Abstract
Previous research has shown that host Cyclophilin A (CyPA) can promote dendritic cell maturation and the subsequent innate immune response when incorporated into an HIV-1 Gag protein to circumvent the resistance of dendritic cells to HIV-1 infection. This led us to hypothesize that CyPA may improve HIV-1 Gag-specific vaccine immunogenicity via binding with Gag antigen. The adjuvant effect of CyPA was evaluated using a DNA vaccine with single or dual expression cassettes. Mouse studies indicated that CyPA specifically and markedly promoted HIV-1 Gag-specific cellular immunity but not an HIV-1 Env-specific cellular response. The Gag/CyPA dual expression cassettes stimulated a greater Gag-specific cellular immune response, than Gag immunization alone. Furthermore, CyPA induced a broad Gag-specific T cell response and strong cellular immunity that lasted up to 5 months. In addition, CyPA skewed to cellular rather than humoral immunity. To investigate the mechanisms of the adjuvant effect, site-directed mutagenesis in CyPA, including active site residues H54Q and F60A resulted in mutants that were co-expressed with Gag in dual cassettes. The immune response to this vaccine was analyzed in vivo. Interestingly, the wild type CyPA markedly increased Gag cellular immunity, but the H54Q and F60A mutants drastically reduced CyPA adjuvant activation. Therefore, we suggest that the adjuvant effect of CyPA was based on Gag-CyPA-specific interactions. Herein, we report that Cyclophilin A can augment HIV-1 Gag-specific cellular immunity as a genetic adjuvant in multiplex DNA immunization strategies, and that activity of this adjuvant is specific, broad, long-term, and based on Gag-CyPA interaction.
Abbreviations
| AIDS | = | Acquired Immune Deficiency Syndrome |
| BMDC | = | Bone Marrow Derived Dendritic Cell |
| CyPA | = | Cyclophilin A |
| DC | = | Dendritic cell |
| EGFP | = | Enhanced Green Fluorescent Protein |
| ELISA | = | Enzyme-Linked Immunosorbent Assay |
| ELISPOT | = | Enzyme-Linked ImmunoSpot |
| SAMHD1 | = | SAM domain and HD domain-containing protein 1. |
Introduction
HIV viral proteins are known to activate host immune responses, and immune response levels are significantly associated with the progression of AIDS disease. More specifically, the anti-HIV-1 Gag antigen-specific cytotoxic T lymphocyte response plays an important role in delaying AIDS progression.Citation1,2 Previous studies have shown that Gag p24 protein-specific CD4+ T cells proliferated in long-term nonprogressors who did not undergo antiretroviral therapy.Citation3 In addition, the magnitude of the Gag CD4+ T cell response is specifically associated with HIV viral load.Citation4 Moreover, the Gag antigen-induced specific CD8+ T cell immune response significantly reduced HIV viral load levels, and enhanced broad Env-specific responses against HIV.Citation5 Accordingly, Gag-specific CD4+ and CD8+ T cells were resistant to HIV viral replication and effectively controlled the disease.Citation6-8 Therefore, how to improve Gag immunogenicity in HIV-1 vaccines is an important goal.
Cyclophilin A (CyPA) is a peptidyl-prolyl cis-trans isomerase, which could catalyze the cis-trans isomerization and accelerate the protein folding in the cytoplasm. Moreover, the interactions between CyPA and the Gag protein have been investigated for decades.Citation9-11 Previous studies demonstrated that CyPA was a specific host factor that was incorporated into HIV-1 virions but not into other primate immunodeficiency viruses (SIV and HIV-2).Citation9 CyPA was found to primarily bind to a critical proline residue in the N-terminal domain of the Gag protein (Pr55gag).Citation12 The detailed crystal structure of CyPA-Gag binding suggested that the sequence Ala88-Gly89-Pro90-Ile91 of the Gag fragment was the major portion to bind with the active site of CyPA, and that the Pro90-Ile91 residues were bound to 2 residues (Pro-Phe) of CyPA.Citation13 Researchers also found that the interaction of HIV-1 Gag with CyPA was necessary for the formation of infectious HIV-1 virions.Citation9,10 One recent report suggested that HIV-1 could induce dendritic cell maturation and a series of innate immune responses, including the induction of an antiviral type I interferon response, activation of the transcription factor IRF3, and stimulation of T cell responses.Citation14 Additionally, this activated immunity was triggered by the interaction of newly synthesized HIV-1 capsids with cellular CyPA.
