ABSTRACT
Stat3 is constitutively activated in several tumor types and plays an essential role in maintaining their malignant phenotype and immunosupression. To take advantage of the promising antitumor activity of Stat3 targeting, it is vital to understand the mechanism by which Stat3 regulates both cell autonomous and non-autonomous processes. Here, we demonstrated that turning off Stat3 constitutive activation in different cancer cell types induces senescence, thus revealing their Stat3 addiction. Taking advantage of the senescence-associated secretory phenotype (SASP) induced by Stat3 silencing (SASP-siStat3), we designed an immunotherapy. The administration of SASP-siStat3 immunotherapy induced a strong inhibition of triple-negative breast cancer and melanoma growth associated with activation of CD4 + T and NK cells. Combining this immunotherapy with anti-PD-1 antibody resulted in survival improvement in mice bearing melanoma. The characterization of the SASP components revealed that type I IFN-related mediators, triggered by the activation of the cyclic GMP-AMP synthase DNA sensing pathway, are important for its immunosurveillance activity. Overall, our findings provided evidence that administration of SASP-siStat3 or low dose of Stat3-blocking agents would benefit patients with Stat3-addicted tumors to unleash an antitumor immune response and to improve the effectiveness of immune checkpoint inhibitors.
Introduction
Stat3 has been recognized as an oncogene as its activation promotes cellular transformation.Citation1 Stat3 constitutive activation contributes to cell survival, angiogenesis, and proliferation in a large variety of cancer types.Citation2,Citation3 In addition, Stat3 also controls innate and adaptive antitumor immune responses by the production of immunosuppressive factors that exert a “feed-forward” mechanism activating Stat3 in different immune cell subtypes.Citation4 Indeed, it drives the differentiation of myeloid-derived suppressor cells (MDSCs) and regulatory T (Tregs) cells and hampers NK cell activation and CD8 + T cell cytoxicity.Citation5,Citation6 Conversely, blockade of Stat3 activation in cancer cells induces an increase in pro-inflammatory mediators that leads to antitumor immunity.Citation4,Citation7 In line with these findings, we previously demonstrated that immunization with Stat3-blocked breast cancer cells induces an antitumor immune response involving CD4 + T and NK cells.Citation8 However, the effect of Stat3 inhibition non-autonomous activity in vivo and the molecular mechanism that regulates the production of cytokines/chemokines able to reinstate immunosurveillance have not been addressed.
We have previously shown that Stat3 blockade in murine breast cancer models in vitro, induces cellular senescence.Citation8 Cellular senescence is a stable cell growth arrest that can be induced by replicative exhaustion, chemotherapy or radiotherapy, or oncogenic stimuli, among others.Citation9-Citation12 Another source of cellular senescence is related to oncogene addiction: cancer cells frequently become “addicted to” the constitutive activation or overexpression of a single oncogene to preserve their malignant phenotype.Citation13 Blockade of oncogene addiction can reactivate cellular senescence, a process known as oncogene inhibition-induced senescence (OIIS).Citation14 Senescent cells produce a secretory profile composed mainly of growth factors, cytokines, and proteinases, a signature termed senescence-associated secretory phenotype (SASP). The SASP can either foster tumorigenesis or stimulate the innate and adaptive antitumor immune response, a process called senescence immunosurveillance.Citation15,Citation16 Thus, we hypothesized that Stat3 blockade in cancer cells triggers an OIIS program able to secrete mediators that induce senescence immunosurveillance.
In this work, we report that targeting Stat3 in Stat3-addicted cancer cells turns them senescent and produces a SASP able to unleash an antitumor immune response against established 4T1 murine triple-negative breast cancer and B16 melanoma. This SASP overcomes melanoma resistance to antiprogrammed cell death protein 1 (PD-1) antibody. We identify type I IFN-induced factors as the main components responsible for the immunosurveillance properties of the SASP and disclose the underlying mechanism of their production.
Materials and methods
Reagents and antibodies
Detailed information regarding the reagents and antibodies used in this study is provided in Supplem. Table 1.
Animals and tumors
Experiments were carried out with virgin female BALB/c mice, raised at the Institute of Biology and Experimental Medicine of Buenos Aires and with male C57BL/6 purchased from The Charles River Laboratories, and were maintained in pathogen-free conditions. All animal studies were conducted in accordance with the highest standards of animal care as outlined by the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals and were approved by the Institute of Biology and Experimental Medicine and Institute Curie Animal Research Committee. Progestin-dependent ductal tumor line C4HD was originated in mice treated with 40 mg medroxyprogesterone acetate (MPA; Gador) every 3 months for 1 year and were maintained by serial transplantation in animals treated with 40 mg s.c. depot–MPA in the opposite flank to tumor inoculum.Citation8
Cell lines and siRNA transfections
T-47D, BT-474, MDA-MB-468, MDA-MB-231, CT-26, HuVEC, YAC-1, and NCI-N87 cells were obtained from the American Type Culture Collection and JIMT-1 cells from the German Resource Center for Biological Material. KPL-4 and cells were kindly provided by Dr. J. Kurebayashi (Kawasaki Medical School, Kurashiki, Japan). B16F10-OVA (B16) melanoma cell line was kindly provided by Dr. K. Rock (University of Massachusetts Medical School, Boston, MA), and MCA101-OVA (MCA101) was kindly provided by Dr. Clothilde Théry (Institut Curie, PSL Research University INSERM U932, Paris, France). Primary cultures of epithelial cells from C4HD tumors growing in MPA treated mice were performed as described previously.Citation8 Experiments and cell maintenance were performed in DMEM:F12 (1:1 v/v) or RPMI1640 medium (Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Internegocios). Cells were routinely tested for Mycoplasma contamination.
siRNAs were synthesized by Dharmacon. Experiments were performed with at least two different siRNA sequences for each protein, but we here presented results obtained with only one of them. A control siRNA oligonucleotide from Dharmacon that does not target any known mammalian gene was used as a negative control. siRNAs were used at a final concentration of 100 nmol/L and were transfected with Dharmafect1 reagent for 48 h according to the manufacturer’s specifications and as we previously described.Citation17
Conditioned medium
4T1, B16, MCA101, and MDA-MB-231 cells plated in 100-mm dishes were transiently transfected with the indicated siRNA or treated with 100 mmol/L doxorubicin (Abbot, Buenos Aires, Argentina). Then, cells were washed three times with PBS and cultured for 24 h with serum-free medium. After incubation, the conditioned medium (CM) was collected and centrifuged at 400 g. This CM was mixed with a complete medium in a proportion of 1 to 1 for in vitro studies. For in vivo administration, CM was collected, filtered through a 0.2-µm pore filter, concentrated with spin-columns of 3kD cutoff (Merck Millipore) and lyophilized. The lyophilized CM was embedded in Elvax particles (DuPont)Citation18 and implanted s.c. as a pellet to the animals.
Tumor experiments
Immunization protocol: BALB/c mice or C57BL/6 animals were injected s.c. with 1 × 104 4T1 or 1 × 105 B16 cells, respectively. When tumors reached 50 mm3, animals were immunized with 1 × 106 4T1 or B16 irradiated cells s.c. Simultaneously, a pellet of CM-siStat3 or CM-siControl was implanted s.c. This immunization was repeated three times according to the detailed experimental condition. In the indicated experiments, anti-PD-1 antibody (10 mg/kg) was administered i.p. Animals were monitored, and tumor width (W) and length (L) were measured with a Vernier caliper three times a week and tumor volume (mm3) was calculated as (L × W2)/2. Endpoint criteria for the survival studies included tumor volume exceeding 1000 mm3 or tumor ulceration.
Treatment with JSI-124: BALB/c or C57BL/6 animals was injected s.c. with 1 × 104 4T1 or 1 × 105 B16 cells, respectively. When tumors reached 100 mm3, animals were treated daily with 1 mg/kg JSI-124 i.p. Animals were monitored, and the tumor volume was measured three times a week.
Evaluation of pulmonary metastasis
BALB/c mice bearing 4T1 tumors were sacrificed, lungs were fixed in Bouin solution, and the number of superficial lung colonies was counted by an investigator who was blind to the experimental arm.
