ABSTRACT
Background: B7-H3 is an immune checkpoint member that belongs to B7-CD28 families and plays a vital role in the inhibition of T-cell function. Importantly, B7-H3 is widely overexpressed on solid tumors, making it become an attractive target for cancer immunotherapy. To clarify the expression panel of B7-H3 in glioma, we explored the clinical and immune features of B7-H3 expression in a large-scale study. Methods and patients: Totally, 1323 glioma samples from Chinese Glioma Genome Atlas (CGGA) dataset, including 325 RNAseq data and 301 mRNA microarray data, and The Cancer Genome Atlas (TCGA) dataset, including 697 RNAseq data, were gathered into our research. The statistical analysis and graphical work were mainly realized by R language. Results: B7-H3 expression was found positively correlated with the grade of malignancy, which might be caused by hypomethylation. The expression level of B7-H3 was consistently up-regulated in IDH wild-type glioma and highly enriched in mesenchymal subtype. GSEA analysis suggested that B7-H3 related genes were more involved in immune response and angiogenesis in glioma. Moreover, B7-H3 showed a consistent positive relationship with stromal and immune cell populations. Further analysis confirmed that B7-H3 played an important role in T-cell-mediated immunity, especially in T-cell-mediated immune response to tumor cell. Circos plots revealed that B7-H3 was tightly associated with most B7 members and other immune checkpoints. Univariate and multivariate cox analysis demonstrated that B7-H3 was an independent prognosticator for glioma patients. Conclusion: B7-H3 represents the malignant phenotype of glioma and independently predicted worse prognosis in glioma patients. Moreover, B7-H3 collaborating with other checkpoint members may contribute to the dysfunctional phenotype of T cell. These findings will be helpful for further optimizing immunotherapies for glioma.
Introduction
Glioma is the most prevalent and devastating primary central nervous system (CNS) tumor, representing 50% of all brain tumors.Citation1 Despite developments in current standard treatment for glioma patients, including neurosurgical resection, radiotherapy, adjuvant chemotherapy and targeted therapy, glioblastoma, the most malignant and aggressive form of glioma, remains a leading cause of death in human cancer, with an overall 5-year survival rate of only 9.8%.Citation2 Considering the poor outcome after standard treatment, new therapeutic approaches are desperately needed, and attention has been increasingly shifted toward immunotherapeutic strategies as a potential promising treatment approach.Citation3
Immune checkpoint inhibitors targeting the B7-CD28 family members, CTLA-4/B7-1/B7-2 and PD-1/PD-L1 have been clinically successful and revolutionized the treatment of a number of advanced cancers, including melanoma and non-small-cell lung cancer.Citation4, Citation5 This great success arouses our increasing interest in whether these immune checkpoints could also be employed in the treatment of glioma. Herein, we performed an explorative analysis based on the expression level of B7 and CD28 family members in glioma, and found that B7-H3 was higher than any other members, reminding us that targeting B7-H3 may have a comparative advantage in glioma.
B7-H3, also known as CD276, is an immune checkpoint molecule belonging to the B7-CD28 pathways.Citation6 Amounts of literature suggest that B7-H3 plays a vital role in the inhibition of T-cell-mediated adaptive immunity, although its receptor(s) has not been discovered.Citation7 Importantly, wide expression of B7-H3 in various human malignancies have been reported, including melanoma, non-small cell lung carcinoma, pancreatic adenocarcinoma, renal cell carcinoma, gastrointestinal stromal tumors, breast invasive carcinoma, etc.,Citation6, Citation8–10 while limited expression is seen on normal tissues. In addition, studies have found that increased B7-H3 expression shows a direct correlation with shortened overall survival (OS).Citation11, Citation12 With a preferential expression on tumor cells and significant clinical significance, B7-H3 becomes an attractive target for cancer immunotherapy.
To investigate the whole expression state of B7-H3 in gloma, we took advantage of Chinese Glioma Genome Atlas (CGGA) dataset, including 325 RNAseq data and 301 mRNA microarray data of whole grade glioma. To further validate what we have revealed in CGGA dataset, 697 RNAseq data of glioma from The Cancer Genome Atlas (TCGA) network were obtained. Simultaneously, protein level of B7-H3 in glioma was separated from the Human Protein Atlas (HPA). This is the first and largest general study characterizing the clinical, molecular and immunological characteristics of B7-H3 expression in whole grade glioma. A better understanding of the B7-H3 state and feature in glioma will surely help to further optimize associated cancer immunotherapies.