In this study, we focused on the potential for CyPA to serve as a genetic adjuvant in HIV-1 Gag vaccines. Given the impact of CyPA incorporated with Gag on the immune response, we hypothesize that delivering Gag and CyPA into the same somatic cells should stimulate intracellular sensors, induce an interferon immune response, and assist Gag in promoting immunogenicity, subsequently promoting a systemic, adaptive immune response. We investigated the efficacy of combined CyPA DNA with an HIV-1 Gag DNA vaccine in inducing an immunological response against HIV-1, and demonstrated that CyPA can improve Gag vaccine immunogenicity in the context of DNA dual expression cassettes.
Results
CyPA specifically enhanced the HIV-1 Gag cellular immune response
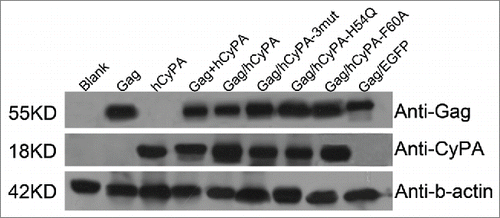
Western blot analysis confirmed the expression of the ∼55-kDa Gag protein and ∼18-kDa CyPA protein at comparable levels () in all of these constructed plasmids.
Figure 1. Vaccine antigens expressed in vitro. Each lane was loaded with the lysed proteins from transfected products. From left to right: blank control, p4.0-Gag, p4.0-hCyPA, p4.0-Gag + p4.0-hCyPA, p4.0-Gag/hCyPA, p4.0-Gag/hCyPA-3mut, p4.0-Gag/hCyPA-H54Q, p4.0-Gag/hCyPA-F60A, and p4.0-Gag/EGFP. All plasmids were transfected into 293T cells separately, and cells were harvested 48 h later for western blot analysis. Anti-Gag, anti-CyPA, and anti-β-actin antibodies were used to test each protein expression. Results are representative of 2 independent experiments.
To verify the species specificity of the CyPA adjuvant, the effects of mouse and human CyPA on Gag immunogenicity were compared. Mice co-immunized with either the mCyPA or hCyPA gene generated a stronger immune response than the Gag-immunized only group (). The nucleotide sequence homology between Mus musculus and Homo sapiens CyPA was as high as 95% (data not shown), which might explain the similar adjuvant effect of mouse and human CyPA.
Figure 2. Assessment of CyPA species-specificity and antigen-specificity of the adjuvant effect. The Gag-specific cellular immune response was assessed by IFN-γ ELISPOT array to investigate the difference between the mouse and human CyPA adjuvant effect when co-administrated with Gag vaccine (A). Ten 6- to 8-week-old female Balb/c mice in each group were administered vaccines under Scheme I in , with CyPA plasmid mixed with Gag vaccine before immunization, and a 50 μg DNA vaccine at a final volume of 100 μl each (50 μl for each tibialis anterior muscle) injected directly. Mice were immunized 3 times at 2-week intervals with Gag DNA plasmid co-formulated with CyPA adjuvant DNA. One week after the last vaccination, animals were sacrificed, and individual mouse splenocytes were isolated and used to assess cellular function by IFN-γ ELISPOT. The CyPA-Gag antigen specific adjuvant activation (B) was tested following Scheme II. We designed a multiplex comparison to verify the CyPA adjuvant effect on Gag antigen specificity. “Gag + hCyPA sep” denotes Gag and hCyPA plasmid injected separately into the tibialis anterior muscles of 2 legs. Moreover, Env DNA, an HIV antigen, was tested for specificity of CyPA adjuvant activation. All the procedures of immunization, timeline, interval time and operation were followed method and material session description. The p values are labeled with asterisks for p < 0.05 (*) and p < 0.0001 (****). In addition, N.S indicates the non-significant difference between the compared groups.