Preparation of cell suspensions
Subcutaneous tumors were collected and cut into small pieces and placed into 2.5 mL of CO2 independent medium (Gibco, Thermo Fisher Scientific, Waltham, MA) containing 0.1 mg/mL DNase I and 0.1 mg/ml Liberase TL. After mechanical dissociation, samples were incubated with shaking at 37°C for 30 min. The cell suspension was then filtered with a 70-μm cell strainer (BD Biosciences) for direct tumor cell analysis and further separated on a Percoll gradient (GE Healthcare Life Sciences) from 40% to 75% interface to recover mononuclear cells for immune cell infiltrate analysis.
Spleens were removed and dissected into small pieces and single-cell suspensions were obtained by mechanical disruption. A red blood cell lysis (5 min, room temperature) was performed using 1 ml ammonium–chloride–potassium lysis buffer.
Flow cytometry
Surface staining was performed at 4°C with the indicated antibodies resuspended in PBS with 2% FBS and 2 mM EDTA. All intracellular staining was performed using intracellular Fixation/Permeabilization buffer set (eBioscience/Thermo Fisher) according to the manufacturer’s instructions. Live-cell detection was performed using the Zombie Aqua Fixable Viability Kit (Biolegend). Flow cytometry was performed on an LSR Fortessa or Canto II flow cytometer (BD Biosciences) and analyzed using FlowJo software (version 10x, BD Biosciences).
NK cell degranulation assay
For degranulation evaluation, splenocytes were cultured alone or with YAC-1 (1x104 cells) overnight at 37°C. During the last 4 h, CD107a antibody was added. Cells were harvested and stained with CD3 and CD49b antibodies to gate NK cells (CD3-CD49b+ cells) and to determine the percentage of CD107a+ cells by flow cytometry.
Detection of Ag-specific T cells by Enzyme-Linked ImmunoSpot Assay (ELISPOT)
IFNϒ-producing antigen-specific CD8 T cells were measured by ELISPOT. Briefly, microplates (MAIPS4510; Merck Millipore) were coated with antimurine IFNϒ (Diaclone). Peripheral blood mononuclear cells (0.3×106) were cultured overnight in the presence of either control medium or class I–restricted OVA-I peptide (257–264, SIINFEKL, 10 mM). Cells and peptides were resuspended in complete medium RPMI supplemented with 10% FBS, 2 mM L-glutamine (Life Technologies), 1% penicillin/streptomycin (Life Technologies) and ß-mercaptoethanol. Detection of IFNϒ-producing cells was performed with biotinylated anti-IFNϒ (matched pairs; Diaclone) followed by streptavidin–alkaline phosphatase (Mabtech) and revealed using the appropriate substrate (Bio-Rad). Spots were counted using an ELISPOT Reader System ELR02 (AID Diagnostika). Results were expressed as the number of cytokine-producing cells per 1 × 106 of total cells.
Cytokine production
For detecting cytokine and chemokine secretion, the CM-siStat3 and CM-siControl form 4T1 and B-16-OVA cells were evaluated using Proteome Profiler Mouse XL Cytokine Array developed according to the manufacturer’s protocol (R&D Systems).
Evaluation of cytokine production (CXCL10, CCL2, CCL5, IL-15, IL-1β, IL-6 and CXCL2) was quantified by sandwich ELISA using paired cytokine-specific antibodies according to the manufacturer’s instructions.
T cell activation and proliferation assays
Splenocytes were isolated from healthy BALB/c or C57BL/6 mice and T cells were purified by negative selection using EasySep™ (Stemcell Technologies) Mouse T Cell Isolation Kit according to the manufacturer’s specifications. For activation and differentiation assay, T cells were stimulated with CD3/CD28 beads (Gibco, Thermo Fisher Scientific) for 72 h in 96-well plates using a 0.1:1 beads:lymphocyte ratio, and cells were stained with the indicated antibodies. For proliferation assay, stimulation with CD3/CD28 beads was performed for 72 h in 96-well plates using a 0.5:1 beads:lymphocyte ratio. In the last 6 h, a 0.5 μCi [3H] thymidine (NEN, DuPont; specific activity: 20 Ci/mmol) was added. The assay was performed by quadruplicate as described previously.Citation8 In some cases, antibodies anti-CCL2, CCL5, CXCR3, IL-15, and IFNAR were added at the beginning of the experiment.
C4HD, 4T1, MDA-MB-231, CT26, B16, MCA101, T-47D, and HuVEC cells were culture during 48 h, and a pulse of 0.5 μCi [3H] thymidine was added in the last 16 h. In other set of experiments, tumor cells were transfected with siRNA Stat3 with or without siRNA p21CIP1 or p16INK4a for 48 h.
Wound healing
A wound assay was employed to study the effect of CM-siStat3 of tumor cell on its cell migration. Wounds 400 μm wide were made in confluent monolayers. Cells were treated with CM-siControl or CM-siStat3 obtained from the corresponding cell line and allowed to migrate into the cell‐free area. At 18 h, cells were viewed and photographed with an inverted Olympus CKX41 microscope with 10×/0.25 objective using Olympus camera Q-Color 5. The cell-free wound area was quantified using ImageJ software (National Institutes of Health).
Sa-β-gal staining
Two methods were performed to assess the SA-β-gal activity at pH 6.0. Transfected cells grown on glass coverslips or frozen tumor tissues (10-μm thickness) mounted on slides were washed in PBS, fixed in 3% formaldehyde, and washed again in PBS and incubated 16 h at 37°C without CO2 with fresh SA-β-gal staining solution: 1 mg/mL 5-bromo-4-chloro-3-indolyl b-D-galactopyranoside (X-Gal) (Sigma Aldrich), 150 mM NaCl, 2 mM MgCl2, 40 mM citric acid/Na phosphate buffer (pH 6), 5 mM potassium ferrocyanide, and 5 mM potassium ferricyanide. The stained cells and tissues were viewed and photographed with an inverted Olympus CKX41 microscope with 20X/0.4 objective using Olympus camera Q-Color 5. For the second method, we used 5-dodecanoylaminofluorescein di-β-d-galactopyranoside, obtained from Sigma-Aldrich as a substrate to detect SA-β-gal activity. Briefly, cells grown on 12-well plates were pretreated with 100 nmol/L bafilomycin A1 (Sigma-Aldrich) for 1 h at 37°C and 5% CO2 humidified environment and then were incubated with 33 μmol/L C12FDG for another 2 h. Cells were washed with PBS and analyzed by flow cytometry with the Canto II flow cytometer. The flow cytometry data were analyzed with FlowJo software.
Immunofluorescence detection of trimethyl K9 histone H3
Cells grown in glass coverslips were fixed and permeabilized with ice-cold methanol and were then blocked in PBS 1% bovine serum albumin (BSA). Trimethyl K9 histone H3 was detected using a rabbit mAb (Merck Millipore), followed by incubation with antirabbit IgG-Alexa 488 (Molecular Probes). Negative controls were carried out using PBS instead of primary antibodies. Nuclei were detected by propidium iodide (PI) staining (5 μg/ml). Cells were analyzed using a Nikon Eclipse E800 confocal laser microscopy system using a 60x/1.4 oil immersion objective. Quantitative analysis of confocal images was performed with ImageJ. We evaluated the integrated fluorescence intensity value for Trimethyl K9 histone H3 for 50 cells from each condition and obtained an average value. The nuclear compartment was defined according to the PI images. Fluorescence background (median) was subtracted in all cases.