Results
B7-H3 was widely expressed in human glioma tissue specimens and correlated with the grade of malignancy
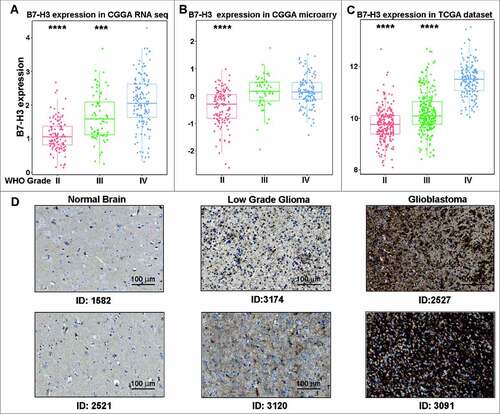
To clarify the landscape of B7 (CD80, CD86, PD-L1, PD-L2, ICOSLG, B7-H3, VTCN1 and HHLA2) and CD28 (CD28, CTLA4, ICOS, PD-1 and TMIGD2) family member Citation13 expression in glioma, the mRNA level of these genes was analyzed in TCGA database. Results showed that B7-H3had a higher mRNA level than any other members (Fig. S1). To further find out B7-H3 expression panel in glioma, 1323 samples from CGGA and TCGA database were analyzed. In CGGA RNA-seq and microarray database, B7-H3 mRNA expression level was the highest in glioblastoma (GBM, WHO grade IV) and showed extremely higher than other grades (A and B). Further analysis of B7-H3 expression in TCGA database showed the same expression pattern (C), indicating that B7-H3 expression was correlated with the grade of malignancy of different gliomas in line with other tumors.Citation6 Simultaneously, protein expression pattern of B7-H3 in glioma was separated from HPA, which showed particularly stronger expression pattern in glioma samples, especially in glioblastoma (D). DNA demethylation is considered to be one of the reasons for some immune checkpoints being up-regulated in tumor sites.Citation14 Consequently, we explored methylation pattern of B7-H3 in whole grade glioma and found that B7-H3 gene promoter was significantly hypomethylated in glioblastoma in both CGGA and TCGA datasets (Fig. S2). These findings indicate that hypomethylation may partly account for the ectopic expression of B7-H3 in glioma.
B7-H3 expression was consistently up-regulated in IDH wild-type glioma
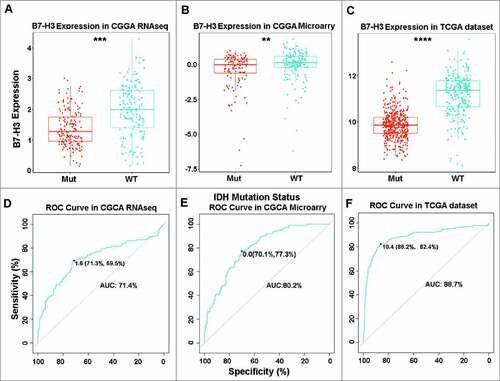
Mutations of the isocitrate dehydrogenase (IDH) enzyme is the most common mutations in 40% glioma and play a crucial role in glioma progression.Citation15 Hence, we focused on the expression pattern of B7-H3 in different IDH states. It turned out that B7-H3 was significantly up-regulated in IDH wild-type glioma in CGGA RNA-seq and microarray datasets (A and B), as well as in TCGA cohort (C). Subsequently, ROC analysis was performed to assess the diagnostic value of B7-H3 as biomarker detecting IDH wild-type glioma. The area under the curves (AUC) of B7-H3 assay in IDH wild-type and IDH mutant-type groups from CGGA RNA-seq and microarray data sets were 71.4% and 80.2%, respectively (D and E). Similarly, the AUC in TCGA cohort was 88.7% (F). Collectively, our results suggest that B7-H3 is selectively distributed and may serve as a biomarker for IDH wild-type glioma.
