariate assessment. Given that positive PD-L1-TC immunostaining oppositely impacted OS in an ARID1A- and pATM-dependent manner, DDR activation status may represent an effect modifier of the relationship between PD-L1-TC and OS in patients with advanced GC.
Table 3. Uni- and multivariate Cox regression models for overall survival (OS) performed in the DDRon group (left columns) and in the DDRoff group (right columns).
Discussion
In the present study, we examined the expression pattern of PD-L1 in a relatively large series of advanced GC patients treated with first-line therapy. Collectively, our results suggest that: i) PD-L1 expression in cancer cells is associated with better PFS exclusively in a subset of patients whose tumors did not harbor biomarkers reflecting the activation of genome-protecting mechanisms (DDRoff), ii) while this pattern was retained when evaluating OS, the relationship between PD-L1-TCs and OS was inverted in the DDRon subgroup, configuring a role of effect modifiers for DDR markers, and iii) the assessment of PD-L1 in the immune infiltrate did not yield any additional information. To our knowledge, this is the first report striving to tie PD-L1, pathways that ensure the correction of DNA lesions and survival outcomes in advanced GC patients, and with special emphasis on PFS. We are aware that this study is hypothesis-generating in nature owing to its retrospective design. Another aspect that needs to be mentioned is that, both in this study and in our previous report,Citation33 we did not evaluate pATR expression levels. This choice is rooted in the fact that tissue samples were characterized for both protein biomarkers and genomic alterations. Beyond DDR-related markers, in the same cohort we also evaluated another oncogenic cooperation, namely the Hippo-Wnt pathway crosstalk.Citation36 Given this fairly wide characterization, we preferred to evaluate other effectors, such as pChk1 and pWee1, which act downstream of pATR and whose pharmacological inhibitors are at a more advanced stage of clinical development, as highlighted by the recent dissemination of final results from phase II trials.Citation37,Citation38 It is also worth mentioning that the model exploited for testing the significance of PD-L1 expression, namely combined pATM expression and ARID1A mutations, beyond being biology-driven, applies to all clinical endpoints (PFS and OS). Indeed, PD-L1 expression did not achieve full statistical significance for both PFS and OS when stratifying by individual DDR biomarkers (PFS data are reported in , OS data are available upon request). Our belief is that this is attributable to the size of analyzed subgroups, that enabled us to exclusively uncover the strongest association. Consistently, in a prospective study with biomarker validation purposes that we have recently initiated and that is mentioned below, we considered an expanded set of DDR- and immune-related markers.
On the other hand, this study has some important strengths, such as that half of the patients were treated in prospective phase II trials.Citation29-Citation32 Moreover, we did not record any marked differences in the distribution of PD-L1 expression when comparing DDR marker positive vs negative tumors (supplementary Table 2). This observation, in association with the results from multivariate Cox models, allows us to exclude potential confounding factors.
From a molecular perspective, the characterization of GC carried out by the Cancer Genome Atlas Research (TCGA) Network assists us in framing these results.Citation39 Indeed, ARID1A mutations were significantly more frequently observed in hypermutated GC as compared with non-hypermutated tumors (44% vs 14%).Citation39 This suggests that ARID1A mutations may be a genomic trait characterizing hypermutated GC, thus enforcing our hypothesis that PD-L1-TCs is a protective factor exclusively in a molecular scenario characterized by defective activation of the DDR machinery, replication infidelity and accumulation of DNA lesions. More recently, Sato and collegues described a model envisioning an increase of PD-L1 expression in cancer cells upon DNA DSBs, in a process that requires ATM/ATR/Chk1 activity.Citation40
When interpreting OS data, as already anticipated some studies reported that patients with PD-L1-positive GC had improved prognosis, whereas other reports did not reveal any association between PD-L1 and OS, or conveyed the opposite message.Citation18-Citation20,Citation41-Citation44 The multivariate Cox regression models for OS pointed to PD-L1-TC as a “Janus-faced” marker, having the potential to be either protective or detrimental depending upon inactivation or activation of DDR pathways, respectively. Thus, our findings might reconcile currently available evidence regarding the prognostic significance of PD-L1 expression in GC. In turn, unfolding the connection between PD-L1 and pathways that protect cancer cells against chemotherapy-induced death stimuli holds two therapeutic implications: i) the design of biomarker-driven interventional trials, envisioning the administration of chemotherapy or PD-1-blocking agents on the basis of the individual molecular portrait, and ii) the possibility of combining immune checkpoint blockade with agents targeting central DDR pathway components, such as ATM, ATR, Chk1 and Wee1, which are currently undergoing clinical development.Citation45 Indeed, the pharmacological abrogation of cell cycle checkpoints/DNA damage repair effectors might increase the mutational load and neoantigen burden, thus enhancing the efficacy of immune-directed therapies. Proof-of-concept clinical trials combining immune-directed therapies and DNA repair-targeting agents (e.g. PARP and ATR inhibitors) are ongoing and will provide evidence on whether such therapeutic combinations fulfill the efficacy criteria to be investigated in larger, randomized trials.Citation46