Western blot analysis
Cells were lysed in a buffer containing 50mM Tris (pH 7.4), 150mM NaCl, 1mM EDTA, 1mM EGTA, 10% glycerol, 1% Nonidet P-40, 0.1% SDS, 1 mM Mg2Cl, 1 mM phenylmethylsulfonylfluoride (PMSF), 10μg/ml leupeptin, 5 μg/ml pepstatin, 5μg/ml aprotinin,1 mM sodium orthovanadate and 25mM NaF. Lysates were centrifuged at 12,000 × g for 30 min at 4°C and protein content in the supernatant was determined using a Bradford Proteins were solubilized in sample buffer (60 mM Tris–HCl, pH 6.8, 2% SDS, 10% glycerol, 0.7mM, 2β mercaptoethanol, and 0.01% bromophenol blue) and subjected to SDS-PAGE. Proteins were electroblotted onto nitrocellulose. Membranes were immunoblotted with the antibodies shown in Supplem. Table 1. Enhanced chemiluminescence was performed according to the manufacturer’s instructions (GE Healthcare). Signal intensities of phosphor-proteins were analyzed by densitometry using ImageJ software and normalized to total protein bands. Experiments assessing total protein content were also repeated three to five times and signal intensities were normalized to β-tubulin bands, used as loading control.
Proteomic studies
SILAC labeling, secretome collection 4T1 cells were cultured for 8 passages in SILAC media Arg- and Lys-free DMEM supplemented with “light” (12C614N2-Lys and 12C614N4-Arg) or “heavy” (12C615N2 Lys and 12C615N4-Arg) isotopes. The stable isotope labeling was confirmed by LC-MS/MS after protein in-gel separation and digestion of blue bands.
4T1 cells labeled with “light” media were transfected with Control siRNA and cells labeled with “heavy” media were transfected with Stat3 siRNA. After 48 h of transfection, cells were washed 5 times with PBS and cultured for another 24 h with serum-free medium.
For secretome collection, three secretomes of independent experiments were collected (total volume: 5 mL/condition), samples mixed in a 1:1 ratio and filtered through a 0.2 µm syringe filter. Samples were concentrated to 500 µl using centrifugal filtration units with 3 kD molecular weight cutoff.
Trypsin digestion Secretomes mixed at a 1:1 ratio were analyzed after a combination of FASP and fractionation through strong anion exchange separation. Ninety percent of each secretome was diluted in 500 µL of 25 mM ammonium bicarbonate before reduction with 5 mM dithiothreitol at 37°C for 1 h and alkylation with 10-mM iodocacetamide for 30 min at RT in the dark. Samples were then processed by a FASP procedure using 3kD Nanosep devices (Pall), according to standard protocols. Briefly, samples were loaded into the filtration devices and centrifuged at 13,000 g for 25 min. 500 µL of 25-mM ammonium bicarbonate was added and concentrated again. This step was repeated twice. The resulting concentrate was diluted to 200 µL with 25-mM ammonium bicarbonate and 2 µg trypsin/LysC was added. After overnight incubation at RT, peptides were collected by centrifugation of the filter units for 5 min. Strong anion exchange separation-based fractionation of peptides was performed as described.Citation19 The six pH eluted fractions were loaded onto a homemade C18 SepPak-packed stage tip for desalting (principle by stacking one 3M Empore SPE Extraction Disk Octadecyl (C18) and beads from SepPak C18 Cartidge Waters into a 200-µL micropipette tip). Desalted samples were reconstituted in injection buffer (2% MeCN, 0.3% TFA) before LC-MS/MS analysis.
LC-MS/MS analysis Online LC was performed with an RSLCnano system (Ultimate 3000, Thermo Scientific) coupled online to an Orbitrap Fusion Tribrid mass spectrometer (MS, Thermo Scientific). Peptides were trapped on a C18 column (75-μm inner diameter × 2 cm; nanoViper Acclaim PepMapTM 100, Thermo Scientific) with buffer A (2/98 MeCN/H2O in 0.1% formic acid) at a flow rate of 4.0 µL/min over 4 min. Separation was performed on a 50 cm × 75 μm C18 column (nanoViper Acclaim PepMapTM RSLC, 2 μm, 100Å, Thermo Scientific) regulated to a temperature of 55°C with a linear gradient of 5% to 25% buffer B (100% MeCN in 0.1% formic acid) at a flow rate of 300 nl/min over 100 min. Full-scan MS was acquired in the Orbitrap analyzer with a resolution set to 120,000, and ions from each full scan were HCD fragmented and analyzed in the linear ion trap.
Data Processing and Protein Identification Data were searched against the UniProtKB/Swiss-Prot Mus musculus database using SequestHT through Thermo Scientific Proteome Discoverer (v 2.1). The mass tolerances in MS and MS/MS were set to 10 ppm and 0.6 Da, respectively. We set carbamidomethyl cysteine, oxidation of methionine, N-terminal acetylation, heavy 13C615N2-Lysine (Lys8) and 13C615N4-Arginine (Arg10), medium 2H4-Lysine (Lys4), and 13C6-Arginine (Arg6) as variable modifications. We set specificity of trypsin digestion and allowed two missed cleavage sites.
The resulting files were further processed by using myProMS (v 3.5).Citation20 The Sequest HT target and decoy search result were validated at 1% false discovery rate with Percolator. For SILAC-based protein quantification, peptides XICs (Extracted Ion Chromatograms) were retrieved from Thermo Scientific Proteome Discoverer. Global MAD normalization was applied on the total signal to correct the XICs for each biological replicate (n = 3). Protein ratios were computed as the geometrical mean of related peptides. To estimate ratio significance, a t-test was performed with the R package limmaCitation21 and the false discovery rate has been controlled, thanks to the Benjamini-Hochberg procedureCitation22 with a threshold set to 0.05. (All quantified proteins have at least two peptides quantified.) Data are available via ProteomeXchange with identifier PXD013948.Citation23 Secreted proteins were identified by Secretome P and Signal P databases using “The Human Cancer Secretome Database”Citation24 and MetaSecKB (http://bioinformatics.ysu.edu/secretomes/animal/index.php).
TCGA data preparation
RNA-seq, Microarray and iTRAQ expression datasets for Breast Cancer were downloaded from TCGACitation25 using TCGA-AssemblerCitation26,RCitation27 software pipeline. Only primary tumor samples were kept, for a total 1095 RNA-seq samples, 528 Microarray samples, and 105 iTRAQ samples. Out of those 105 iTRAQ samples, 28 were discarded (77 were kept) after a variance quality control.Citation28 Data were then normalized so that the expression had similar distributions across samples.
Samples were classified into breast-cancer intrinsic subtypes according to PAM50 molecular signature,Citation29 using PBCMCCitation30 R package with 10000 permutations, a 0.01 significance threshold, and a 0.1 correlation cutoff. To match TCGA data with the protein annotation obtained from myProMSoutput, MyGene.infoCitation31 annotation services were used.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 6.0. The identity of the statistical test performed, p values and n values are stated in the figure legends. Longitudinal tumor growth data were analyzed using two-way ANOVA to assess overall group differences followed by Tukey post hoc comparisons. Comparison of the number of lung metastasis among different groups was done by the nonparametric Mann–Whitney U test. Survival was analyzed using the Kaplan-Meier model and log-rank Mantel-Cox test were used to compare two treatment groups. Differential expression analysis between breast cancer intrinsic subtypes in TCGA data was carried out using limma R packageCitation21 between basal-like and the other subtypes (Luminal A, Luminal B and Her2-enriched) and p values were adjusted using Benjamini-Hochberg procedureCitation22 with a threshold set to 0.05. Parallelisms between CM-siStat3 treatment and breast cancer subtypes were studied through contingency tables and Pearson’s chi-squared test. A p value < 0.05 was accepted as statistically significant.
