ABSTRACT
The formation of paraspeckle, a stress-induced nuclear body, increases in response to viral infection or proinflammatory stimuli. Paraspeckle consists of lncRNA (nuclear paraspeckle assembly transcript 1, NEAT1) and protein components including NONO, SFPQ, PSPC1, etc., which are shown to be involved in viral infection and cancer. Both NEAT1 and NONO expression increase in human hepatocellular carcinoma (HCC) samples according to TCGA data. However, the role of paraspeckle in HCC progression needs further identification. IL-6 signaling is well known to contribute to HCC progression. Here we reported that IL-6 signaling increased paraspeckle formation in HCC cells. Destruction of paraspeckle formation by silencing the paraspeckle essential components NEAT1_2 or NONO could suppress IL-6-induced STAT3 phosphorylation in HCC cells, and consequently repressed IL-6-promoted in vitro HCC cell invasion, cell cycle progression and survival. Mechanistically, paraspeckle promotes IL-6-induced STAT3 phosphorylation by binding and trapping peroxiredoxin-5 (PRDX5) mRNA in nucleus, decreasing protein level of PRDX5 which can directly interact with STAT3 and inhibit STAT3 phosphorylation. Besides, glutathione S-transferase P (GSTP1) protein, which inhibits DNA damage and apoptosis through its detoxification and anti-oxidation function, was also trapped within paraspeckles under IL-6 stimulation. Paraspeckle-trapping of both PRDX5 mRNA and GSTP1 protein contributes to IL-6-increased DNA damage in HCC cells. Our results demonstrate that paraspeckle can nuclear entrap the inhibitors of IL-6/STAT3 signaling as well as DNA damage, and then strengthen the promoting effect on HCC progression by IL-6. Therefore, paraspeckle contributes to the inflammation-related HCC progression and might be a potential therapeutic target for HCC.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors and the third leading cause of cancer-related mortalities in the world. Although multiple treatments had been developed to control HCC in clinic, the therapeutic approaches have limited efficacy, and the poor prognosis of HCC patients is mainly ascribed to high risk of metastasis and high recurrence rate.Citation1 Therefore, further exploring the molecular mechanisms of HCC progression and developing more effective treatments for HCC are urgently needed. Furthermore, HCC is one representative example of inflammation- and infection-associated cancers with continuous hepatocyte death, long-lasting local infiltration, reactive oxygen and nitrogen species accumulation. MyD88-dependent production of IL-6 is in the centre of the inflammation, HCC and sex triangle.Citation2 Besides, IL-6 signaling promotes the development of HCC via preventing DNA-damage-induced hepatocytes apoptosis and promoting tumor angiogenesis.Citation3 Tumor-associated macrophages promote expansion of HCC stem cells by producing IL-6.Citation4 Constitutive STAT3 activation has been found in various cancers including HCC, promoting tumor development by enhancing cell proliferation and preventing apoptosis.Citation5–Citation8 Targeting IL-6 or its downstream transcription factor STAT3 have been proved to suppress tumor growth and metastasis of HCC,Citation9,Citation10 suggesting the important role of IL-6/STAT3 signaling in HCC tumorigenesis. Identifying regulators which are responsible for overactivation of IL-6/STAT3 signaling in HCC cells would be helpful for revealing the molecular mechanism for inflammation-induced tumorigenesis and progression of HCC as well as providing new potential targets for treating HCC.
Compared with messenger RNAs, there exist a variety of large and small non-coding RNAs which do not encode proteins.Citation11 Beside microRNAs,Citation12 long non-coding RNAs (lncRNAs) which are defined with length over 200 nucleotides have been revealed to play important roles in HCC tumorigenesis and progression.Citation13,Citation14 With the development of lncRNA microarray and whole-genome transcriptome sequencing platforms, deregulation of intergenic, antisense, and other lncRNAs such as circular RNAs and competitive endogenous RNAs have been also observed in HCC.Citation15
Both cytoplasmic and nuclear lncRNAs are suggested to be involved in the regulation of signaling pathways such as WNT and STAT3 signaling which are overactivated in HCC, and epithelial to mesenchymal transition (EMT) and determination and maintenance of hepatic cancer stem cells (CSCs).Citation16 For the molecular mechanisms, lncRNAs can mediate epigenetic dysregulation in HCC, e.g. lncTCF7, highly expressed in liver cancer tissues and CSCs, recruits the SWI/SNF chromatin remodeling complex to promote the TCF7 gene transcription, leading to activation of the Wnt signaling pathway;Citation17 LncRNAs can mediate post-transcription regulation of mRNA levels, e.g. lncRNA-ATB activated by TGF-β in HCC upregulates mRNA levels of ZEB1, ZEB2 and IL-11 through competitively binding microRNAs and other mechanisms;Citation18 lncRNAs can regulate post-translational modification of proteins, e.g. lnc-β-Catm, highly expressed in human HCC tumors and liver CSCs, promotes methylation of β-catenin mediated by methyltransferase EZH2, thereby stabilizing β-catenin;Citation19 lncRNAs can recruits transcription factors, e.g. LncSox4, highly expressed in HCC tissues and in liver tumour-initiating cells, interacts with and recruits STAT3 to activate Sox4 promoter.Citation20 However, there are large numbers of lncRNAs which are highly expressed in HCC, the roles of which in HCC progression are largely unknown. Therefore, new molecular mechanisms regulating HCC-related pathways mediated by specific lncRNAs remains to be revealed.
LncRNA NEAT1 (nuclear paraspeckle assembly transcript 1) exists in two isoforms: NEAT1_1 (human 3.7kb) and NEAT1_2 (human 22.7kb). Through arresting binding of CPSF6-NUDT21 complex in the vicinity of alternative polyadenylation site of NEAT1_1, heterogeneous nuclear ribonucleoprotein K promotes the production of NEAT1_2. Thus, NEAT1_1 and NEAT1_2 have the same promoter and NEAT1_1 sequence is consistent to the front sequence of NEAT1_2.Citation21 Paraspeckle is a kind of highly structured and organized nonmembranous nuclear body.Citation22 LncRNA NEAT1_2 is essential for assembling of paraspeckles, acting as the scaffold for enrichment of RNA-binding proteins in the Drosophila behavior and human splicing (DBHS) family including NONO, SFPQ and PSPC1 which are also essential for the structural maintenance of paraspeckles.Citation23 Through structural analysis of paraspeckles using structured illumination microscopy, paraspeckle is found to consist of core-shell structures with 3ʹ region and 5ʹ region of NEAT1_2 distributing in the shell, and the middle region of NEAT1_2 and its core protein components including NONO, SFPQ and PSPC1 arranging in the core of paraspeckle spheres.Citation24 Previous data showed that paraspeckle could alter nuclear retention of mRNAs containing inverted repeats of Alu sequences in their 3ʹ-untranslated regions (UTRs) by NONO.Citation25 Upon immune stimuli, splicing factor proline/glutamine-rich (SFPQ) is relocated from Il8 gene promoter to the paraspeckles, leading to transcriptional activation of IL-8.Citation26 Notably, based on TCGA database, expression of NEAT1 and NONO increases in HCC samples. However, the role of paraspeckles in HCC progression is still unknown.