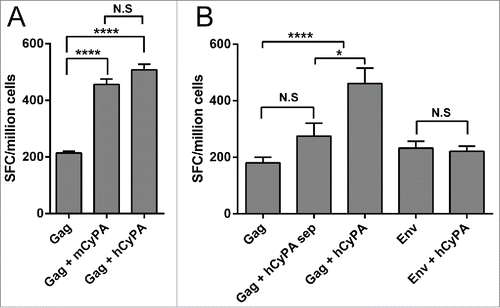
Table 1. The mouse immunization strategy
To confirm the specificity of the CyPA adjuvant effect in a mice model, we compared an HIV-1 Gag DNA vaccine with HIV-1 Env DNA vaccine co-immunized with CyPA (). The resulting cellular immune responses demonstrated that CyPA prompted Gag-specific but not HIV-1 Env-specific immunogenicity. The magnitude of CyPA-enhanced cellular immunity was approximately 2-fold higher when compared with the Gag-only immunized group. More interestingly, if Gag and CyPA DNA vaccines were inoculated into 2 separate legs, the CyPA-enhanced Gag DNA vaccine immune response disappeared. This result suggested that CyPA might exert its adjuvant effect and promote HIV Gag-specific immunogenicity due to physical interactions between CyPA and Gag.
Gag/hCyPA dual expression cassettes optimized the Gag cellular immune response and the adjuvant effect of hCyPA based on interaction with Gag
To optimize the procedure for DNA vaccine manufacturing and immunization, we constructed dual expression cassettes that included Gag and CyPA genes, which were under the control of 2 independent CMV promoters in a chimeric DNA vaccine. The results of ELISPOT assay showed that both the one-plasmid scheme and the pre-mixed 2-plasmid scheme increased the strength of the Gag-specific cellular immune response (). This result not only confirmed the effectiveness of the Gag/hCyPA dual expression cassette strategy but also confirmed the specificity of the CyPA adjuvant effect.
Figure 3. Gag/CyPA dual expression cassette strategy augments Gag cellular immunity and the CyPA mutant effect on adjuvant activation. To investigate the efficiency of the Gag/CyPA dual expression cassette vaccine and identify the mechanism of CyPA adjuvantivity on the Gag vaccine with CyPA mutants, the Gag-specific cellular immune response was assessed by IFN-γ ELISPOT array. Ten 6- to 8-week-old female Balb/c mice in each group were vaccinate according to Scheme III in . p4.0-Gag/hCyPA (68 μg) was used instead of a 50 μg dose to keep the same Gag gene copy number in the different vaccines, and was compared with 50 μg of p4.0-Gag DNA to assess the immunogenicity of the Gag/CyPA chimeric vaccine. Multi-site mutations of H54Q/R55A/F60A CyPA markedly induced a Gag-specific cellular immune response. To explore this finding, we determined the effect of 2 single-site CyPA mutants (H54Q and F60A) in a DNA vaccine. All mice were immunized 3 times at a 2-week intervals, and sacrificed one week after the last vaccination. The IFN-γ ELISPOT array was used to investigate the antigen-specific cellular immune response induced by the vaccination regimens. The p values were labeled with asterisks for p < 0.05 (*), p < 0.01(**), p < 0.001 (***), and p < 0.0001 (****).