Results
Stat3 silencing induces senescence in Stat3-addicted cancer cells
To investigate the underlying cancer cell characteristics that dictate whether it can undergo senescence by Stat3 blockade, we silenced Stat3 by siRNA transfection in several cancer cell lines (Supplem. Figure 1(a)). Stat3 knockdown induced a sharp rise in senescence-associated ß-galactosidase (SA-ß-gal) activity and in heterochromatin formation trimethyl K9 histone H3, markers of cellular senescence,Citation32,Citation33 in HER2-positive breast cancer cells, triple-negative breast cancer, colon cancer, and melanoma () and Supplem. Figure 1(b,c)). However, no modulation of SA-ß-gal was observed in fibrosarcoma MCA101 cells and in the luminal breast cancer BT-474 and T-47D cells () and Supplem. Figure 1(b)). Strikingly, all cell lines that experienced senescence after Stat3 knockdown had high levels of Stat3 phosphorylation at Tyr705. In contrast, senescence was absent in the cell lines in which Stat3 phosphorylation was barely detected ()). Proliferation of C4HD, 4T1, MDA-MB-231, CT-26, and B16-OVA (hereafter abbreviated B16) cells was largely decreased after Stat3 silencing; meanwhile, MCA101 and T-47D cell proliferation was not affected ()). Furthermore, senescence induced by Stat3 knockdown was dependent on the cyclin-dependent kinase inhibitors p16INK4 or p21CIP1 (Supplem. Figure 1(d–f)).Citation34
Figure 1. Stat3 silencing induces senescence in cancer cells addicted to Stat3 signaling pathway
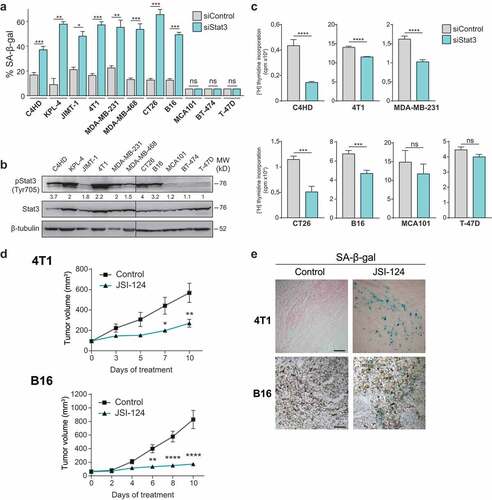
To study whether Stat3 inhibition-induced senescence also takes place in vivo, mice bearing 4T1 or B16 tumors were treated with the pharmacological inhibitor of Stat3 activation, JSI-124. Notably, the growth of both tumors was greatly suppressed by JSI-124 treatment ()). The number of micrometastasis was decreased in mice bearing 4T1 tumor treated with the Stat3 inhibitor vs control counterpart (Supplem. Figure 2(a)). We observed strong SA-ß-gal staining, and blockade of Stat3 phosphorylation in 4T1 and B16 tumors under JSI-124 treatment () and Supplem. Figure 2(b)). Altogether, our results demonstrate that Stat3 inhibition promotes a senescence program in vitro and in vivo in cancer cells that depend on Stat3 activation to maintain their malignant phenotype, thus revealing that they are Stat3-addicted. Therefore, Stat3 should be considered an oncogene whose inactivation leads to OIIS, as it has been demonstrated for MYC, RAS, and BCR-ABL1.Citation35,Citation36
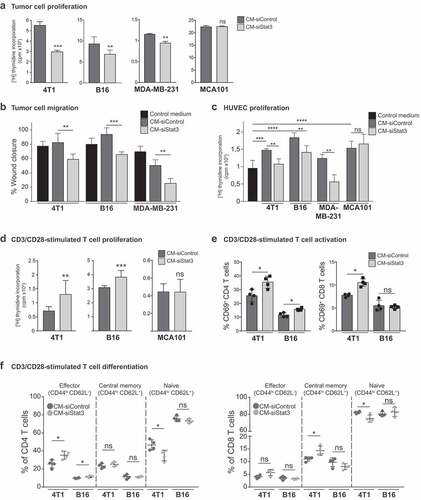
SASP from Stat3-silenced cells has anti-tumor and immune-stimulating activity in vitro
To evaluate the paracrine activity of the SASP induced by Stat3 silencing (SASP-siStat3) we used different cells of the tumor microenvironment to perform functional assays. To test this, we obtained the conditioned medium (CM) from 4T1, B16 and MDA-MB-231 cells transfected with Stat3 siRNA (CM-siStat3) or with control siRNA (CM-siControl). CM from MCA101 cells that did not undergo senescence upon Stat3 silencing ()) was included as control. We observed that 4T1, B16, and MDA-MB-231 cells cultured with CM-siStat3 exhibited a decrease in proliferation and migration when compared to cells cultured with CM-siControl (,b), respectively). When studying CM-siStat3 angiogenic potential, we revealed that CM-siStat3 from 4T1, B16, and MDA-MB-231 cells do not increase HUVEC proliferation vs. CM-siControl ()). Finally, to investigate the immune-modulatory activities of the SASP-siStat3, we performed functional assays of murine T cells stimulated with anti-CD3/CD28-coated beads. CM-siStat3 from 4T1 and B16 cells enhanced T cell proliferation in comparison with CM-siControl ()). The CM-siStat3 from MCA101 cells showed similar behavior than CM-siControl in the above-mentioned assays ()). As other control, we evaluated therapy-induced senescence by doxorubicin treatment (Supplem. Figure 3(a,b)). The CM from 4T1 and B16 cells treated with doxorubicin (CM-Doxorubicin) did not modify CD3/CD28-induced T cell proliferation vs control medium (Supplem. Figure 3(c)). Activation of CD4 + T cells, determined by CD69 expression, increased in the presence of CM-siStat3 vs CM-siControl from 4T1 and B16 cells ()). In order to track the fate of naïve T cells exposed to CM-siStat3, CD62L, and CD44 expression was determined. CM-siStat3 from 4T1 and B16 cells induced a significant increase of effector cells, a decrease of naïve cells and no change in central memory CD4 + T cell subpopulation vs CM-siControl ()). The CD8 + T cell subpopulation analysis revealed a significantly skew toward central memory cells and away from naïve cells in CM-siStat3 vs CM-siControl only from 4T1 cells ()). Collectively, these results indicate that the SASP-siStat3 has multiple paracrine antitumor effect, impairing tumor cell proliferation, migration, and angiogenic activity, while enhancing T cell proliferation and CD4 + T cell differentiation to an effector phenotype.
Figure 2. Antitumor activity of SASP from Stat3-silenced cancer cells
SASP-siStat3 is an adjuvant for a cellular cancer immunotherapy
The numerous in vitro antitumor effects of the SASP induced by Stat3 silencing described above encouraged us to study its application as a potential immunotherapy. Then, the CM-siStat3 and CM-siControl from 4T1 cells were concentrated, lyophilized, and embedded in Elvax particles.Citation18 We designed a therapeutic administration protocol consisting of immunizations with wild type 4T1 irradiated tumor cells simultaneously with the administration of CM-siStat3 (SASP-siStat3 immunotherapy) or CM-siControl ()). In animals treated with CM-siStat3, tumors grew strikingly more slowly and had fewer lung metastases than in the CM-siControl group (,c)). CM-siStat3 immunized animals showed similar leukocyte and T-cell tumor infiltration to the CM-siControl group ()), but CM-siStat3 immunized animals showed an increased percentage of effector CD4 + T cells (but not CD8 + T cells) in tumor and spleen ()). NK cells from CM-siStat3 immunized animals also showed an increase in activation, measured by CD69 staining, both in tumor and spleen, compared to the CM-siControl group ()). In regard to suppressor immune cells, CM-siStat3 immunized animals exhibited a significant decrease of Treg cell proportions in the tumor (but not in the spleen) ()). The presence of monocytic MDSC (M-MDSC) was lower in the tumor of CM-siStat3 vs CM-siControl group, while for the granulocytic (MDSC) G-MDSC population, a trend was observed ()). The antitumor effect of the SASP-siStat3 immunotherapy was dependent on the concomitant presence of an antigen source, provided by the irradiated cells, since the immunization with the CM-siStat3 alone did not impact tumor growth (data not shown).
Figure 3. Immunotherapy of SASP from Stat3-silenced 4T1 cells with irradiated wild type cells promotes an antitumor immune response
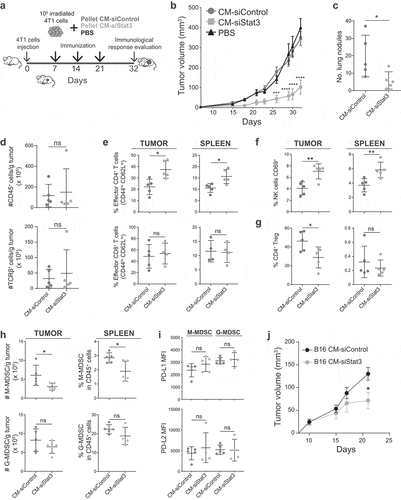
To evaluate the clinical applicability of the treatment, we had analyzed the potential use of CM-siStat3 from cancer cells as an universal preparation suitable for treating different tumors. Thus, we injected 4T1 tumor-bearing mice with CM-siStat3 from a different cell type, the melanoma B16 cells, together with the irradiated 4T1 cells. ) shows that CM-siStat3 from B16 decreases 4T1 tumor growth with respect to animals treated with CM-siControl from B16. These results suggest the CM-siStat3 from cancer cells can have therapeutic potential.