In the present study, we provide evidence that paraspeckle is engaged in IL-6/STAT3 signaling-involved HCC progression. Through nuclear trapping of mRNAs and proteins, paraspeckle strengthens IL-6-induced STAT3 phosphorylation and DNA damage in HCC. These data illustrate the importance of paraspeckles in HCC progression and its potential as the target for the control of HCC.
Results
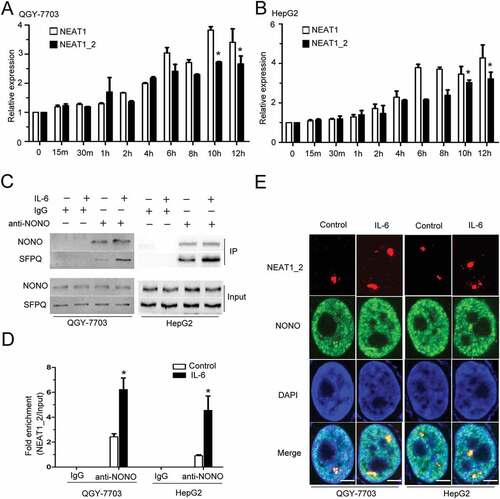
IL-6 increases paraspeckle formation in HCC cells
We first analyzed the expression levels of lncRNA NEAT1 and paraspeckle assemble-related protein NONO in normal liver and hepatocellular carcinoma tissues based on TCGA data. As shown in Fig. S1A and B, both NEAT1 and NONO mRNAs are more abundant in primary HCC tumor samples compared with the normal liver controls, indicating that paraspeckle sub-structure may be involved in HCC progression. We found that IL-6 stimulation could elevate the expression of both NEAT1_2 and NEAT1 (contains both NEAT1_1 and NEAT1_2) in HepG2 and QGY-7703 HCC cells (), while the expression of paraspeckle assemble-related proteins NONO and SFPQ remained unchanged following IL-6 stimulation (Fig. S2A). It has been shown that NEAT1_2 but not NEAT1_1 is indispensable for paraspeckle formation.Citation21 NEAT1_1 alone is unable to maintain paraspeckle integrity.Citation27 And, NEAT1_2 (but not Neat1_1) acts as a scaffold for RNA/DNA binding proteins and its transcription is essential for paraspeckle formation.Citation28,Citation29 co-IP and RIP assays showed that interactions between SFPQ and NONO or between NONO and NEAT1_2 were both strengthened under IL-6 stimulation (). We further used super-resolution microscopy assay to confirm the effect of IL-6 on paraspeckle formation by showing the dramatically increased co-localizations between NONO and NEAT1_2 in nucleus of HepG2 and QGY-7703 HCC cells (). These results indicate that IL-6 signaling contributes to the increased expression of paraspeckle subunits in HCC, and increases NEAT1_2-dependent paraspeckle formation in the nucleus of HCC cells.
Figure 1. IL-6 increases paraspeckle formation in HCC cells
IL-6 increases NEAT1_2 transcription in HCC cells through STAT3 and histone modifications
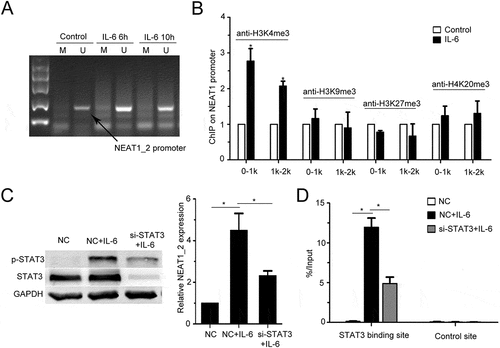
To reveal the mechanism of IL-6-mediated upregulation of NEAT1_2 transcription, we detected the DNA methylation level of the CpG island and histone methylation level at NEAT1_2 promoter, both of which were reported to play important roles in regulating NEAT1_2 transcription.Citation30–Citation32 By methylation specific polymerase chain reaction (MSP) assay, we found that IL-6 could not affected DNA methylation level in the CpG island at NEAT1_2 promoter in QGY-7703 cells (). Through ChIP assay using antibodies against several types of histone methylation, we found that IL-6 stimulation increased the level of histone H3 lysine 4 trimethylation (H3K4me3) at NEAT1_2 promoter (). As STAT3 is a major transcription factor activated by IL-6, and STAT3 is involved in NEAT1 transcription regulation during HSV-1 infection in HeLa cells,Citation33 we found that STAT3 silencing decreased IL-6-promoted NEAT1_2 transcription () and ectopic expression of STAT3 strengthened IL-6-induced NEAT1_2 transccription (Fig. S2D), suggesting that STAT3 mediates IL-6-increased NEAT1_2 transcription in QGY-7703 cells. Furthermore, we also observed direct binding of STAT3 to NEAT1_2 promoter region in QGY-7703 cells upon IL-6 stimulation (). These results indicate that IL-6/STAT3 signaling increases transcription of NEAT1_2 in HCC cells.
Figure 2. IL-6 promotes NEAT1 transcription through STAT3 and H3K4me3
Paraspeckle participates in il-6-promoted invasion and survival of HCC cells
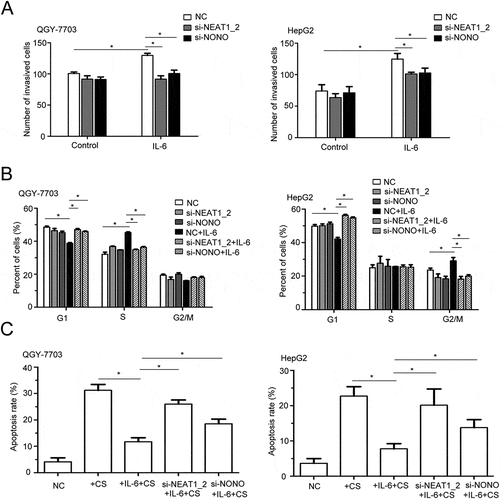
To explore the role of paraspeckle in IL-6-promoted HCC progression in vitro, we silenced expression of NEAT1_2 or NONO (Fig. S2B, C) to destroy the paraspeckle formation under IL-6 stimulation.Citation34 In Matrigel invasion assay, silencing of NEAT1_2 or NONO alone could not changed the migration of QGY-7703 and HepG2 cells, but significantly decreased IL-6-promoted migration of both cells (). Furthermore, silencing of NEAT1_2 or NONO also suppressed IL-6-promoted cell cycle progression, with decreased S phase and increased G1 phase in NEAT1_2- or NONO-silenced QGY-7703 cells, and increased G1 phase and decreased G2/M phase in NEAT1_2- or NONO-silenced HepG2 cells, as compared with control cells after IL-6 stimulation (). Moreover, silencing of NEAT1_2 or NONO alleviated the IL-6-mediated inhibition of cis platinum-induced apoptosis of QGY-7703 cells and HepG2 cells (). We also detected the level of 8-OHDG (8-hydroxy-2-deoxy guanosine), produced by oxidative damage of DNA caused by reactive oxygen and nitrogen species, under IL-6 stimulation.Citation35,Citation36 As shown in Fig. S3A and C, IL-6 significantly increased 8-OHDG levels in nuclear lysis; whereas silencing of NEAT1_2 or NONO suppressed IL-6-increased 8-OHDG levels in nuclear lysis of QGY-7703 cells and HepG2 cells (Fig. S3B, D). Therefore, paraspeckle is involved in IL-6-induced invasion and survival of HCC cells.