The strategy we are pursuing to achieve an efficient translation of this knowledge into the clinical setting deserves a final mention. Indeed, we are striving to prospectively validate a previously identified DDR-linked signature associated with adverse survival outcomes in advanced GC patients.Citation33 This trial also envisions the assessment of immune-related biomarkers along with an extensive genetic characterization. This latter encompasses the evaluation of a number of alterations, including both mutations and copy number variations, fuelling basal activation of the DDR machinery or denoting an inefficient processing of DNA lesions. In particular, we are focusing on alterations involved in deregulated G1-S transition (e.g. TP53 and CCNE1), oncogene-induced replication stress (e.g. MYC and PTEN), and ATM/ATR-initiated DNA repair (e.g. ATM and ARID1A). Results from this study will provide further knowledge on how two intertwined phenomena, namely the anticancer immune response and DNA repair deficiency, impact survival outcomes in advanced GC.
In conclusion, our data point to PD-L1-TC expression as a potential biomarker of better PFS in advanced GC patients treated with chemotherapy in the first-line setting. This link relies on the activation of genome-protecting pathways, being exclusively observed when tumors did not carry biomarkers denoting activation of these mechanisms. Conversely, PD-L1-TC expression apparently influences OS in a more complex, DDR-dependent, way.
Patients and methods
For this analysis, 72 patients with histologically confirmed, inoperable locally advanced or metastatic GC or esophagogastric junction (EOJ) cancer who received first-line chemotherapy (August 2001-August 2014) were included. Median follow-up was 12 months (IQR 5–23 months). Patients were considered eligible on the following basis: i) complete data on clinical features and treatment outcomes, ii) complete data on PD-L1 immunostaining, and iii) complete data on DDR biomarkers, including the expression of DDR kinases/DNA damage markers and mutations in the TP53 and ARID1A genes. Chemotherapy regimens and schedules were already detailed elsewhere.Citation33 Tumor responses were evaluated by Response Evaluation Criteria in Solid Tumors (RECIST) criteria v.1.1. PFS was calculated as the time between the first cycle of chemotherapy until radiological evidence of disease progression or death due to any cause. OS was computed as the time from the first cycle of chemotherapy to death due to any cause. Written informed consents were obtained from all the participants. The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the “Regina Elena” National Cancer Institute of Rome.
Study procedures
The immunohistochemical assessment of PD-L1 was performed in formalin-fixed paraffin-embedded (FFPE) tissues with the monoclonal mouse anti-PD-L1 antibody clone 22C3 (1:20). PD-L1 expression was evaluated both in tumor cells and in the immune infiltrate (identified by morphological assessment), and considered positive when expressed in >1% of the respective cellular compartment. A detailed description of reagents and procedures used for evaluating DDR markers (proteins and genes) was already provided.Citation33 Shortly, DNA damage markers (γ-H2AX and pRPA32) were classified as positive/negative using the median score of all tumors as the cut-off points, whereas DDR kinases were considered positive when ≥20% of the neoplastic cells showed a distinct nuclear immunoreactivity of any intensity. Targeted DNA deep sequencing was carried out on a NextSeq 500 (Illumina, Inc., San Diego, CA, USA). TP53 and ARID1A mutations were filtered on the basis of variant allele frequencies (≥10%), and considering the established or predicted oncogenicity of the detected mutations.Citation33
Statistical analysis
Descriptive statistics were computed for all the variables of interest. The relationship between categorical variables was investigated with the Pearson's Chi-squared test of independence (2-tailed) or the Fisher Exact test, depending upon the size of the groups compared. Survival curves were estimated with the Kaplan-Meier product-limit method and compared by log-rank test. Variables potentially affecting PFS and OS were tested in univariate Cox proportional hazard models. Multivariate Cox models were built by stepwise regression through backward elimination. The related estimates were reported as hazard ratio (HR) and 95% confidence interval (CI). Level of significance was defined as p < 0.05. Statistical analyses were carried out using SPSS version 21.0 (SPSS Inc., Chicago, Illinois, USA).