B7-H3 was a potential biomarker for mesenchymal molecular subtype glioma
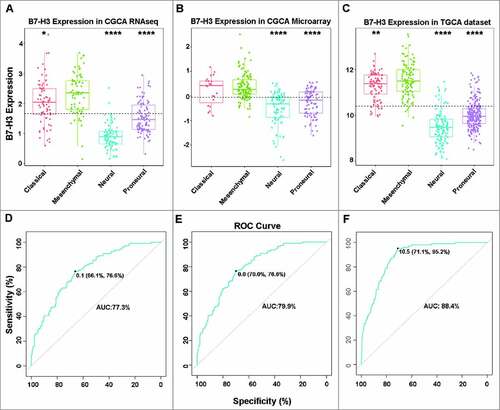
The expression levels of B7-H3 in different glioma subtypes were further analyzed. The results indicated that B7-H3 expression levels in the four subtypes were quite different in both CGGA and TCGA datasets. Compared with three other subtypes, mesenchymal subtype showed a concordant strong expression (A, B and C). To further confirm this result, ROC analysis was used to explore the potential diagnostic value of B7-H3 expression in mesenchymal subtype glioma. As expected, the AUCs of B7-H3 in CGGA RNA-seq, microarray data and TCGA RNA-seq datasets were 77.3%, 79.9% and 88.4%, respectively (D, E and F). These results suggest that B7-H3 may act as a potential biomarker for mesenchymal.
Figure 3. B7-H3 showed a strong expression pattern in mesenchymal molecular subtype glioma. (A, B, C) B7-H3 was highly enriched in mesenchymal molecular subtype glioma. (D, E, F) B7-H3 could serve as a biomarker to predict mesenchymal molecular subtype glioma. #, ## and #### represent p < 0.05, p < 0.01 and p < 0.0001, respectively
B7-H3 was closely associated with immune related biological process and angiogenesis in glioma
Since B7-H3 expression in glioma was strongly associated with malignancy, we inferred that B7-H3 may have important biologic functions in glioma. To find out the biologic role of B7-H3 in glioma, GSEA analysis was used.Citation16 First, genes strongly correlated with B7-H3 expression were selected (Top 600 genes ranked by Pearson |R|) from CGGA RNA-seq and TCGA RNA-seq datasets, respectively. After GSEA analysis, we found that genes positively correlated with B7-H3 expression were more involved in immune system process, immune response and angiogenesis in both CGGA and TCGA cohorts (Figs. S3A and B). In line with previous studies,Citation17 these results remind us that B7-H3 may broadly overexpress not only on cancer cells but also on tumor-infiltrating blood vessels, facilitating the immunosuppression and angiogenesis in glioma.
B7-H3 related immune signatures in glioma
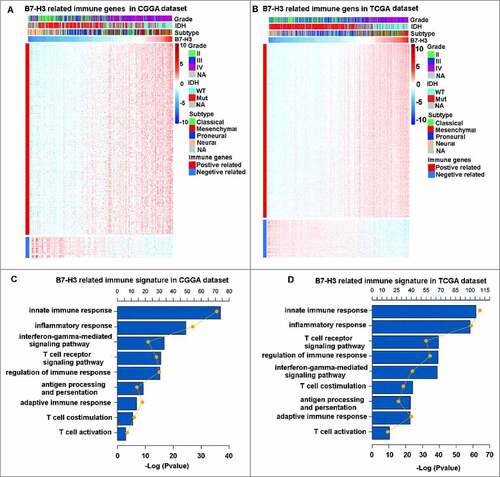
To identify the B7-H3 associated immune signature in glioma, genesets related to the immune response (http://amigo.geneontology.org/amigo/landing) were selected. The optimal related genes to B7-H3 (Pearson |R|>0.4) were identified, which contains 466 and 748 genes in CGGA RNA-seq and TCGA RNA-seq datasets, respectively (A and B). GO analysis in DAVID Bioinformatics Resources 6.8 was used to clarify the biofunctions of these genes. Results showed that genes positively related to B7-H3 were significantly enriched in innate immune response, inflammatory response, T cell costimulation and T cell activation both in CGGA and TCGA cohorts (C and D). These results suggest that B7-H3 plays a crucial role in immune functions in glioma, especially in T cell mediated responses.
Figure 4. B7-H3 related immune genes and signatures in glioma. (A, B) B7-H3 showed a strong positive correlation with most immune genes. (C, D) B7-H3 was closely related to immune response in glioma, especially in T cell mediated immunity. The yellow plots mean the number of genes for each column and the lines mean the tendency of changes in gene number
B7-H3 expression was tightly associated with immune cell infiltration and special T cell mediated immunity in glioma
To fully understand the relationship between B7-H3 expression with immune cell infiltration, ESTIMATE algorithm analyze was used.Citation18 In CGGA RNA-seq and TCGA RNA-seq datasets, B7-H3 showed a consistent significant positive relationship with immune score (A and B, upper panel), which further suggested the important role of B7-H3 in immune response. As the most important immune cells in glioma microenvironment, macrophage plays a crucial role in the evading immune destruction.Citation19 Therefore, the expression level of B7-H3 in macrophage was analyzed. Interestingly, compared with healthy donors and GBM patients’ peripheral monocytes, we firstly found that B7-H3 was specifically expressed in GBM-infiltrating macrophages (Fig. S4). Meanwhile, B7-H3 also showed a strong relationship with stromal cell infiltration, further reminding us that B7-H3 may widely expressed on the integral part of stromal cells, such as endothelial cells in glioma.