Figure 3. The effects of paraspeckle destruction on HCC cell invasion, proliferation and apoptosis under IL-6 stimulation
Paraspeckle promotes IL-6/STAT3 signaling through trapping PRDX5 mrna in nucleus of HCC cells
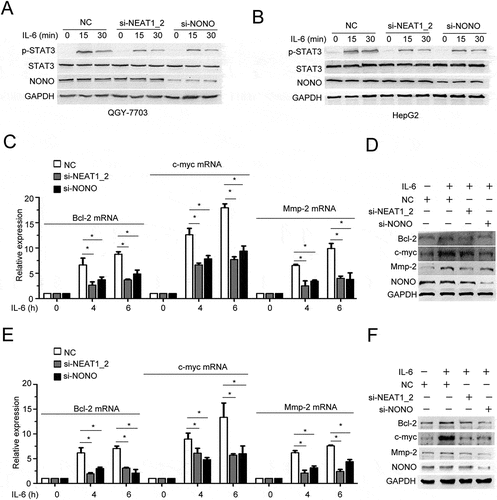
We further investigated whether paraspeckles are capable of regulating IL-6/STAT3 signaling in HCC cells. Silencing of NEAT1_2 or NONO impaired IL-6-induced Tyr705 phosphorylation of STAT3 (). In addition, ectopic expression of mNEAT1v2 or NONO strengthened it (Fig. S4A, B). The mRNA and protein expression of downstream targets of IL-6/STAT3 signaling pathway, including Bcl-2, c-myc and Mmp2, were also decreased in NEAT1_2- or NONO-silenced HCC cells (). To investigate the mechanism involved in paraspeckle-promoted IL-6/STAT3 activation, we performed RIP-seq using anti-NONO antibody to precipitate protein-RNA complexes in QGY-7703 cells with or without IL-6 stimulation. Specific and common transcripts that bound NONO before and after IL-6 stimulation were identified (Fig. S5A). The transcriptomic distributions of NONO-binding RNAs were similar between the control group and IL-6 group, and about 60% were within introns (Fig. S5B). In mature RNA elements, peaks are more distributed in CDS (Fig. S5C). KEGG pathway analysis showed that RNA-associated genes were most enriched in three cancer-related pathways, with highest rich factors in these three pathways, further indicating a role of NONO in regulating cancer-related gene transcripts (Fig. S5D).
Figure 4. Paraspeckle destruction inhibits IL-6-induced STAT3 phosphorylation
Among the transcripts in RIP-seq results of the IL-6 group, we noticed peroxiredoxin-5 (PRDX5) which may be involved in regulating STAT3 activation.Citation37 We found that ectopic overexpression of PRDX5 suppressed IL-6-induced STAT3 phosphorylation (). IL-6 stimulation increased the association between NONO and PRDX5 mRNA (), which was decreased by NEAT1_2 knockdown (), further confirming that PRDX5 mRNA was a paraspeckle-associated RNA. We then measured the mRNA and protein expression of PRDX5 and found that IL-6 stimulation did not change PRDX5 mRNA level but decrease PRDX5 protein level (), indicating that IL-6 stimulation may promote paraspeckle-mediated nuclear trapping of PRDX5 mRNA, inhibiting protein translation from mRNA. Through fractionation of nuclear and cytoplasmic RNAs, we found that IL-6 stimulation indeed increased mRNA level of PRDX5 in nuclear fraction, and destruction of paraspeckles by silencing of NEAT1_2 or NONO impaired IL-6 stimulation-promoted nuclear retention of PRDX5 mRNA (). Moreover, knockdown of PRDX5 partly canceled the inhibitory effect of NEAT1_2 or NONO silencing on IL-6-induced STAT3 activation (). These results indicate that paraspeckle can promote IL-6/STAT3 signaling through nuclear trapping of PRDX5 mRNA, therefore decreasing protein level of PRDX5 and then the interaction of PRDX5 and STAT3.
Figure 5. Paraspeckle promotes IL-6-induced STAT3 phosphorylation through trapping PRDX5 mRNA in nucleus
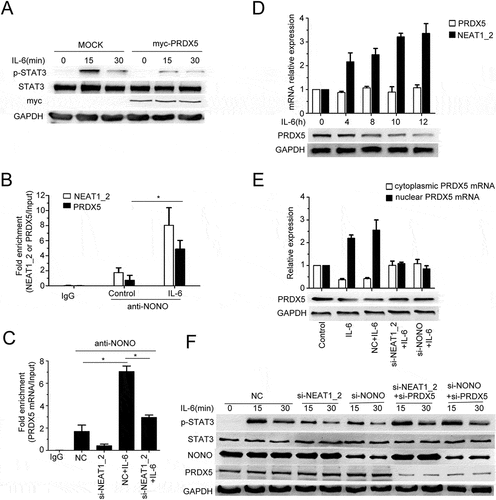
PRDX5 interacts with STAT3 and represses STAT3 phosphorylation in HCC cells
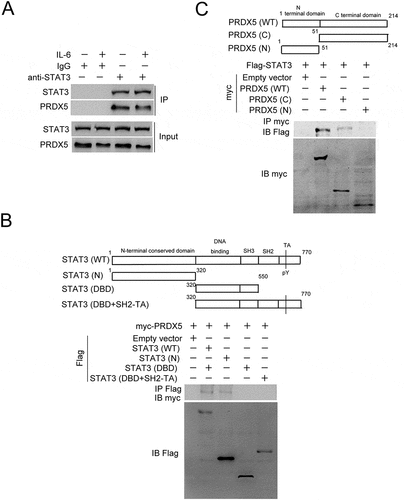
We further investigated the mechanism of the PRDX5-mediated repression of STAT3 activation, and found that PRDX5 could interact with STAT3, which was decreased by IL-6 stimulation (). By performing co-immunoprecipitation assay with various constructs containing different functional domains of STAT3 and PRDX5, we found that the N-terminal conserved domain of STAT3 and C-terminal domain of PRDX5 were responsible for the interaction between STAT3 and PRDX5, respectively (). Together with the results from to F, the data indicate that IL-6 represses the PRDX5-STAT3 interaction through increasing paraspeckle formation, releasing the repression of STAT3 activation by PRDX5.