List of abbreviations
| ATM | = | ataxia-telangiectasia mutated |
| ATR | = | ataxia telangiectasia and Rad3-related protein |
| Chk1 | = | checkpoint kinase 1 |
| Chk2 | = | checkpoint kinase 2 |
| DDR | = | DNA damage and repair |
| DSBs | = | double-strand breaks |
| GC | = | gastric cancer |
| MMR | = | mismatch-repair |
| OS | = | overall survival |
| γ-H2AX | = | phosphorylated H2A Histone Family Member X |
| PFS | = | progression-free survival |
| PD-1 | = | programmed cell death protein-1 |
| PD-L1 | = | programmed cell death ligand-1 |
| PD-L1-TCs | = | PD-L1 expression in tumor cells |
| PD-L1-TILs | = | PD-L1 expression in tumor-infiltrating lymphocytes |
| pRPA32 | = | phosphorylated replication protein A2 |
| SSBs | = | single- strand breaks |
| TILs | = | tumor-infiltrating lymphocytes |
| Wee1 | = | wee1-like protein kinase. |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
supp_data.zip
Download Zip (4.9 MB)Acknowledgments
We thank Tania Merlino for technical assistance.
Additional information
Funding
References
- Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, et al. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372:2521–32. doi:10.1056/NEJMoa1503093. PMID:25891173.
- Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C, Kalinka-Warzocha E, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372:320–30. doi:10.1056/NEJMoa1412082. PMID:25399552.
- Wolchok JD, Chiarion-Sileni V, Gonzalez R, Rutkowski P, Grob JJ, Cowey CL, Lao CD, Wagstaff J, Schadendorf D, Ferrucci PF, et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2017;377:1345–56. doi:10.1056/NEJMoa1709684. PMID:28889792.
- Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe S, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375:1823–33. doi:10.1056/NEJMoa1606774. PMID:27718847.
- Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373:123–35. doi:10.1056/NEJMoa1504627. PMID:26028407.
- Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373:1627–39. doi:10.1056/NEJMoa1507643. PMID:26412456.
- Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G, Plimack ER, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373:1803–13. doi:10.1056/NEJMoa1510665. PMID:26406148.
- Maleki Vareki S, Garrigós C, Duran I. Biomarkers of response to PD-1/PD-L1 inhibition. Crit Rev Oncol Hematol. 2017;116:116–24. doi:10.1016/j.critrevonc.2017.06.001. PMID:28693793.
- Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275–87. doi:10.1038/nrc.2016.36. PMID:27079802.
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–13. doi:10.1126/science.aan6733. PMID:28596308.
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–20. doi:10.1056/NEJMoa1500596. PMID:26028255.
- Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–661. doi:10.1038/363558a0. PMID:8505985.
- Blake C, Tsao JL, Wu A, Shibata D. Stepwise deletions of polyA sequences in mismatch repair-deficient colorectal cancers. Am J Pathol. 2001;158:1867–70. doi:10.1016/S0002-9440(10)64143-0. PMID:11337385.
- Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–9. doi:10.1126/science.8484122. PMID:8484122.
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi:10.1038/nature11252. PMID:22810696.
- Germano G, Lamba S, Rospo G, Barault L, Magrì A, Maione F, Russo M, Crisafulli G, Bartolini A, Lerda G, et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature. 2017;552:116–20. PMID:29186113.
- Wang Q, Liu F, Liu L. Prognostic significance of PD-L1 in solid tumor: An updated meta-analysis. Medicine (Baltimore). 2017;96:e6369. doi:10.1097/MD.0000000000006369. PMID:28471952.
- Liu YX, Wang XS, Wang YF, Hu XC, Yan JQ, Zhang YL, Wang W, Yang RJ, Feng YY, Gao SG, et al. Prognostic significance of PD-L1 expression in patients with gastric cancer in East Asia: a meta-analysis. Onco Targets Ther. 2016;9:2649–54. PMID:27226727.