Figure 5. B7-H3 was tightly associated with immune score and T cell specific immunity. (A and B, upper panel) B7-H3 was positive related with immune score and stromal score. (A and B, lower panel) B7-H3 was closely related to T cell specific immunity
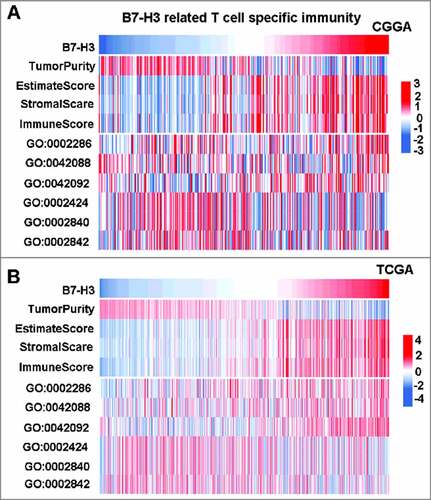
At the same time, relationship between B7-H3 expression and T cell mediated immunity was explored. T cell related special biological process genesets were selected from AmiGO 2 Web portal. Then GSVA analysis Citation20 was performed to evaluate the relationship. Results showed that, in both CGGA and TCGA cohorts, B7-H3 was positively correlated with T cell activation involved in immune response (GO:0002286) and negatively correlated with T cell mediated immune response to tumor cell (GO:0002424) ( A and B, lower panel), indicating that B7-H3 acts as an immune suppressor when T cells are activated in glioma.
Correlation between B7-H3 and immune checkpoints
The B7 family is a big family including many immune checkpoints.Citation13 Hence, the relationship between B7-H3 and B7 family checkpoints were first analyzed. B7-H3 was tightly associated with CD80, CD86, PD-L1 and PD-L2 both in CGGA microarray and TCGA RNA-seq datasets (A and B), indicating that synergistic effects of these markers.
Figure 6. Relationship between B7-H3 and other immune checkpoints. (A, B) Correlation of B7-H3 and other B7 family members. (C, D) Correlation of B7-H3 and other immune checkpoints
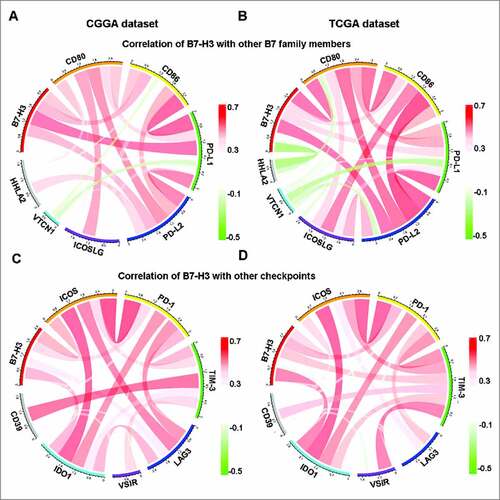
Numerous immune checkpoint members have been emerged as
therapeutic targets in clinical application or clinical trial.Citation21 Therefore, the relationship between B7-H3 and other immune checkpoints, including ICOS, PD-1, TIM-3, LAG3, VISR, IDO1 and CD39,Citation22-25 were analyzed. Surprisingly, B7-H3 showed strong positive correlations with ICOS, PD-1, TIM-3, LAG3 and IDO1 (C and D), indicating that multi-checkpoint blockade is a promising direction in glioma treatment.
Prognostic impact of B7-H3 in glioma
As B7-H3 showed significant relationship with malignancy and immune suppressor in glioma, we additionally evaluated the prognostic impact of B7-H3 expression in whole grade glioma. Log-rank (Mantel-Cox) analysis of survival times revealed that patients with higher B7-H3 expression in their tumors had a significantly shorter survival than patients in the B7-H3 lower group, both in CGGA and TCGA dataset (A, B and C). Next, the potential association between expression level of B7-H3 and survival time of GBM patients were studied. Interestingly, we found the same prognostic significance of B7-H3 in GBM patients in CGGA and TCGA dataset (D, E and F). Importantly, univariate and multivariate cox analysis based on CGGA and TCGA dataset demonstrated that B7-H3 is an independent prognosticator for glioma patients ().