Figure 6. Protein-protein interaction analysis
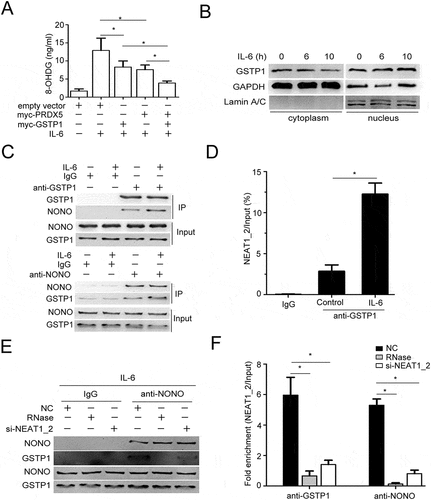
Paraspeckle promotes il-6-increased DNA damage through trapping GSTP1 protein in the nucleus of HCC cells
As described above, IL-6 stimulation increased the level of 8-OHDG in HCC cells, partially dependent of paraspeckles. As a member of peroxiredoxin family of antioxidant enzymes, PRDX5 catalyzes the reduction of hydrogen peroxide, which may contribute to oxidation-induced DNA damage. We found overexpression of PRDX5 could partially repress IL-6 increased 8-OHDG level in HCCs, indicating other proteins may be involved in IL-6-induced oxidation DNA damage (). Paraspeckle can trap not only mRNAs but also proteins in nucleus, to this end, we further analyzed the NONO-associated proteins identified by co-immunoprecipitation combined with protein electrophoresis and subsequent mass spectra analysis and noticed GSTP1 (glutathione S-transferase P). As previously reported, loss of GSTP1, which protected the cells against genomic damage mediated by oxidants and electrophiles, led to accumulation of oxidative DNA products in prostatic epithelium under chronic intraprostatic inflammation, and increased intracellular production of ROS.Citation35,Citation36 We found that ectopic expression of GSTP1 indeed could partially decrease IL-6-increased 8-OHDG production in QGY-7703 cells, and notably in a synergistic manner with PRDX5 (). Furthermore, IL-6 stimulation decreased cytoplasmic GSTP1 protein levels but increased nuclear GSTP1 protein levels (). Through co-immunoprecipitation using antibodies against NONO or GSTP1, we demonstrated that IL-6 promoted the interaction between NONO and GSTP1 () as well as GSTP1 and NEAT1_2 () in QGY-7703 cells. To evaluate whether the interaction between NONO and GSTP1 depends on paraspeckle formation, RNase digestion or si-NEAT1_2 transfection were used to destroy paraspeckle structure.Citation34 The results showed that destruction of paraspeckle structure suppressed GSTP1 binding to NONO or NEAT1_2 in IL-6-stimulated QGY-7703 cells (). The data suggest that, besides trapping PRDX5 mRNA, IL-6-increased paraspeckle formation traps more GSTP1 proteins in the nucleus of HCC cells, leading to increased oxidative DNA damage.
Figure 7. Paraspeckle promotes IL-6-induced DNA damage through trapping GSTP1
Discussion
Recently, lncRNAs have been identified as potential key regulators of the inflammatory response, largely involved in regulating activation of pro-inflammatory pathways and transcription induction of inflammatory genes during innate immune response. Furthermore, dysregulation of lncRNAs which contributes to overexpression of pro-inflammatory cytokines and overactivation of pro-inflammatory pathways are also largely implicated in the pathogenesis of inflammatory diseases, including cancers.Citation38 Revealing more inflammation-related lncRNAs and their function, especially for maintaining chronic inflammation will be helpful for unveiling the pathogenic mechanisms of inflammatory disease. lncRNA – NEAT1 has been found to promote HCC by sponging microRNA, modulating abnormal lipolysis or affecting the epithelial-mesenchymal transition.[Citation39-Citation41]Moreover, NEAT1 positively relates to tumorgenesis and metastasis of HCC.Citation42 While these studies focus on NEAT1_1 and HCC, the role of NEAT1_2 in HCC is still unknown. Our study identified lncRNA NEAT1_2-mediated paraspeckle as a new regulator of IL-6/STAT3 proinflammatory signaling pathway, although in HCC cells, which might also play important roles in innate immune cells and other IL-6 signaling-responsive cells.
Chronic inflammation leads to genotoxicity, aberrant tissue repair, proliferative responses, invasion and metastasis.Citation43 Besides nuclear factor-κB, STAT3 is the key transcription factor which is converged by major inflammatory pathways that are involved in inflammation-induced liver carcinogenesis.Citation44 Although the main cause of STAT3 activation in HCC may be due to the elevated expression of pro-inflammatory IL-6 and related cytokines, such as IL-11 and IL-22 in cancer microenvironment, the detailed mechanisms underlying chronic and constant IL-6/STAT3 overactivation in transformed hepatocytes remain elusive. In the present study, we found that lncRNA NEAT1_2-mediated nuclear sub-structure paraspeckle is involved in promoting IL-6/STAT3 activation via trapping negative regulators of IL-6 signaling and tumor repressors in nucleus of HCC cells. These findings indicate that destruction of paraspeckle formation may offer interesting opportunities to therapeutic intervention as well as prevention.
NEAT1_2 have been found to constitute the crucial structural framework of organizing paraspeckle.Citation45,Citation46 As one of nonmembraneous sub-nuclear structures found in mammalian cells, paraspeckle could retain kinds of mRNAs and proteins, e.g. sequestrating transcription factors from the promoters of target genes.Citation47,Citation48 Paraspeckle play important roles in both post-transcription and transcription regulation in multiple biological and pathological processes. And our observations in this study suggest that paraspeckles are functional sub-nuclear structures involved in IL-6/STAT3 activation in HCC cells, and increased formation of paraspeckles is a positive feedback loop of IL-6/STAT3 signaling to maintain overactivation of STAT3 in HCCs. Activated STAT3 is responsible for the increased paraspeckle formation, and paraspeckle promotes activation of STAT3 which activates transcription of pro-survival, inflammation, EMT and CSC associated genes.Citation49 Thus, paraspeckle may be largely involved in STAT3-related tumorigenesis and progression of kinds of cancers in addition to IL-6 signaling in HCC. Furthermore, paraspeckle was reported to regulate sensing of DNA viruses and induction of antiviral genes,Citation26,Citation50 indicating that paraspeckle may be also take part in the regulating of HBV infection and HBV-promoted progression of HCCs.
Through transcriptomic analysis of paraspeckle-associated RNAs, we identified that PRDX5, the protein expression of which was repressed by paraspeckles, is a negative regulator of STAT3 phosphorylation. Our data showed direct interaction between PRDX5 and STAT3, but the detailed mechanism underlying PRDX5-meidated repression of STAT3 Tyr705 phosphorylation was waiting to be revealed in the future. As a peroxiredoxin, PRDX5 has antioxidative and cytoprotective functions during oxidative stress, indicating a reciprocal regulation between antioxidation and STAT3 activation. Decreased expression of PRDX5 may also contribute to increased oxidative DNA damage after IL-6 stimulation. Furthermore, besides PRDX5, there may be other potential negative regulators of IL-6/STAT3 signaling lying low in our RIP-seq data.