We further evaluated the effectiveness of our immunotherapy protocol in the B16 preclinical model ()). We observed a strong decrease of tumor growth in mice immunized with B16 irradiated cells and CM-siStat3 ()) and an increase in the survival of these mice ()), when compared to CM-siControl. To improve the efficacy of SASP-siStat3 immunotherapy, we used the checkpoint blocking antibody against PD-1 ()). Notably, anti-PD1 antibody plus the CM-siStat3 combination showed a trend toward suppressed B16 tumor growth and synergistically improved mice survival with respect to the CM-siStat3 alone (,c)). In addition to the experiments testing tumor growth ()), tumor regression was only observed in the combination group (2 out of 12 animals). It is noteworthy that anti-PD-1 monotherapy did not provide a measurable effect on survival or antitumor effect, nor did its combination with CM-siControl (,c)). In addition, PD-L1 levels did not change in B16 cells cultured with CM-siStat3 with respect to CM-siControl (Supplem. Figure 4(a)). Similar results were found in 4T1 cells and tumors (Supplem. Figure 4(a,b)).
Figure 4. Immunotherapy with SASP from Stat3-silenced cancer cells acts in synergy with anti-PD-1 antibodies
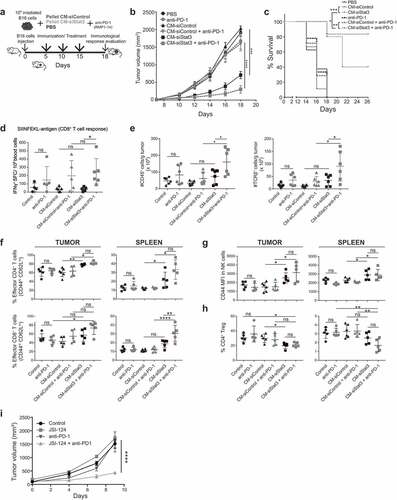
Administration of CM-siStat3 together with anti-PD1 antibody-induced antitumor specific CD8 + T cell response of about fivefold compared to CM-siStat3, measured by IFN-γ production ()). The CM-Control+anti-PD-1 group tends to have more IFN-γ production than CM-Control group making it likely that anti-PD-1 might account for this effect. In addition, the combination therapy revealed an influx in the tumor microenvironment of leukocytes and T cells compared with the CM-siStat3 alone and control groups ()). In accordance with the results obtained with 4T1 tumors, an increase in the proportion of effector CD4 + T cells in tumor and spleen was observed in the CM-siStat3 group, and this was in line with the results obtained in vitro ()). NK cells were also more activated in the tumors and spleen of mice treated with CM-siStat3 vs. CM-siControl ()). The addition of anti-PD-1 antibody to CM-siStat3 led to CD8 + T cell activation and differentiation to an effector phenotype in the tumor and spleen ()). A decrease in Tregs infiltrating the tumor was also evident in CM-siStat3 ()). Addition of anti-PD-1 antibody did not further decrease tumor-infiltrating Tregs. These data suggest that the SASP-siStat3 immunotherapy mobilized and activated CD4 + T and NK cells and acted in synergy with anti-PD-1 antibody, which, in turn, enhanced the expansion and activation of tumor-specific CD8 + T cells to obtain tumor clearance.
Then, we reasoned that combining a Stat3 inhibitor and PD-1 blockade could be another feasible antitumor strategy. Considering the toxicity of the Stat3 inhibitors in the clinical setting, we decided to administer JSI-124 every other day instead of every day ()) together with anti-PD-1; the latter at the same dose as described in that showed to be ineffective to control B16 tumor growth. We observed that JSI-124 or anti-PD-1 alone has no antitumor effect. However, the combination of JSI-124 and anti-PD-1 was able to significantly inhibit B16 tumor growth ()).
The SASP-siStat3 is enriched in chemokines and type I IFN-associated genes
To characterize the immunomodulatory composition of the SASP-siStat3, a multiplex membrane-based antibody array was used. We observed that 11 and 7 molecules were upregulated and 4 and 7 were downregulated in CM-siStat3 vs CM-siControl from 4T1 and B16 cells, respectively () and Supplem. Figure 5(a,b)). Among the downregulated cytokines, we detected vascular endothelial growth factor (VEGF), leukemia inhibitor factor (LIF) and regenerating islet-derived protein 3-gamma (Reg3G), all Stat3-regulated genes () and Supplem. Figure 5). In the repertoire of up-regulated factors in CM-siStat3 from 4T1 and B16 cells we observed an increase in IL-15 and the type I IFN-induced chemokines CXCL10/interferon inducible protein 10, C-C motif chemokine ligand 2 (CCL2) and CCL5 ()), which are potent T cell- and NK cell-activating cytokine/chemokines.Citation37 These results were confirmed by ELISA, while these cytokine/chemokine levels in CM-siStat3 from MCA101 cells were similar to those obtained from CM-siControl ()). The hallmark of SASPs induced by Ras activation, irradiation, or replicative senescence is the upregulation of IL-6, IL-1ß, and CXCL2/macrophage inflammatory protein 2, the mouse orthologous of human IL-8.Citation38 The ELISA determination of IL-1ß and IL-6 revealed that CM-siStat3 from 4T1, B16 and MCA101 cells had the same levels of these proinflammatory cytokines than CM-siControl ()). CXCL2 levels were downregulated in CM-siStat3 in 4T1 cells, and undetectable in B16 and MCA101 cells. On the other hand, the CM-Doxorubicin exhibits an increase in CXCL10 in B16 cells and of CCL5 and CXCL2 in 4T1 cells (Supplem. Figure 3(d)). These data highlight the striking divergence of the SASP induced by Stat3 silencing in contrast with senescence induced by other types of insults.
Figure 5. CCL2, CCL5, IL-15, and CXCL10 are mediators of the immune-stimulating activity of the SASP from Stat3-silenced cancer cells
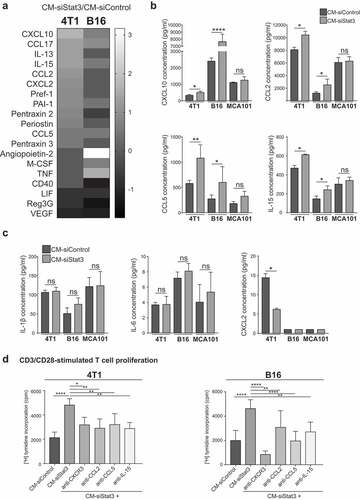
To identify which cytokine/chemokine that emerged from the antibody array studies mediates the senescence immunosurveillance activity of the SASP-siStat3, we performed neutralization assays on T cells stimulated with anti-CD3/CD28-coated beads. ) shows that blockade of CCL2, CCL5, IL-15 and CXCR3 (CXCL10 receptor) reduced T cell proliferation exerted by CM-siStat3 from 4T1 and B16 cells compared to CM-siControl levels. These data suggest that multiple factors secreted by Stat3-silenced cells promote T cell proliferation.