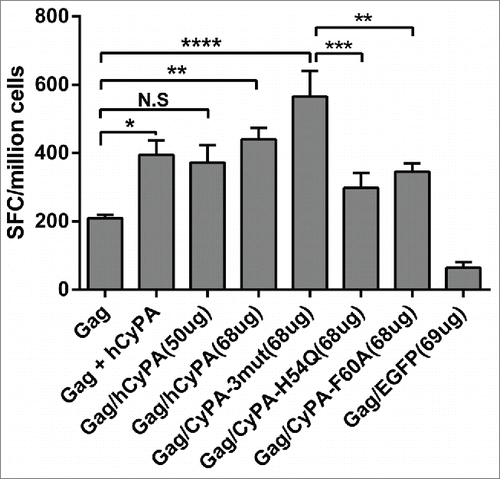
According to previous reports, CyPA active-site mutants with amino acid substitutions located in the Gag-CyPA binding loop could reduce the efficiency of interactions with Gag. Therefore, we selected several critical sites that were previously recognized to play key roles in the CyPA-Gag interaction. Our study suggested that 2 CyPA mutants (H54Q and F60A) were critical for the incorporation of CyPA into HIV-1 virions, and could drastically decrease the Gag-induced cellular immune response. However, multiple mutations (H54Q/R55A/F60A) in CyPA did not reduce its adjuvant effect in vivo (), but markedly and unexpectedly increased the Gag-specific cellular immunological response. The mechanism underlying this odd effect remains unexplained. However, when we compared the effects of vaccination with Gag/CyPA-3mut against the other vaccine groups, the Gag alone group and the CyPA single-site mutation groups showed a statistically significant difference in cellular response. Moreover, similar results were seen by multi-color flow cytometry assay (Fig. S1). The FACS data documented a CyPA adjuvant-stimulated CD8+ T cell response and showed that secreted functional anti-virus cytokines were not enhanced by the CD4+ T cell response.
CyPA induced a broad cellular immune response
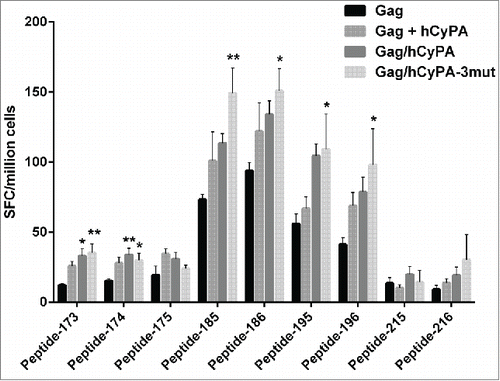
To determine the epitopes in the Gag antigen critical to stimulating an immune response, we designed a peptide mapping experiment using an ELISPOT array. Peptides, 121 in all, covering the entire length of the Gag protein, were split into 22 pools, with each pool containing 11 peptides. As the results showed (), mice vaccinated with Gag DNA alone failed to mount a broad T cell response in contrast to Gag/hCyPA or Gag+hCyPA-immunized mice. Interestingly, 8 pools (the 6th, 7th, 8th, 9th, 10th, 17th, 18th, and 20th pools) were recognized by T cells from immunized mice and induced robust responses with CyPA. Moreover, responses to the 6th, 7th, and 20th pools only appeared in the groups co-inoculated with CyPA. We narrowed down these epitopes in high level response peptide pools, and only 4 peptides (pep185, pep186, pep195, and pep196) provoked stronger Gag cellular responses than CyPA-free groups (). However, these epitopes did not localize to the Gag-CyPA binding loop (PVHAGPIAP).
Figure 4. Identification of dominant Gag epitopes in various vaccination strategies by ELISPOT mapping assay. The figure indicates the T-cell responses of splenocytes from immunized mice to 22 pools, each consisting of 11 Gag 15-aa peptides overlapping by 11-aa, that were measured by ELISPOT assay 2 weeks after the last of 3 DNA immunizations. The black box highlights pools (p value < 0.05) that induced significantly greater cellular immune responses in the CyPA groups, compared with the Gag-immunized only group. The dashed line at 20 spots indicates the cut-off value of this assay.