GSTP1, a well-known tumor repressor protecting cells against DNA damage by catalyzing the conjugation of glutathione to a wide variety of exo- and endogenous electrophilic substrates, is frequently silenced by CpG island promoter hypermethylation in several types of cancers including HCC, especially HBV-related HCC.Citation51,Citation52 GSTP1 also regulates normal cellular functions through interacting with critical cellular proteins, such as those involved in MAPK signaling cascade.Citation53,Citation54 In our study, we found paraspeckle GSTP1 protein in nucleus, however, whether catalytic activity of GSTP1 can be influenced by being trapped to paraspeckle needs further investigation.Citation55 Interestingly, GSTP1 may also inhibit STAT3 activation in HepG2 cells.Citation56 Sequestering GSTP1 proteins in nuclear paraspeckles may be an oncogenic mechanism of IL-6-promoted HCC tumorigenesis before DNA methylation-mediated GSTP1 silencing during HCC progression. Furthermore, other tumor repressors may be also sequestered in nuclear paraspeckles. Thus, components in IL-6-increased paraspeckles identified in our transcriptomic and proteomic assays will provide valuable data for future studies for further revealing new oncogenic role of both paraspeckle and IL-6/STAT3 signaling.
Materials and methods
Cell culture and transfection
HEK293T cells and the human HCC cell lines QGY-7703, HepG2 cells were from American Type Culture Collection (ATCC). HEK293T cells were maintained in DMEM media containing 10% fetal bovine serum (FBS) at 37°C with 5% CO2 in a humidified incubator. QGY-7703 and HepG2 cells were cultured in RPMI-1640 supplemented with 10% fetal bovine serum (FBS). Sequence of siRNA against lncRNA NEAT1_2 (si-NEAT1_2): 5ʹ-GGAACAUUCUCAUUUAAUAtt-3ʹ and 5ʹ-UAUUAAAUGAGAAUGUUCCat-3ʹ;Citation26 Sequence of siRNA against NONO (si-NONO): 5ʹ-GGGGUGGUAUUAAACAAGUCA-3ʹ and 5ʹ-ACUUGUUUAAUACCACCCCUC-3ʹ.Citation26 Sequence of siRNA against PRDX5 (si-PRDX5): 5ʹ-GCAAGAAGGGUGUGCUGUUTT-3ʹ and 5ʹ-AACAGCACACCCUUCUUGCTT-3ʹ. Sequence of siRNA against STAT3 (si-STAT3): 5ʹ-GAAGGAGGCGUCACUUUCA-3ʹ and 5ʹ-UGAAAGUGACGCCUCCUUC-3ʹ.Citation33 Sequence of scrambled siRNA (NC): 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ and 5ʹ-ACGUGACACGUUCGGAGAATT-3ʹ. siRNAs were synthesized by Shanghai GenePharma Co., Ltd. pCMV-mNEAT1_2 (mouse NEAT1_2) plasmid was a gift from Tatsuhiko Kodama.Citation26 Full-length cDNAs of human NONO and STAT3 were amplified using KOD DNA polymerase (TOYOBO, Japan). NONO cDNA was amplified by using forward primer (GATATCGTACGACAGATATGCAGAGTAATAAAACTTT) and reverse primer (TTACTTATCGTCGTCATTAGTATCGGCGACGTTTGT). STAT3 cDNA was amplified by using forward primer (GATATCGTACGACAGATATGGCCCAATGGAATCAGCT) and reverse primer (TTACTTATCGTCGTCATCACTGGGGGAGCTAGCGCA). pCMV-Flag-NONO/STAT3 vector was generated by ligating the NONO/STAT3 cDNA with the pCMV-Flag vector using a ClonExpress II one step cloning kit (Vazyme, China). The transfections were performed with jetPRIME reagent (Polyplus-transfection, Illkirch, France) according to the manufacturer’s instructions.
Quantitative real-time PCR (qrt-PCR)
Cells were treated with or without human IL-6 (50ng/ml, Peprotech, USA) for the indicated time, and then harvested. Total RNA was extracted using Trizol Reagent (Invitrogen, USA). RNA from each sample was reverse-transcribed into cDNA using the PrimeScript RT reagent kit (TOYOBO, Japan). qRT-PCR was performed using the Q7 real-time PCR system (Applied Biosystems, USA), with SYBR Green Master Mix (TOYOBO, Japan). The obtained data were normalized to GAPDH expression levels in each sample. The primers for qRT-PCR were as follows: NEAT1_1: F: 5ʹ-GAGAACCAAAGGGAGGGGTG-3ʹ, R: 5ʹ-TGCTGCGTATGCAAGTCTGA-3ʹ. NEAT1_2: F: 5ʹ-ACATTGTACACAGCGAGGCA-3ʹ, R: 5ʹ-CATTTGCCTTTGGGGTCAGC-3ʹ. GAPDH: F: 5ʹ-GGAGCGAGATCCCTCCAAAAT-3ʹ. R: 5ʹ-GGCTGTTGTCATACTTCTCATGG-3ʹ. Bcl-2: F: 5ʹ-GGTGGGGTCATGTGTGTGG-3ʹ, R: 5ʹ-CGGTTCAGGTACTCAGTCATCC-3ʹ. C-myc: F: 5ʹ-GGCTCCTGGCAAAAGGTCA-3ʹ, R: 5ʹ-CTGCGTAGTTGTGCTGATGT-3ʹ. Mmp-2: F: 5ʹ-TACAGGATCATTGGCTACACACC-3ʹ, R: 5ʹ-GGTCACATCGCTCCAGACT-3ʹ. PRDX5: F: 5ʹ-GCAAGACGGTGCAGTGAAG-3ʹ, R: 5ʹ-ATGGCATCTCCCACCTTGATT-3ʹ.
Western blot
Cells were harvested and lysed using cell lysis buffer (CST, USA). After measuring the protein concentration using BCA kit (Thermofisher, USA), protein was loaded into 10% polyacrylamide SDS gel. After transferred onto nitrate membranes (Millipore, USA) and incubated with primary antibodies at 4°C overnight. Nitrate membranes were washed three times with PBS for a total of 15 min and incubated with specific secondary antibodies (1:10000, CST, USA) for 1 h at room temperature. Later the signals were detected using ECL kit (Pierce, USA). Primary antibodies used were as follows: rabbit anti-NONO (1:1000, Abcam, USA), rabbit anti-SFPQ (1:1000, Abcam, USA), rabbit anti-p-STAT3 (1:1000, CST, USA), rabbit anti-STAT3 (1:1000, Abcam, USA), rabbit anti-GAPDH (1:2000, Abcam, USA), mouse anti-Bcl-2 (1:1000, CST, USA), mouse anti-c-myc (1:1000, CST, USA), rabbit anti-Mmp-2 (1:1000, Abcam, USA), rabbit anti-PRDX5 (1:1000, Abcam, USA), rabbit anti-Flag (1:2000, CST, USA), mouse anti-myc (1:2000, CST, USA), rabbit anti-GSTP1 (1:1000, Abcam, USA) and mouse anti-LaminA/C (1:1000, CST, USA). Co-immunoprecipitation was performed as previously described.Citation57
Rna-immunoprecipitation (RIP)
In brief, after treatments, cells were harvested and lysed in Polysome lysis buffer (100mM KCl, 5mM MgCl2, 10mM HEPES pH 7.0, 0.5% NP40, 1mM DTT, 80U/ml RNase Inhibitors, 400uM VRC and protease inhibitors cocktail) for 30 min on ice. Lysates were sonicated to fragment chromatins and RNAs, and centrifuged, and protein concentration in the supernatant was measured with BCA assay (Thermofisher, USA). Proteins were incubated with Protein G-coupled Magnetic Dynabeads (Life Technologies, USA) pre-coated with rabbit anti-NONO antibody, anti-GSTP1 antibody or IgG control for 4 hr at 4°C. Before the incubation, 1/10 of the supernatant was put aside to be used as input. After incubation, samples were washed 5 times with NT buffer (50 mM, Tris-HCl pH 7.4, 150mM NaCl, 1 mM MgCl2, 1% Triton X-100). Immunoprecipitated RNAs and input RNAs were extracted using Trizol reagent (Invitrogen). For the RIP-seq analysis, reads uniquely mapped to the genome were subjected to calling the peaks in the enriched regions with a fold enrichment of at least 2 over input reads.