We next sought to further decode the secreted proteins of SASP-siStat3 by quantitative proteomic profiling using stable isotope labeling of amino acids in cell culture (SILAC)-based proteomics on CM-siControl and CM-siStat3 from 4T1 cells (Supplem. )). Among the 248 proteins found to be differentially regulated (greater than or equal to two-log fold change, p < .05) between CM-siStat3 and CM-siControl, 129 were upregulated and 119 were downregulated (Supplem. Figure 6(a) and Supplem. Table 2). The clustergram obtained using Wikipathway enrichment analysis on the upregulated and downregulated proteins identified processes involved in senescence and agingCitation39,Citation40 (Supplem. Figure 6(b)). Among the downregulated proteins we also found Stat3 and Stat3-regulated proteins such as CXCL1 (chemokine that recruits neutrophils and promotes angiogenesis) and CYR61 (growth and angiogenic factor) (Supplem. 6(b)), both proteins represented in the oncostatin/LIF signaling pathway.Citation41
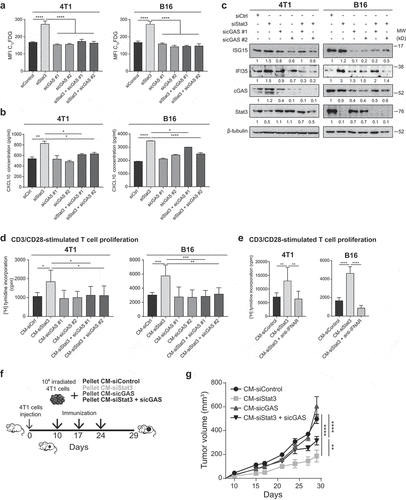
Figure 6. Senescence and type I IFN-related protein production induced by Stat3 silencing is dependent on cGAS
To further evaluate the clinical relevance of silencing Stat3, we studied whether the modulated proteins obtained from proteomic studies in CM-siControl compared to CM-siStat3 (CM-siStat3/CM-siControl) were also modulated in human basal-like subtype compared to other breast cancer subtypes. The comparison was centered in basal-like subtype because 60% of triple-negative breast carcinomas present that molecular profile. The analysis was carried out retrieving RNA-seq, Microarray and iTRAQ data from TCGA.Citation25 We observed a significant direct correlation between the up and downregulated proteins in CM-siStat3/CM-siControl and their transcriptomic (RNA-seq and Microarray) and proteomic levels (iTRAQ) in luminal A/basal-like (Supplem. Figure 6(c)). Basal tumors stand out as aggressive and highly metastatic, while luminal A tumors stand out as more benign,Citation29 therefore the protein regulation observed in CM-siStat3 is concordant to a shift toward a more benign phenotype.
The induction of senescence and of type I IFN response induced by Stat3 silencing relies on cGAS
Most of the upregulated cytokines/chemokines found in the SASP that have an important role in T cell proliferation belong to the type I IFN response ()). Indeed, proteomic data using Reactome showed that the term “immune system” was upregulated (P = .004) and IFN-induced protein 35 (IFI35) and IFN-stimulated gene 15 (ISG15) were represented.Citation42 Recently, it was demonstrated that chromatin fragments in the cytosol of senescent cells trigger the cytosolic DNA sensor cGAS, which produces the second messenger cyclic-GMP–AMP that activates stimulator of interferon gene protein (STING), thus engaging the production of inflammatory cytokines, chemokines, and type I IFNs.Citation43–Citation47 Then, we studied the potential contribution of cGAS on senescence induced by Stat3 silencing and on immunosurveillance characteristic of its SASP. We observed that silencing cGAS did not affect senescence in 4T1 and B16 cells measured by 5-dodecanoylaminofluorescein di-β-D-galactopyranoside staining ()). Blockade of cGAS expression completely inhibited senescence induced by Stat3 silencing. Remarkably, the production of type I IFN-related proteins, CXCL10, ISG15, and IFI35, produced by Stat3 depletion, was greatly compromised in the absence of cGAS (,c)). Then, we observed that CM-siStat3+ sicGAS from 4T1 and B16 cells completely inhibited the CD3/CD28 T cell proliferation exerted by CM-siStat3 ()) suggesting the involvement of type I IFN-related proteins in the stimulating activity in CM-siStat3. In addition, anti-IFN alpha receptor antibodies inhibited the ability of CM-siStat3 from 4T1 and B16 cells to enhance T cell proliferation ()). To study the contribution of cGAS pathway on the antitumor immune response elicited by CM-siStat3, we went back to the immunization protocol using 4T1 tumor model but now including CM-siStat3+ sicGAS ()). We observed that the simultaneous depletion of cGAS and Stat3 in 4T1 cells generates a CM that provides a partial protection from 4T1 tumor growth with respect to CM-siStat3 ()). The CM-sicGAS exhibited no antitumor effect ()). Together, these findings suggest that cGAS is a key player for senescence and for the expression of inflammatory mediators in Stat3-silenced senescent cells, thus implying its involvement in the generation of immunosurveillance upon targeting Stat3.
Discussion
Stat3 has long been recognized as an attractive target in oncology but, despite many efforts, chronic treatment of solid tumors with Stat3 inhibitory drugs in humans faced severe side effects. Therefore, to take advantage of the antitumor activity of Stat3 blockade, it is key to understand the mechanism by which Stat3 regulates both cell autonomous and non-autonomous processes, including host immunity. Here, we disclose that inhibition of Stat3 in cancer cells addicted to this oncogene induces cellular senescence and generates a SASP that has two main antitumor properties: one, linked to the downregulation of downstream targets of Stat3 that promotes angiogenesis and tumorigenesis; and the other, related to the up-regulation of mediators associated with the immunosurveillance properties of OIIS. Here, we prove that the SASP-siStat3 immunotherapy is able to inhibit breast cancer and melanoma growth and enhances the efficacy of anti-PD-1 antibody. In addition, we uncover that the cGAS DNA sensing pathway modulates the production of type I IFN-inducible genes triggered by Stat3 inhibition thus revealing a mechanism of the immunosurveillance properties of the SASP.
The SASP can have either pro or antitumor activities.Citation15,Citation48–Citation51 Seminal works of Campisi´s lab described that SASP from human fibroblast cell lines subjected to ionizing radiation, oxidative stress, oncogenic RAS expression, or loss of p53, had the ability to induce epithelial–mesenchymal transition and to stimulate invasion of tumor cells. In addition, chemotherapy-induced senescence can promote tumor clearance but the persistence of senescent cells can cause chronic inflammation, drug resistance, cancer relapse, and bone marrow suppression.Citation52,Citation53 Interestingly, tumor senescence induced by radiation, chemotherapy, and HER2 activation among others, has Stat3 activation as a hallmark, and Stat3-driven cytokines are the main mediators of the SASP with protumorigenic activity.Citation51,Citation54,Citation55 In contrast, here we targeted Stat3 and observed a strong downregulation of LIF, CXCL2, Reg3G, and VEGF, thus obtaining a SASP with a completely different composition from that described under the above-mentioned insults. Toso and colleagues demonstrated that the SASP of Pten-deficient prostate tumors has a protumorigenic and chemoresistant activity due to constitutive activation of the Jak2/Stat3 pathway.Citation56 In line with our findings, they “reprogramed” this SASP phenotype by blocking Jak2, and observed tumor regression when docetaxel was simultaneously administrated.Citation56 Based on these findings and our results, Stat3 inhibition could be an attractive strategy to reprogram the negative effects of the SASP induced by different insults. Our results highlight the potential translatability of the administration of CM-Stat3 from tumor cells in combination with radiotherapy, as well as low dose of Stat3 inhibitors together with anti-PD-1 strategies in the oncology arena. The observation that CM-siStat3 from B16 cells inhibits 4T1 tumor growth confirmed our previous data where we demonstrated that immunization with irradiated 4T1 cells transfected with a dominant negative plasmid of Stat3 (Stat3Y705F) inhibited CT-26 (colon cancer) and C4HD (ErbB-2+ breast cancer) tumor growth.Citation8 These data suggest that a “universal” CM could be obtained or formulated from Stat3-addicted tumors. In particular, the application of clinical grade CM is an active field of research in the area of regenerative medicine. A phase I trial based on administration of CM from mesenchymal stromal stem cells in multiple sclerosis proved to be safe.Citation57
The SASP from OIIS has been poorly explored. Data available show upregulation of the antitumor factors IFN-γ, TNFα, eotaxin, IL-5, and CCL5 and downregulation of the protumoral factors IL-6 and VEGF, in Myc-inactivated T-cell acute lymphoblastic lymphoma grown in immunocompetent animals.Citation49 The secretion of the above-mentioned cytokines and chemokines relied on the presence of CD4 + T cells, and tumor clearance was dependent on NK cell activity.Citation49 In agreement with this report, we have previously demonstrated that immunization with irradiated Stat3-blocked tumor cells triggers an antitumor immune response dependent on CD4 + T and NK cells.Citation8 Here, we re-create a similar antitumor immune response by using the SASP-siStat3 immunotherapy. Neutralization of CXCR3, CCL2, CCL5, and IL-15 proves that all these cytokines/chemokines are required for the enhanced activity of the SASP on T cell proliferation. Recently, it has been described that the novel cytokine-like 1a (CYTL1) overexpression reduces spontaneous lung metastasis reducing Stat3 phosphorylation in 4T1 tumor and in the lung tissue.Citation58 These findings lead to another strategy feasible to block Stat3. Interestingly, proteomic studies of the SASP also reveal the upregulation of the type I IFN-regulated genes IFI35 and ISG15. It has already been described that Stat3 inhibition in cancer cells upregulates type I IFN improving immunosurveillance.Citation4,Citation59 In particular, Yang and colleagues reported that type I IFNs were involved in the antitumor effect of a combined therapy of Stat3 inhibitors and chemotherapy.Citation59
Additionally, senescent cells have aberrant cytosolic chromatin fragments that trigger innate DNA recognition and induce type I IFN production through the activation of the cGAS–STING pathway.Citation43-Citation47 Our current work refines this scenario by including cGAS as one of the principal mediators of OIIS, the production of cytokines and chemokines and the antitumor effect of the CM of cancer cells upon Stat3 silencing. The knowledge of the interaction of PD-1 with PD-L1 activates a critical immune checkpoint leading to T cell inhibition has been exploited by the therapeutic intervention of antibodies against PD-1 or PD-L1.Citation60-Citation62 However, the clinical effectiveness of these antibodies relies on the presence of a T cell-infiltrated tumor microenvironment.Citation63 Our results obtained in the melanoma model unveil a novel therapeutic strategy for tumors with poor immune cell infiltration, by combining SASP-siStat3 immunotherapy with anti-PD-1 antibodies. We demonstrate that the SASP-siStat3 contains chemokines able to recruit T cells and activate T and NK cells in the tumor bed, allowing the anti-PD-1 effect.