Figure 7. Survival analysis of B7-H3 in glioma. (A, B and C) Survival analysis of B7-H3 in whole grade glioma. (D, E and F) Survival analysis of B7-H3 in glioblastoma
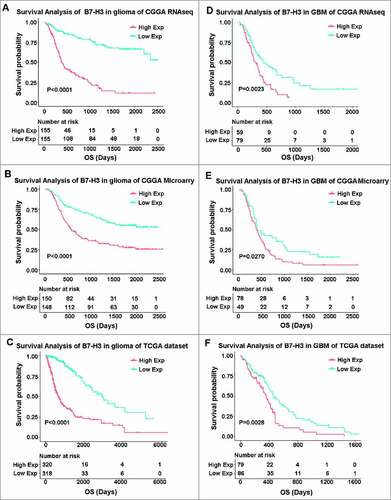
Table 1. Univariate and multivariate regression analyses for predicting overall survival in CGGA and TCGA datasets
Discussion
For decades, despite many efforts have been made to decease the mortality, glioma is still the most common and lethal type of tumor in the CNS. Hence, exploring new therapeutic approaches to prolong survival of glioma patients are burning problems needed to be settled. Emerging evidence has point out that, one of the most important signatures in glioma microenvironment is its ability to suppress the immune system resulting in an impaired recognition and attack of the tumor cells by the immune system, indicating that immunotherapy may be the revolutionary approach to suppress tumor progression.Citation26 Recently, remarkable progress has been made in the clinical trial of immune checkpoint inhibitors targeting B7-CD28 family members in glioma, lighting up the bright future of glioma treatment.Citation27, Citation28
B7-H3 is a member of the B7 family and shows great potential for cancer immunotherapy. More importantly, compared to other B7 and CD28 family members in glioma, B7-H3 showed a significant higher expression, indicating that targeting B7-H3 may have a better affinity. Aberrant expression of B7-H3 in various human malignancies has been reported, but the expression detail of B7-H3 in glioma remains unclear.Citation29, Citation30 Therefore, the expression profile of B7-H3 in glioma needs to be explored in a large-scale study. Here, to clarify the expression panel of B7-H3 in whole grade glioma, we firstly carried out the largest and most comprehensive research characterizing the expression pattern and distribution of B7-H3 in 1323 glioma samples by using CGGA and TCGA datasets. We found that B7-H3 expression was up-regulated in GBM both in mRNA and protein levels. To date, there is no report concerning the relationship between methylation and B7-H3 expression, and we firstly speculated that the abnormal expression of B7-H3 expression in GBM may be partially caused by demethylation. Simultaneously, B7-H3 expression in glioma was found enriched in mesenchymal subtype, which suggest that B7-H3 expression was associated with malignant phenotype and tumor progression. Additionally, B7-H3 was significantly down-regulated in IDH mutation glioma, indicating that IDH may act as a regulator of B7-H3.
Although the receptor(s) for B7-H3 has not been identified, several studies showed that B7-H3 predominantly causes T-cell co-inhibition.Citation7 However, the specific function of B7-H3 in glioma is still not very clear. By GSEA analyses, we found that B7-H3 related genes were more involved in immune response and angiogenesis in glioma. Moreover, the ESTIMATE algorithm results showed that B7-H3 expression was positive correlation with immune cells and stromal cells in glioma samples. Hence, the function of B7-H3 in glioma may realized by the wide expression of B7-H3 in glioma cells and endothelial cells. Additionally, up-regulation of B7-H3 expression in macrophages has been reported in several cancers,Citation31, Citation32 but the expression panel of B7-H3 in glioma associated macrophage is unknown. Here, for the first time, we showed that B7-H3 expression was significantly up-regulated in GBM infiltrating macrophages, indicating that B7-H3 may act as a potent adjunct to facilitating the immune evasion function of macrophages. Moreover, the fact that about 40 #x0025; of the cells in glioma are macrophages raises the intriguing possibility that targeting B7-H3 might emerge as an adjuvant therapy for glioma.Citation19 By analyzing the relationship between B7-H3 expression and T cell specific function, the immune suppressive role of B7-H3 was further confirmed. Collectively, we can speculate that, on the one hand, B7-H3 acts as an innate and adaptive immunity regulator: expressed on cancer cell and tumor infiltrating macrophage, all to accomplish the task of promoting the formation of a serious dysfunctional phenotype in T cells in glioma. On the other hand, B7-H3 acts as a nonimmunological regulator: broadly over-expressed on tumor-infiltrating blood vessels, facilitating angiogenesis and tumor invasion. We have also noticed Dieter Lemke et al. reported that B7-H3 could suppress NK lysis and enhance glioma cell invasiveness,Citation29 in consistence with some other studies previously.Citation33 However, the relationship between B7-H3 expression and NK cell function is not that attractive in our system, which may interfered by some noise. Therefore, the specific role of B7-H3 in glioma still needs further studies.