- Zhang M, Dong Y, Liu H, Wang Y, Zhao S, Xuan Q, Wang Y, Zhang Q. The clinicopathological and prognostic significance of PD-L1 expression in gastric cancer: a meta-analysis of 10 studies with 1,901 patients. Sci Rep. 2016;6:37933. doi:10.1038/srep37933. PMID:27892511.
- Gu L, Chen M, Guo D, Zhu H, Zhang W, Pan J, Zhong X, Li X, Qian H, Wang X. PD-L1 and gastric cancer prognosis: A systematic review and meta-analysis. PLoS One. 2017;12:e0182692. doi:10.1371/journal.pone.0182692. PMID:28796808.
- Jeggo PA, Pearl LH, Carr AM. DNA repair, genome stability and cancer: a historical perspective. Nat Rev Cancer. 2016;16:35–42. doi:10.1038/nrc.2015.4. PMID:26667849.
- Weber AM, Ryan AJ. ATM and ATR as therapeutic targets in cancer. Pharmacol Ther. 2015;149:124–38. doi:10.1016/j.pharmthera.2014.12.001. PMID:25512053.
- Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–9. doi:10.1016/S1535-6108(03)00110-7. PMID:12781359.
- Vriend LE, De Witt Hamer PC, Van Noorden CJ, Würdinger T. WEE1 inhibition and genomic instability in cancer. Biochim Biophys Acta. 2013;1836:227–35. PMID:23727417.
- Scott TL, Rangaswamy S, Wicker CA, Izumi T. Repair of oxidative DNA damage and cancer: recent progress in DNA base excision repair. Antioxid Redox Signal. 2014;20:708–26. doi:10.1089/ars.2013.5529. PMID:23901781.
- Gaillard H, García-Muse T, Aguilera A. Replication stress and cancer. Nat Rev Cancer. 2015;15:276–89. doi:10.1038/nrc3916. PMID:25907220.
- Dixon H, Norbury CJ. Therapeutic exploitation of checkpoint defects in cancer cells lacking p53 function. Cell Cycle. 2002;1:362–8. doi:10.4161/cc.1.6.257. PMID:12548006.
- Maugeri-Saccà M, Bartucci M, De Maria R. Checkpoint kinase 1 inhibitors for potentiating systemic anticancer therapy. Cancer Treat Rev. 2013;39:525–33. doi:10.1016/j.ctrv.2012.10.007. PMID:23207059.
- Di Lauro L, Vici P, Belli F, Tomao S, Fattoruso SI, Arena MG, Pizzuti L, Giannarelli D, Paoletti G, Barba M, et al. Docetaxel, oxaliplatin, and capecitabine combination chemotherapy for metastatic gastric cancer. Gastric Cancer. 2014;17:718–24. doi:10.1007/s10120-013-0321-3. PMID:24318671.
- Di Lauro L, Giacinti L, Arena MG, Sergi D, Fattoruso SI, Giannarelli D, Lopez M. Phase II study of epirubicin, oxaliplatin and docetaxel combination in metastatic gastric or gastroesophageal junction adenocarcinoma. J Exp Clin Cancer Res. 2009;28:34. doi:10.1186/1756-9966-28-34. PMID:19267943.
- Di Lauro L, Nunziata C, Arena MG, Foggi P, Sperduti I, Lopez M. Irinotecan, docetaxel and oxaliplatin combination in metastatic gastric or gastroesophageal junction adenocarcinoma. Br J Cancer. 2007;97:593–7. doi:10.1038/sj.bjc.6603917. PMID:17667920.
- Di Lauro L, Belli F, Arena MG, Carpano S, Paoletti G, Giannarelli D, Lopez M. Epirubicin, cisplatin and docetaxel combination therapy for metastatic gastric cancer. Ann Oncol. 2005;16:1498–502. doi:10.1093/annonc/mdi281. PMID:15956036.
- Ronchetti L, Melucci E, De Nicola F, Goeman F, Casini B, Sperati F, Pallocca M, Terrenato I, Pizzuti L, Vici P, et al. DNA damage repair and survival outcomes in advanced gastric cancer patients treated with first-line chemotherapy. Int J Cancer. 2017;140:2587–95. doi:10.1002/ijc.30668. PMID:28233295.