Our findings highlight that Stat3 addiction confers vulnerabilities to cancer cells and that targeting Stat3 triggers a senescence program that unleashes an antitumor immune response mediated by its SASP. This can open a potential clinical application of our findings, through stratification of tumors by Stat3 activation levels. Cancer patients with constitutive activation of Stat3 are candidates for a short-term treatment with Stat3-blocking strategies prior to or along with immune checkpoint inhibitors.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download ()Acknowledgments
We thank A. Molinolo (UCSD, San Diego, CA) for his constant help and the mouse facility, mass spectrometry platform, and flow cytometry core from Institut Curie. We are grateful for V. Chiauzzi´s technical assistance. G. González from Radiation Oncology SA, Buenos Aires, Argentina, is acknowledged for his expert support in cell irradiation. We are grateful to Fundación René Baron and Fundación Williams for their institutional support to IBYME-CONICET.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE Jr. Stat3 as an oncogene. Cell. 1999;98:295–16. doi:10.1016/S0092-8674(00)81959-5.
- Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. doi:10.1038/sj.onc.1203527.
- Proietti CJ, Izzo F, Diaz Flaque MC, Cordo RR, Venturutti L, Mercogliano MF, De Martino M, Pineda V, Munoz S, Guzman P, et al. Heregulin co-opts PR transcriptional action via Stat3 role as a coregulator to drive cancer growth. Mol Endocrinol. 2015;29:1468–1485. doi:10.1210/me.2015-1170.
- Wang T, Niu G, Kortylewski M, Burdelya L, Shain K, Zhang S, Bhattacharya R, Gabrilovich D, Heller R, Coppola D, et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004;10:48–54. doi:10.1038/nm976.
- Vasquez-Dunddel D, Pan F, Zeng Q, Gorbounov M, Albesiano E, Fu J, Blosser RL, Tam AJ, Bruno T, Zhang H, et al. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J Clin Invest. 2013;123:1580–1589. doi:10.1172/JCI60083.
- Gotthardt D, Putz EM, Straka E, Kudweis P, Biaggio M, Poli V, Strobl B, Muller M, Sexl V. Loss of STAT3 in murine NK cells enhances NK cell-dependent tumor surveillance. Blood. 2014;124:2370–2379. doi:10.1182/blood-2014-03-564450.
- Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi:10.1038/nri1995.
- Tkach M, Coria L, Rosemblit C, Rivas MA, Proietti CJ, Diaz Flaque MC, Beguelin W, Frahm I, Charreau EH, Cassataro J, et al. Targeting Stat3 induces senescence in tumor cells and elicits prophylactic and therapeutic immune responses against breast cancer growth mediated by NK cells and CD4+ T cells. J Immunol. 2012;189:1162–1172. doi:10.4049/jimmunol.1102538.
- Johnson LM, Price DK, Figg WD. Treatment-induced secretion of WNT16B promotes tumor growth and acquired resistance to chemotherapy: implications for potential use of inhibitors in cancer treatment. Cancer Biol Ther. 2013;14:90–91. doi:10.4161/cbt.22636.
- Jones KR, Elmore LW, Jackson-Cook C, Demasters G, Povirk LF, Holt SE, Gewirtz DA. p53-Dependent accelerated senescence induced by ionizing radiation in breast tumour cells. Int J Radiat Biol. 2005;81:445–458. doi:10.1080/09553000500168549.
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi:10.1016/S0092-8674(00)81902-9.
- Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi:10.1016/0014-4827(65)90211-9.
- Weinstein IB. Cancer. Addiction to oncogenes–the achilles heal of cancer. Science. 2002;297:63–64. doi:10.1126/science.1073096.
- Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi:10.1016/S1097-2765(00)80367-6.
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi:10.1038/nature05529.
- Kang TW, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, Hohmeyer A, Gereke M, Rudalska R, Potapova A, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479:547–551. doi:10.1038/nature10599.
- Mercogliano MF, De Martino M, Venturutti L, Rivas MA, Proietti CJ, Inurrigarro G, Frahm I, Allemand DH, Deza EG, Ares S, et al. TNFalpha-induced mucin 4 expression elicits trastuzumab resistance in HER2-positive breast cancer. Clin Cancer Res. 2017;23:636–648. doi:10.1158/1078-0432.CCR-16-0970.
- Kumamoto T, Huang EK, Paek HJ, Morita A, Matsue H, Valentini RF, Takashima A. Induction of tumor-specific protective immunity by in situ langerhans cell vaccine. Nat Biotechnol. 2002;20:64–69. doi:10.1038/nbt0102-64.
- Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi:10.1038/nmeth.1322.
- Poullet P, Carpentier S, Barillot E. myProMS, a web server for management and validation of mass spectrometry-based proteomic data. Proteomics. 2007;7:2553–2556. doi:10.1002/pmic.200600784.
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi:10.1093/nar/gkv007.
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi:10.1016/s0166-4328(01)00297-2.
- Vizcaino JA, Csordas A, Del Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:11033. doi:10.1093/nar/gkv1145.
- Feizi A, Banaei-Esfahani A, Nielsen J. HCSD: the human cancer secretome database. Database. 2015. doi:10.1093/database/bav051.
- Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi:10.1038/ng.2764.
- Wei L, Jin Z, Yang S, Xu Y, Y Z, Ji Y. TCGA-assembler 2: software pipeline for retrieval and processing of TCGA/CPTAC data. Bioinformatics. 2018;34:1615–1617. doi:10.1093/bioinformatics/btx812.
- R: A Language and Environment for Statistical Computing. R Core Team organization = R Foundation for Statistical Computing. Vienna, Austria; 2018. https://www.R-project.org/
- Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, Wang X, Qiao JW, Cao S, Petralia F, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi:10.1038/nature18003.