As a immune checkpoint in B7 family, the expression panel between B7-H3 and other family members is still unknow. We firstly analyzed the relationship between B7-H3 and B7 family checkpoints. Same as CD80 and CD86, B7-H3 was reported to have co-stimulatory and co-inhibitory activities.Citation6 Therefore, it seems logical that the expression profile of B7-H3 was similar with CD80 and CD86. Meanwhile, many studies show that B7-H3 predominantly acts as a co-inhibitory molecular in tumor sites,Citation34 cooperate with PD-L1, PD-L2, ICOSLG, ICOS, PD-1, TIM-3, LAG3 and IDO1 together to built an immune suppressive microenvironment. Great achievements have been made in the immunotherapy targeting immune checkpoints, and blocking monoclonal antibodies (mAbs), including Tecentriq, Nivolumab and Ipilimumab have been approved by the FDA and are now widely used clinically.Citation35, Citation36 Despite the similar antibody to B7-H3 is currently unavailable used in clinical, the good relationship between B7-H3 and other immune checkpoints draw a bright blueprint for the future combined treatment of glioma.
Besides, we found higher B7-H3 expression level predicted significantly worse survival for glioma patients, as well as GBM patients. Importantly, B7-H3 was a significantly independent prognosticator in glioma. The significant prognostic significance of B7-H3 gives us more confidence that B7-H3 blocking mAbs may significantly improve prognosis of glioma patients, especially GBM patients.
In summary, this is the first study exploring the clinical and immune features of B7-H3 in a large-scale glioma samples. Better elucidating the involvement of B7-H3 pathway in immune responses and cancer development will greatly help with the design of more effective therapeutic agents for glioma. However, our analysis has its limitation because the results and conclusions may be interfered by some noise, and further molecular biological experiments are required to validate our findings.
Methods
Sample and data collection
B7-H3 transcriptional level data of glioma were obtained from CGGA (http://www.cgga.org.cn/) and TCGA (https://genome-cancer.ucsc.edu/). In total, 325 samples of RNA sequencing data and 301 samples of mRNA microarray data from CGGA dataset were enrolled into this study. In order to intensify the findings that we have revealed in CGGA dataset, 697 glioma samples of all grades from TCGA dataset were used as a validation cohort. Protein level of B7-H3 was obtained from the Human Protein Atlas (https://www.proteinatlas.org/). The expression level of B7-H3 in GBM infiltrating macrophages was downloaded from Gene Expression Omnibus (GEO) (GSE77043). T cell specific genesets were download from AmiGO 2 Web portal (http://amigo.geneontology.org/amigo/landing), including GO:0002286: T cell activation involved in immune response, GO:0042088: T-helper 1 type immune response, GO:0042092: T-helper 2 type immune response, GO:0002424: T cell mediated immune response to tumor cell, GO:0002840: regulation of T cell mediated immune response to tumor cell and GO:0002842: positive regulation of T cell mediated immune response to tumor cell.
Gene set enrichment analysis (GSEA) analysis
After Spearman correlation analysis, GSEA was performed by the GSEA software and gene sets used in this work were downloaded from the Molecular Signatures Database (http://software. broadinstitute.org/gsea/msigdb/index.jsp).
Statistical analysis
The prognostic value of B7-H3 was estimated by Kaplan–Meier analysis and Cox proportional hazard model analysis using SPSS statistical software (version 19). Other statistical computations and figures drawing were performed with several packages (ggplot2, pheatmap, pROC and corrgram) in the statistical software environment R, version 3.4.1 (http://www.r-project. org). For all statistical methods, p < 0.05 was considered as significant difference.