RNA FISH and immunofluorescence microscopy
NONO and the NEAT1_2 RNA FISH assays were performed using the ViewRNA ISH Cell Assay kit (Affymetrix, USA) following the protocol of the manufacturer. Detailed procedures were as previously described.Citation58
Cell invasion assay
The invasive abilities of HCC cells were determined using 24-well transwell chambers coated with Matrigel (BD Pharmingen). Treated cells (5 × 104 per group) in serum-free medium were seeded in the top chamber. The bottom chamber was filled with RPMI-1640 supplement with 10% FBS. After incubated at 37°C in a humidified incubator containing 5% CO2 for 24 hour, cells that migrated to the underside of the membrane were fixed with 4% paraformaldehyde (Sangon, Shanghai, CHINA), stained with crystal violet (Beyotime, Shanghai, China), imaged, and counted with a microscope (Olympus, Japan).
Cell cycle assay
Cells seeded into a 12-well plate were transfected with siRNAs and later treated with or without IL-6. After 24h, the cells were collected and washed using PBS, fixed using pre-cold 70% ethanol for 24 h. Centrifuged and stained with Propidium Iodide (PI), cells were washed using PBS and cell cycle was analyzed by flow cytometry (LSRII, BD, USA).
Apoptosis assay
In brief, cells were seeded in 6-well plates at 8 × 105 per well. Seventy-two hours after treatments, cells in the suspension and that were adhered were both harvested and labeled with AnnexinV (BD, USA) for 15 minutes in dark. PI was added to each sample before the cell apoptosis distribution was analyzed by flow cytometry (LSRII, BD, USA).
8-OHDG detection
Cells were treated and lysed using NP40 lysis buffer (Beyotime, Shanghai, China) on ice. After centrifugation, undissolved nucleus debris was washed and lyzed using cell lysis buffer (CST, USA). Supernatant of nuclear lysis was collected by centrifugation and 8-OHDG level was detected using a 8-hydroxy 2-deoxyguanosine ELISA kit (Abcam, USA) according to manufacturer’s instructions.
Nuclear/cytosol RNA fractionation
To isolate cytosolic and nuclear RNAs from treated cells, we used nuclear/cytosol fractionation kit (Biovision, USA). In brief, cells were homogenized in Cell Fractionation Buffer thoroughly before centrifuge for 5 min at 1500rpm. Supernatant was collected as cytosolic fraction, while nuclear pellet was washed and lysed by Cell Disruption Buffer. Such samples were mixed with 2X Lysis/Binding Solution before extracting RNA according to the manufacturer’s protocol.
Chromatin immunoprecipitation (chip)
ChIP was performed as follows: cells were crosslinked with 1% formaldehyde, lyzed on ice and supernatant was carefully removed and nuclear pellets were re-suspended in 0.5ml of nuclear lysis buffer, then sonicated to create appropriately sized chromatin fragments. After centrifuged to remove insoluble materials, supernatant was transferred to clean microfuge tubes in 50μl aliquots. Then 3μg anti-H3K4me3 antibody (Abcam, USA), anti-H3K9me3 antibody (Abcam, USA), anti-H3K27me3 antibody (Abcam, USA), anti-H4K20me3 antibody (Abcam, USA), or IgG was added to each nuclear extract, and incubated at 4°C overnight. Nuclear extracts were later incubated with magnetic protein A/G beads for 2 h at 4°C to capture protein/DNA complexes, then beads were sequentially washed with low salt buffer, high salt buffer, LiCl wash buffer and TE buffer, then protein/DNA complexes were eluted and reverse cross-linked to free the DNA. Purified DNA was analyzed by qPCR. The primers for ChIP were as below: 0-1k: F: 5ʹ-GTGTTTCCCAGGCTGGTCTCG-3ʹ, R: 5ʹ-CTCCCTGGCGCCTGCTTAGCCC-3ʹ; 1k-2k: F: 5ʹ-GGGAGCAAGCCTGGGCTTGC-3ʹ, R: 5ʹ-CGCCGACCCTGCCCGGAGAG-3ʹ; STAT3 binding site: F: 5ʹ-TGCCACATCACCACCTTCTG-3ʹ, R: 5ʹ-GAAGACATTTCGCCTGCGTC-3ʹ; control-site: F: 5ʹ-GAGGGGACGTGTTTCCTGAG-3ʹ, R: 5ʹ-TGTCCCTCGGCTATGTCAGA-3ʹ.
Cpg island methylation measurement
QGY-7703 cells were treated with or without IL-6 for the indicated times. Cellular DNA was extracted using DNA extracting kits (Sangon, Shanghai, CHINA). Firstly, bisulfite conversion of the extracted DNA was performed. Then, methylation specific polymerase chain reaction (MSP) was performed to measure DNA methylation level. All these processes were performed in strict accordance with kit instructions. Methylated (M) and Unmethylated (U) primers of MSP were designed to amplify CpG rich regions using online software, MethPrimer (www.urogene.org/methprimer/) and used for the amplification. The primer sequences were listed as below: M: F: 5ʹ-GGTTTTTGGGAAGAGTTAAAATTAC-3ʹ, R: 5ʹ-CAAAAAACCAACGAAATACTACGA-3ʹ; U: F: 5ʹ-TTTTTGGGAAGAGTTAAAATTATGA-3ʹ, R: 5ʹ-CAAAAAACCAACAAAATACTACAAA-3ʹ.
Statistical analysis
Statistical significance between two groups was determined by unpaired two-tailed Student’s t test. And, multiply groups data was analyzed using one-way ANOVA. Differences were considered to be significant when p < 0.05. *, p < 0.05.
Abbreviations
| GSTP1: | = | Glutathione S-?transferase P |
| HCC: | = | Hepatocellular carcinoma |
| MSP: | = | Methylation specific polymerase chain reaction |
| NEAT1_2: | = | Nuclear paraspeckle assembly transcript 1_2 |
| PRDX5: | = | Peroxiredoxin-5 |
| SFPQ: | = | splicing factor proline/glutamine-rich |
| STAT3: | = | Signal transducer and activator of transcription 3. |
Supplemental Material
Download MS Word (868.2 KB)Acknowledgments
We thank Prof. Chunmei Wang, Drs. Yiquan Xue, Qingzhu Shi, Yang Shi and Wanwan Huai for helpful discussions. This work was supported by grants from the National Key R&D program of China (2018YFA0507401), the National Natural Science Foundation of China (81672798, 81671564, 81701566), and the National Science and Technology Major Project of China (2017ZX10203206).