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi:10.1200/JCO.2008.18.1370.
- Fresno C, Gonzalez GA, Merino GA, Flesia AG, Podhjacer OL, Llera AS, Fernández E. A novel non-parametric method for uncertainty evaluation of correlation-based molecular signatures: its application on PAM50 algorithm. Bioinformatics. 2017;33:693–700. doi:10.1093/bioinformatics/btw704.
- Xin J, Mark A, Afrasiabi C, Tsueng G, Juchler M, Gopal N, Stupp GS, Putman TE, Ainscough BJ, Griffith OL, et al. High-performance web services for querying gene and variant annotation. Genome Biol. 2016;17(1):1–7. doi:10.1186/s13059-016-0953-9.
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9367. doi:10.1073/pnas.92.20.9363.
- Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi:10.1016/S0092-8674(03)00401-X.
- Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al Regaiey K, Su L, Sharpless NE. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi:10.1172/JCI22475.
- Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, Sundberg CD, Bishop JM, Felsher DW. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102–104. doi:10.1126/science.1071489.
- Tharkar-Promod S, Johnson DP, Bennett SE, Dennis EM, Banowsky BG, Jones SS, Shearstone JR, Quayle SN, Min C, Jarpe M, et al. HDAC1,2 inhibition and doxorubicin impair Mre11-dependent DNA repair and DISC to override BCR-ABL1-driven DSB repair in Philadelphia chromosome-positive B-cell precursor acute lymphoblastic leukemia. Leukemia. 2018;32:49–60. doi:10.1038/leu.2017.174.
- Rauch I, Muller M, Decker T. The regulation of inflammation by interferons and their STATs. JAKSTAT. 2013;2:e23820. doi:10.4161/jkst.23820.
- Freund A, Orjalo AV, Desprez PY, Campisi J. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol Med. 2010;16:238–246. doi:10.1016/j.molmed.2010.03.003.
- Hernandez-Segura A, de Jong TV, Melov S, Guryev V, Campisi J, Demaria M. Unmasking transcriptional heterogeneity in senescent cells. Curr Biol. 2017;27:2652–2660. doi:10.1016/j.cub.2017.07.033.
- Uthus EO, Brown-Borg HM. Altered methionine metabolism in long living Ames dwarf mice. Exp Gerontol. 2003;38:491–498. doi:10.1016/S0531-5565(03)00008-1.
- Chen Y, Du XY. Functional properties and intracellular signaling of CCN1/Cyr61. J Cell Biochem. 2007;100:1337–1345. doi:10.1002/jcb.21194.
- de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912–920. doi:10.1189/jlb.69.6.912.
- Yang H, Wang H, Ren J, Chen Q, Chen ZJ. cGAS is essential for cellular senescence. Proc Natl Acad Sci U S A. 2017;114:E4612–E4620. doi:10.1073/pnas.1705499114.
- Dou Z, Ghosh K, Vizioli MG, Zhu J, Sen P, Wangensteen KJ, Simithy J, Lan Y, Lin Y, Zhou Z, et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature. 2017;550:402–406. doi:10.1038/nature24050.
- Gluck S, Guey B, Gulen MF, Wolter K, Kang TW, Schmacke NA, Bridgeman A, Rehwinkel J, Zender L, Ablasser A. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat Cell Biol. 2017;19:1061–1070. doi:10.1038/ncb3586.
- Galluzzi L, Vanpouille-Box C, Bakhoum SF, Demaria S. SnapShot: CGAS-STING Signaling. Cell. 2018;173:276. doi:10.1016/j.cell.2018.03.015.
- Vanpouille-Box C, Demaria S, Formenti SC, Galluzzi L. Cytosolic DNA sensing in organismal tumor control. Cancer Cell. 2018;34:361–378. doi:10.1016/j.ccell.2018.05.013.
- Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi:10.1371/journal.pbio.0060301.
- Rakhra K, Bachireddy P, Zabuawala T, Zeiser R, Xu L, Kopelman A, Fan AC, Yang Q, Braunstein L, Crosby E, et al. CD4(+) T cells contribute to the remodeling of the microenvironment required for sustained tumor regression upon oncogene inactivation. Cancer Cell. 2010;18:485–498. doi:10.1016/j.ccr.2010.10.002.
- Liu D, Hornsby PJ. Senescent human fibroblasts increase the early growth of xenograft tumors via matrix metalloproteinase secretion. Cancer Res. 2007;67:3117–3126. doi:10.1158/0008-5472.CAN-06-3452.
- Zacarias-Fluck MF, Morancho B, Vicario R, Luque GA, Escorihuela M, Villanueva J, Rubio IT, Arribas J. Effect of cellular senescence on the growth of HER2-positive breast cancers. J Natl Cancer Inst. 2015:107. doi:10.1093/jnci/djv020.
- Ewald JA, Desotelle JA, Wilding G, DF J. Therapy-induced senescence in cancer. J Natl Cancer Inst. 2010;102:1536–1546. doi:10.1093/jnci/djq364.
- Demaria M, O’Leary MN, Chang J, Shao L, Liu S, Alimirah F, Koenig K, Le C, Mitin N, Deal AM, et al. Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov. 2017;7:165–176. doi:10.1158/2159-8290.CD-16-0241.
- Ohanna M, Cheli Y, Bonet C, Bonazzi VF, Allegra M, Giuliano S, Bille K, Bahadoran P, Giacchero D, Lacour JP, et al. Secretome from senescent melanoma engages the STAT3 pathway to favor reprogramming of naive melanoma towards a tumor-initiating cell phenotype. Oncotarget. 2013;4:2212–2224. doi:10.18632/oncotarget.1143.
- Yu YC, Yang PM, Chuah QY, Huang YH, Peng CW, Lee YJ, Chiu SJ. Radiation-induced senescence in securin-deficient cancer cells promotes cell invasion involving the IL-6/STAT3 and PDGF-BB/PDGFR pathways. Sci Rep. 2013;3:1675. doi:10.1038/srep01675.
- Toso A, Revandkar A, Di Mitri D, Guccini I, Proietti M, Sarti M, Pinton S, Zhang J, Kalathur M, Civenni G, et al. Enhancing chemotherapy efficacy in Pten-deficient prostate tumors by activating the senescence-associated antitumor immunity. Cell Rep. 2014;9:75–89. doi:10.1016/j.celrep.2014.08.044.
- Dahbour S, Jamali F, Alhattab D, Al Radaideh A, Ababneh O, Al Ryalat N, Al Bdour M, Hourani B, Msallam M, Rasheed M, et al. Mesenchymal stem cells and conditioned media in the treatment of multiple sclerosis patients: clinical, ophthalmological and radiological assessments of safety and efficacy. CNS Neurosci Ther. 2017;23:866–874. doi:10.1111/cns.12759.
- Wang X, Li T, Cheng Y, Wang P, Yuan W, Liu Q, Yang F, Liu Q, Ma D, Ding S, et al. CYTL1 inhibits tumor metastasis with decreasing STAT3 phosphorylation. Oncoimmunology. 2019;8:e1577126. doi:10.1080/2162402X.2019.1577126.
- Yang H, Yamazaki T, Pietrocola F, Zhou H, Zitvogel L, Ma Y, Kroemer G. STAT3 inhibition enhances the therapeutic efficacy of immunogenic chemotherapy by stimulating type 1 interferon production by cancer cells. Cancer Res. 2015;75:3812–3822. doi:10.1158/0008-5472.CAN-15-1122.
- Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–1034. doi:10.1084/jem.192.7.1027.
- Wang C, Thudium KB, Han M, Wang XT, Huang H, Feingersh D, Garcia C, Wu Y, Kuhne M, Srinivasan M, et al. In vitro characterization of the anti-PD-1 antibody nivolumab, BMS-936558, and in vivo toxicology in non-human primates. Cancer Immunol Res. 2014;2:846–856. doi:10.1158/2326-6066.CIR-14-0040.
- Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi:10.1126/science.aaa8172.
- Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, Alaparthy S, Berman D, Jure-Kunkel M, Siemers NO, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother. 2012;61:1019–1031. doi:10.1007/s00262-011-1172-6.