Disclosure of potential conflicts of interest
The authors declare no potential conflicts of interest.
KONI_A_1461304_Supple.zip
Download Zip (3.2 MB)Supplementary Material
Supplemental data for this article can be accessed on the publisher's website.
Additional information
Funding
References
- Prosniak M, Harshyne LA, Andrews DW, Kenyon LC, Bedelbaeva K, Apanasovich TV, Heber-Katz E, Curtis MT, Cotzia P, Hooper DC. Glioma grade is associated with the accumulation and activity of cells bearing M2 monocyte markers. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2013;19:3776–86.
- Tran B, Rosenthal MA. Survival comparison between glioblastoma multiforme and other incurable cancers. Journal of Clinical Neuroscience: Official Journal of the Neurosurgical Society of Australasia. 2010;17:417–21.
- Finocchiaro G, Pellegatta S. Immunotherapy for glioma: getting closer to the clinical arena? Curr Opin Neurol. 2011;24:641–7.
- Mahoney KM, Freeman GJ, McDermott DF. The Next Immune-Checkpoint Inhibitors: PD-1/PD-L1 Blockade in Melanoma. Clin Ther. 2015;37:764–82.
- He J, Hu Y, Hu M, Li B. Development of PD-1/PD-L1 Pathway in Tumor Immune Microenvironment and Treatment for Non-Small Cell Lung Cancer. Sci Rep. 2015;5:13110.
- Janakiram M, Shah UA, Liu W, Zhao A, Schoenberg MP, Zang X. The third group of the B7-CD28 immune checkpoint family: HHLA2, TMIGD2, B7x, and B7-H3. Immunol Rev. 2017;276:26–39.
- Picarda E, Ohaegbulam KC, Zang X. Molecular Pathways: Targeting B7-H3 (CD276) for Human Cancer Immunotherapy. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2016;22:3425–31.
- Wang J, Chong KK, Nakamura Y, Nguyen L, Huang SK, Kuo C, Zhang W, Yu H, Morton DL, Hoon DS. B7-H3 associated with tumor progression and epigenetic regulatory activity in cutaneous melanoma. J Invest Dermatol. 2013;133:2050–8.
- Altan M, Pelekanou V, Schalper KA, Toki M, Gaule P, Syrigos K, Herbst RS, Rimm DL. B7-H3 Expression in NSCLC and Its Association with B7-H4, PD-L1 and Tumor-Infiltrating Lymphocytes. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2017;23:5202–9.
- Chen Y, Sun J, Zhao H, Zhu D, Zhi Q, Song S, Zhang L, He S, Kuang Y, Zhang Z, et al. The coexpression and clinical significance of costimulatory molecules B7-H1, B7-H3, and B7-H4 in human pancreatic cancer. Onco Targets Ther. 2014;7:1465–72.
- Sun TW, Gao Q, Qiu SJ, Zhou J, Wang XY, Yi Y, Shi JY, Xu YF, Shi YH, Song K, et al. B7-H3 is expressed in human hepatocellular carcinoma and is associated with tumor aggressiveness and postoperative recurrence. Cancer Immunol Immunother: CII. 2012;61:2171–82.
- Zang X, Thompson RH, Al-Ahmadie HA, Serio AM, Reuter VE, Eastham JA, Scardino PT, Sharma P, Allison JP. B7-H3 and B7x are highly expressed in human prostate cancer and associated with disease spread and poor outcome. PNAS. 2007;104:19458–63.
- Janakiram M, Chinai JM, Zhao A, Sparano JA, Zang X. HHLA2 and TMIGD2: new immunotherapeutic targets of the B7 and CD28 families. Oncoimmunology. 2015;4:e1026534.
- Goltz D, Gevensleben H, Dietrich J, Ellinger J, Landsberg J, Kristiansen G, Dietrich D. Promoter methylation of the immune checkpoint receptor PD-1 (PDCD1) is an independent prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. Oncoimmunology. 2016;5:e1221555.
- Branzoli F, Di Stefano AL, Capelle L, Ottolenghi C, Valabregue R, Deelchand DK, Bielle F, Villa C, Baussart B, Lehéricy S, et al. Highly specific determination of IDH status using edited in vivo magnetic resonance spectroscopy. Neuro Oncol. 2018;20:907-916.
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102:15545–50.