Supplementary Material
Supplementary data can be accessed here
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018 Jan;68(1):7–30. doi:10.3322/caac.21442. Epub 2018 Jan 4. PubMed PMID: 29313949.
- Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM, Karin M. Gender disparity in liver cancer due to sex differences in Myd88-dependent IL-6 production. Science. 2007;37(5834):121–124. PMID:17615358. doi:10.1126/science.1140485
- Bergmann J, Muller M, Baumann N, Reichert M, Heneweer C, Bolik J, Lucke K, Gruber S, Carambia A, Boretius S, et al. IL-6 trans-signaling is essential for the development of hepatocellular carcinoma in mice. Hepatology 2017. 2017;65(1):89–103. PMID:27770462. doi:10.1002/hep.28874
- Wan S, Zhao E, Kryczek I, Vatan L, Sadovskaya A, Ludema G, Simeone DM, Zou W, Welling TH. Tumor-associated macrophages produce interleukin 6 and signal via STAT3 to promote expansion of human hepatocellular carcinoma stem cells. Gastroenterology. 2014;147(6):1393–1404. PMID:25181692. doi:10.1053/j.gastro.2014.08039
- Lee H, Herrmann A, Deng JH, Kujawski M, Niu G, Li Z, Forman S, Jove R, Pardoll DM, Yu H. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell. 2009;15:283–293. PMID:19345327. doi:10.1016/j.ccr.2009.02.015
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE Jr. Stat3 as an oncogene. Cell. 1999;98:295–303. PMID:10458605.
- Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Kloppel G, Yoshimura A, Reindl W, Sipos B, Akira S. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19:456–469. PMID:21481788. doi:10.1016/j.ccr.2011.03.009
- Fukuda A, Wang SC, Morris JP 4th, Folias AE, Liou A, Kim GE, Akira S, Boucher KM, Firpo MA, Mulvihill SJ, et al. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell. 2011;19:441–455. PMID:21481787. j.ccr.2011.03.002.
- Yang X, Liang L, Zhang XF, Jia HL, Qin Y, Zhu XC, Gao XM, Qiao P, Zheng Y, Sheng YY, et al. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology. 2013;58(1):158–170. PMID:23398948. doi:10.1002/hep.26305
- Wang H, Su X, Yang M, Chen T, Hou J, Li N, Cao X. Reciprocal control of miR-197 and IL-6/STAT3 pathway reveals miR-197 as potential therapeutic target for hepatocellular carcinoma. Oncoimmunology. 2015;4(6):e1031440. PMID:26451302. doi:10.1080/2162402X.2015.1008371
- Khorkova O, Hsiao J, Wahlestedt C. Basic biology and therapeutic implications of lncRNA. Adv Drug Deliv Rev. 2015;87:15–24. PMID:26024979. doi:10.1016/j.addr.2015.05012
- Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, Qin L, Wu X, Zheng Y, Yang Y, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19(2):232–243. PMID:21316602. doi:10.1016/j.ccr.2011.01.001
- Li Z, Zhang J, Liu X, Li S, Wang Q, Chen D, Hu Z, Yu T, Ding J, Li J, et al. The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat Commun. 2018;20:9(1):1572. PMID:29679004. doi:10.1038/s41467-018-04006-0
- Yuan JH, Liu XN, Wang TT, Pan W, Tao GF, Zhou WP, Wang F, Sun SH. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 2017;19(7):820–832. PMID:28553938. doi:10.1038/ncb3538
- Wong CM, Tsang FH, Ng IO. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol. 2018;15(3):137–151. PMID:29317776. doi:10.1038/nrgastro.2017.169
- Klingenberg M, Matsuda A, Diederichs PT. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67(3):603–618. PMID:28438689. doi:10.1016/j.jhep.2017.04.009
- Wang Y, He L, Du Y, Huang G, Luo J, Yan X, Ye B, Li C, Xia P, Zhang G, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16(4):413–425. PMID:25842979. doi:10.1016/j.stem.2015.03.003
- Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. PMID:24768205. doi:10.1016/j.ccr.2014.03.010
- Zhu P, Wang Y, Huang G, Ye B, Liu B, Wu J, Du Y, He L, Fan Z. lnc-β-Catm elicits EZH2-dependent β-catenin stabilization and sustains liver CSC self-renewal. Nat Struct Mol Biol. 2016;23(7):631–639. PMID:27239797. doi:10.1038/nsmb.3235
- Chen ZZ, Huang L, Wu YH, Zhai WJ, Zhu PP, Gao YF. LncSox4 promotes the self-renewal of liver tumour-initiating cells through Stat3-mediated Sox4 expression. Nat Commun. 2016;7:12598. PMID:27553854. doi:10.1038/ncomms12598
- Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T. Alternative 3ʹ-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31(20):4020–4034. PMID:22960638. doi:10.1038/emboj.2012.251
- Naganuma T, Hirose T. Paraspeckle formation during the biogensis of long non-coding RNAs. RNA Biol. 2013;10(3):456–461. PMID:23324609. doi:10.4161/rna.23547
- Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell. 2005;16:5304–5315. PMID:16148043. doi:10.1091/mbc.E05-06-0587
- West JA, Mito M, Kurosaka S, Takumi T, Tanegashima C, Chujo T, Yanaka K, Kingston RE, Hirose T, Bond C, et al. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J Cell Biol. 2016;214(7):817–830. PMID:27646274. doi:10.1083/jcb.201601071
- Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35(4):467–478. PMID:19716791. doi:10.1016/j.molcel.2009.06.027
- Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53(3):393–406. PMID:24507715. doi:10.1016/j.molcel.2014.01.009
- Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106(8):2525–2530. PMID:19188602. doi:10.1073/pnas.0807899106
- Li R, Harvey AR, Hodgetts SI, Fox AH. Functional dissection of NEAT1 using genome editing reveals substantial localization of the NEAT1_1 isoform outside paraspeckles. RNA. 2017;23(6):872–881. PMID:28325845. doi:10.1261/rna.059477.116
- Jiang L, Shao C, Qj W, Chen G, Zhou J, Yang B, Li H, Gou LT, Zhang Y, Wang Y, et al. NEAT1 scaffolds RNA-binding proteins and the microprocessor to globally enhance pri-miRNA processing. Nat Struct Mol Biol. 2017;24(10):816–824. PMID:28846091. doi:10.1038/nsmb.3455
- Lee CJ, Evans J, Kim K, Chae H, Kim S. Determining the effect of DNA methylation on gene expression in cancer cells. Methods Mol Biol. 2014;1101:161–178. PMID:24233782. doi:10.1007/978-1-62703-1_9
- Zhao S, Zhong Y, Fu X, Wang Y, Ye P, Cai J, Liu Y, Sun J, Mei Z, Jiang Y, et al. H3K4 methylation regulates LPS-induced proinflammatory cytokine expression and release in macrophages. Shock. 2018. [Epub ahead of print. PMID:29570119. doi:10.1097/SHK.0000000000001141.