- Shen J, Peng Y, Wei L, Zhang W, Yang L, Lan L, Kapoor P, Ju Z, Mo Q, Shih IeM, et al. ARID1 A deficiency impairs the DNA Damage checkpoint and sensitizes cells to PARP inhibitors. Cancer Discov. 2015;5:752–67. doi:10.1158/2159-8290.CD-14-0849. PMID:26069190.
- Watanabe R, Ui A, Kanno S, Ogiwara H, Nagase T, Kohno T, Yasui A. SWI/SNF factors required for cellular resistance to DNA damage include ARID1A and ARID1B and show interdependent protein stability. Cancer Res. 2014;74:2465–75. doi:10.1158/0008-5472.CAN-13-3608. PMID:24788099.
- Melucci E, Casini B, Ronchetti L, Pizzuti L, Sperati F, Pallocca M, De Nicola F, Goeman F, Gallo E, Amoreo CA, et al. Expression of the Hippo transducer TAZ in association with WNT pathway mutations impacts survival outcomes in advanced gastric cancer patients treated with first-line chemotherapy. J Transl Med. 2018;16:22. doi:10.1186/s12967-018-1385-y. PMID:29402328.
- Leijen S, van Geel RM, Sonke GS, de Jong D, Rosenberg EH, Marchetti S, et al. Phase II study of WEE1 inhibitor AZD1775 plus carboplatin in patients with TP53-mutated ovarian cancer refractory or resistant to first-line therapy within 3 months. J Clin Oncol. 2016;34:4354–61. doi:10.1200/JCO.2016.67.5942. PMID:27998224.
- Wehler T, Thomas M, Schumann C, Bosch-Barrera J, Viñolas Segarra N, Dickgreber NJ, Dalhoff K, Sebastian M, Corral Jaime J, Alonso M, et al. A randomized, phase 2 evaluation of the CHK1 inhibitor, LY2603618, administered in combination with pemetrexed and cisplatin in patients with advanced nonsquamous non-small cell lung cancer. Lung Cancer. 2017;108:212–6. doi:10.1016/j.lungcan.2017.03.001. PMID:28625637.
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. doi:10.1038/nature13480. PMID:25079317.
- Sato H, Niimi A, Yasuhara T, Permata TBM, Hagiwara Y, Isono M, Nuryadi E, Sekine R, Oike T, Kakoti S, et al. DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat Commun. 2017;8:1751. doi:10.1038/s41467-017-01883-9. PMID:29170499.
- Böger C, Behrens HM, Mathiak M, Krüger S, Kalthoff H, Röcken C. PD-L1 is an independent prognostic predictor in gastric cancer of Western patients. Oncotarget. 2016;7:24269–83. doi:10.18632/oncotarget.8169. PMID:27009855.
- Kim JW, Nam KH, Ahn SH, Park DJ, Kim HH, Kim SH, Chang H, Lee JO, Kim YJ, Lee HS, et al. Prognostic implications of immunosuppressive protein expression in tumors as well as immune cell infiltration within the tumor microenvironment in gastric cancer. Gastric Cancer. 2016;19:42–52. doi:10.1007/s10120-014-0440-5. PMID:25424150.
- Xing X, Guo J, Wen X, Ding G, Li B, Dong B, Feng Q, Li S, Zhang J, Cheng X, et al. Analysis of PD1, PDL1, PDL2 expression and T cells infiltration in 1014 gastric cancer patients. Oncoimmunology. 2017;7:e1356144. doi:10.1080/2162402X.2017.1356144. PMID:29399387.
- Tamura T, Ohira M, Tanaka H, Muguruma K, Toyokawa T, Kubo N, Sakurai K, Amano R, Kimura K, Shibutani M, et al. Programmed Death-1 Ligand-1 (PDL1) Expression Is Associated with the Prognosis of Patients with Stage II/III Gastric Cancer. Anticancer Res. 2015;35:5369–76. PMID:26408698.
- Manic G, Obrist F, Sistigu A, Vitale I. Trial Watch: Targeting ATM-CHK2 and ATR-CHK1 pathways for anticancer therapy. Mol Cell Oncol. 2015;2:e1012976. doi:10.1080/23723556.2015.1012976. PMID:27308506.
- Mouw KW, Goldberg MS, Konstantinopoulos PA, D'Andrea AD. DNA Damage and Repair Biomarkers of Immunotherapy Response. Cancer Discov. 2017;7:675–93. doi:10.1158/2159-8290.CD-17-0226. PMID:28630051.