- Seaman S, Zhu Z, Saha S, Zhang XM, Yang MY, Hilton MB, Morris K, Szot C, Morris H, Swing DA, et al. Eradication of Tumors through Simultaneous Ablation of CD276/B7-H3-Positive Tumor Cells and Tumor Vasculature. Cancer Cell. 2017;31:501–15.e8.
- Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612.
- Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19:20–7.
- Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7.
- Torphy RJ, Schulick RD, Zhu Y. Newly Emerging Immune Checkpoints: Promises for Future Cancer Therapy. Int J Mol Sci. 2017;18:e2642.
- Li J, Wang L, Chen X, Li L, Li Y, Ping Y, Huang L, Yue D, Zhang Z, Wang F, et al. CD39/CD73 upregulation on myeloid-derived suppressor cells via TGF-beta-mTOR-HIF-1 signaling in patients with non-small cell lung cancer. Oncoimmunology. 2017;6:e1320011.
- Garg AD, Vandenberk L, Van Woensel M, Belmans J, Schaaf M, Boon L, et al. Preclinical efficacy of immune-checkpoint monotherapy does not recapitulate corresponding biomarkers-based clinical predictions in glioblastoma. Oncoimmunology. 2017;6:e1295903.
- Li G, Wang Z, Zhang C, Liu X, Cai J, Wang Z, Hu H, Wu F, Bao Z, Liu Y, et al. Molecular and clinical characterization of TIM-3 in glioma through 1,024 samples. Oncoimmunology. 2017;6:e1328339.
- Yang L, Wang L, Zhang Y. Immunotherapy for lung cancer: advances and prospects. Am J Clin Exp Immunol. 2016;5:1–20.
- Cheng W, Ren X, Zhang C, Cai J, Liu Y, Han S, Wu A. Bioinformatic profiling identifies an immune-related risk signature for glioblastoma. Neurology. 2016;86:2226–34.
- Xue S, Hu M, Iyer V, Yu J. Blocking the PD-1/PD-L1 pathway in glioma: a potential new treatment strategy. J Hematol Oncol. 2017;10:81.
- Filley AC, Henriquez M, Dey M. Recurrent glioma clinical trial, CheckMate-143: the game is not over yet. Oncotarget. 2017;8:91779–94.
- Lemke D, Pfenning PN, Sahm F, Klein AC, Kempf T, Warnken U, Schnölzer M, Tudoran R, Weller M, Platten M, et al. Costimulatory protein 4IgB7H3 drives the malignant phenotype of glioblastoma by mediating immune escape and invasiveness. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2012;18:105–17.
- Baral A, Ye HX, Jiang PC, Yao Y, Mao Y. B7-H3 and B7-H1 expression in cerebral spinal fluid and tumor tissue correlates with the malignancy grade of glioma patients. Oncol Lett. 2014;8:1195–201.
- Lee YH, Martin-Orozco N, Zheng P, Li J, Zhang P, Tan H, Park HJ, Jeong M, Chang SH, Kim BS, et al. Inhibition of the B7-H3 immune checkpoint limits tumor growth by enhancing cytotoxic lymphocyte function. Cell Res. 2017;27:1034–45.
- Mao Y, Chen L, Wang F, Zhu D, Ge X, Hua D, Sun J. Cancer cell-expressed B7-H3 regulates the differentiation of tumor-associated macrophages in human colorectal carcinoma. Oncol Lett. 2017;14:6177–83.
- Castriconi R, Dondero A, Augugliaro R, Cantoni C, Carnemolla B, Sementa AR, Negri F, Conte R, Corrias MV, Moretta L, et al. Identification of 4Ig-B7-H3 as a neuroblastoma-associated molecule that exerts a protective role from an NK cell-mediated lysis. PNAS. 2004;101:12640–5.
- Suh WK, Gajewska BU, Okada H, Gronski MA, Bertram EM, Dawicki W, Duncan GS, Bukczynski J, Plyte S, Elia A, et al. The B7 family member B7-H3 preferentially down-regulates T helper type 1-mediated immune responses. Nat Immunol. 2003;4:899–906.
- Wolchok JD, Rollin L, Larkin J. Nivolumab and Ipilimumab in Advanced Melanoma. N Engl J Med. 2017;377:2503–4.
- Bouffet E, Larouche V, Campbell BB, Merico D, de Borja R, Aronson M, Durno C, Krueger J, Cabric V, Ramaswamy V, et al. Immune Checkpoint Inhibition for Hypermutant Glioblastoma Multiforme Resulting From Germline Biallelic Mismatch Repair Deficiency. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2016;34:2206–11.