- Krishnan S, Horowitz S, Trievel RC. Structure and function of histone H3 lysine 9 methyltransferases and demethylases. Chembiochem. 2011;12(2):254–263. PMID:21243713. cbic.201000545.
- Wang Z, Fan P, Zhao Y, Zhang S, Lu J, Xie W, Jiang Y, Lei F, Xu N, Zhang Y. NEAT1 modulates herpes simplex virus-1 replication by regulating viral gene transcription. Cell Mol Life Sci. 2017;74(6):1117–1131. PMID:27783096. doi:10.1007/s00018-016-2398-4
- Anantharaman A, Jadaliha M, Tripathi V, Nakagawa S, Hirose T, Jantsch MF, Prasanth SG, Prasanth KV. Paraspeckles modulate the intranuclear distribution of paraspeckle-associated Ctn RNA. Sci Rep. 2016;6:34043. PMID:27665741. doi:10.1038/srep34043
- Kanwal R, Pandey M, Bhaskaran N, Maclennan GT, Fu P, Ponsky LE, Gupta S. Protection against oxidative DNA damage and stress in human prostate by glutathione S-transferasse P1. Mol Carcinog. 2014;53(1):8–18. PMID:22833520. doi:10.1002/mc.21939
- MacLennan GT, Eisenberg R, Fleshman RL, Taylor JM, Fu P, Resnick MI, Gupta S. The influence of chronic inflammation in prostatic carcinogenesis: a 5-year followup study. J Urol. 2006;176(3):1012–1016. PMID:16890681. j.juro.2006.04.033.
- Tu J, Zhao Z, Xu M, Lu X, Chang L, Ji J. NEAT1 upregulates TGF-β1 to induce hepatocellular carcinoma progression by sponging has-mir-139-5p. J Cell Physiol. 2018. Epub ahead of print. PMID:29797561. doi:10.1002/jcp.26524
- Choi HI, Ma SK, Bae EH, Lee J, Kim SW. Peroxiredoxin 5 protects TGF-β induced fibrosis by inhibiting Stat3 activation in rat kidney interstitial fibroblast cells. PloS One. 2016;11(2):e0149266. PMID:26872211. doi:10.1371/journal.pone.0149266
- Mowel WK, Kotzin JJ, McCright SJ, Neal VD, Henao-Mejia J. Control of immune cells homeostasis and function by lncRNAs. Trends Immunol. 2018;39(1):55–69. PMID:28919048. doi:10.1016/j.it.2017.08.009
- Liu X, Liang Y, Song R, Yang G, Han J, Lan Y, Pan S, Zhu M, Liu Y, Wang Y, et al. Long non-coding RNA NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular carcinoma proliferation. Mol Cancer. 2018;17(1):90. PMID:29764424. doi:10.1186/s12943-018-0838-5
- Zheng X, Zhang Y, Liu Y, Fang L, Li L, Sun J, Pan Z, Xin W, Huang P. HIF-2α activated lncRNA NEAT1 promotes hepatocellular carcinoma cells invasion and metastasis by affecting the epithelial-mesenchymal transition. J Cell Biochem. 2018;119(4):3247–3256. PMID:29091312. doi:10.1002/jcb.26481
- Guo S, Chen W, Luo Y, Ren F, Zhong T, Rong M, Dang Y, Feng Z, Chen G. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int J Clin Exp Pathol. 2015;8(5):5395–5402. PMID:26191242. eCollection2015.
- Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumors, immune cells and microorganisms. Nat Rev Cancer. 2013;13(11):759–771. PMID:24154716. doi:10.1038/nrc3611
- He G, Karin M. NF-kB and STAT3 – key players in liver inflammation and cancer. Cell Res. 2011;21(1):159–168. PMID:21187858. doi:10.1038/cr.2010.183
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA, NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. PMID:19217333. doi:10.1016/j.molcel.2009.01.026
- Yamazaki T, Hirose T. The building process of the functional paraspeckle with long non-coding RNAs. Front Biosci(Elite Ed). 2015;1(7):1–41. PMID:25553361.
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. PMID:16239143. doi:10.1016/j.cell.2005.08.033
- Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S, Benard M, Fox AH, et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell. 2014;25:169–183. PMID:24173718. doi:10.1091/mbc.E13-09-0558
- Yu H, Lee H, Herrmann A, Buettner R, JoveR. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–746. PMID:25342631. doi:10.1038/nrc3818
- Morchikh M, Cribier A, Raffel R, Amraoui S, Cau J, Severac D, Dubois E, Schwartz O, Bennasser Y, Benkirane M. HEXM1 and NEAT1 long non-coding RNA form a muti-subunit complex that regulates DNA-mediated innate immune response. Mol Cell. 2017;67(3):387–399. PMID:28712728. doi:10.1016/j.molcel.2017.06.020
- Schnekenburger M, Karius T, Diederich M. Regulation of epigenetic traits of the glutathione S-transferase P1 gene: from detoxification toward cancer prevention and diagnosis. Front Pharmacol. 2014;16(5):170. PMID:25076909. doi:10.3389/fphar.2014.00170
- Zhong S, Tang MW, Yeo W, Liu C, Lo YM, Johnson PJ. Silencing of GSTP1 gene by CpG island DNA hypermethylation in HBV-associated hepatocellular carcinomas. Clin Cancer Res. 2002;8(4):1087–1092. PMID:11948118.
- Ruscoe JE, Rosario LA, Wang T, Gate L, Arifoglu P, Wolf CR, Henderson CJ, Ronai Z, Tew KD. Pharmacologic or genetic manipulation of glutathione S-transferase P1-1 (GSTpi) influences cell proliferation pathways. J Pharmacol Exp Ther. 2011;298:339–345. PMID:11408560.
- Wu Y, Fan Y, Xue B, Luo L, Shen J, Zhang S, Jiang Y, Yin Z. Human glutathione S-transferase P1-1 interacts with TRAF2 and regulates TRAF2-ASK1 signals. Oncogene. 2006;25(42):5787–5800. PMID:16636664. sj.onc.1209576.
- Kong KH, Takasu K, Inoue H, Takahashi K. Tyrosine-7 in human class Pi glutathione S-transferase is important for lowering the pKa of the thiol group of glutathione in the enzyme-glutathione complex. Biochem Biophys Res Commun. 1992;184(1):194–197. PMID:1567427.
- Kou X, Chen N, Feng Z, Luo L, Yin Z. GSTP1 negatively regulates Stat3 activation in epidermal growth factor signaling. Oncol Lett. 2013;5(3):1053–1057. PMID:2346146. doi:10.3892/ol.2012.1098
- Wang C, Wang Q, Xu X, Xie B, Zhao Y, Li N, Cao X. The methyltransferase NSD3 promotes antiviral innate immunity via direct lysine methylation of IRF3. J Exp Med. 2017;214(12):3597–3610. PMID:29101251. doi:10.1084/jem.20170856
- Wang P, Xu J, Wang Y, Cao X. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science. 2017;24;358(6366):1051–1055. PMID:29074580. doi:10.1126/science.aao0409