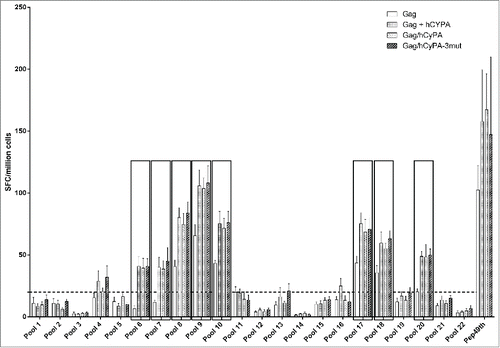
Figure 5. Individual epitopes identified by ELISPOT mapping following distinct immunization regimens. To narrow down the dominant epitopes determined by pool mapping, several individual peptides were selected through cross-referencing of Gag peptides mapped in the pool matrix (Table S1). The IFN-γ ELISPOT assay was performed to verify the efficiency of each single peptide in inducing Gag-specific cellular immunity. The asterisk denotes significant differences (* indicates p < 0.05 and ** indicates p < 0.01) compared with the Gag only group.
In our long-term assessment, both Gag/hCyPA and Gag/hCyPA-3mut chimeric vaccines induced cellular immune responses still showed protection at 150 days after the last immunization, but the protective effect was less in the 2-plasmid scheme (Gag + hCyPA). At 60 days post vaccination, the dual expression chimeric vaccines showed a trend toward greater cellular immune responses, but the difference was insignificant when compared with others (). Even up to 210 days post immunization, vaccination with the dual expression cassettes showed a trend of strong cellular immunity, but one that was not statistical different from that seen with the Gag only group.
Figure 6. Long-term immunity monitored up to 7 months after the last vaccination. Long-term cellular immune response detected by IFN-γ ELISPOT assay (A). We monitored the dynamic changes of vaccine-induced Gag cellular immunity at 15, 30, 60, 150, and 210 days after the last immunization. The asterisks indicate significant differences, with p < 0.05 (*) and p < 0.01(**), in comparison with the Gag only group at the same time point. Binding antibody titer analyzed by ELISA (B). The dynamic changes in vaccine-induced Gag-specific binding antibody were analyzed at 30, 60, 150, and 210 days after the last immunization in each vaccine group.
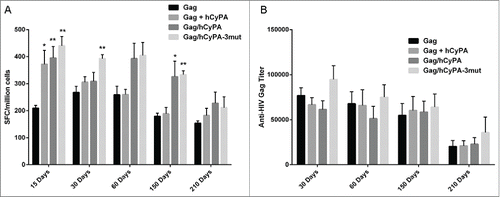
These results indicated that Gag combined with CyPA was indeed an effective approach to inducing broad and long-term T cell responses, and that the dual expression cassette strategy also resulted in long-lasting cellular immunity.
CyPA did not improve the Gag antibody titer
To determine whether the CyPA was effective in enhancing the Gag-specific humoral immune response, we tested the Gag-specific antibody titer by ELISA. Neither Gag co-immunized with hCyPA nor the Gag/hCyPA dual cassette vaccine could elicit high Gag-specific antibody titers when compared with Gag DNA alone (). Only the Gag/hCyPA-3mut chimera induced slightly but not significantly stronger humoral immunity than the hCyPA adjuvant-free scheme at day 30. Furthermore, up to half a year after the last immunization, the hCyPA adjuvant did not appear to promote a Gag-specific antibody response. These results demonstrated that CyPA promoted an excellent cellular and Gag-specific adjuvant effect, but that it did not impact the antibody response.
Discussion
In several HIV-1 vaccine preclinical or clinical trials, Gag antigen did not perform well.Citation15-17 When Gag was pre-mixed with HIV-1 Env antigen for co-immunization, it failed to stimulate robust Gag-specific immunity. In this study, we investigated the immunogenicity of a Gag/CyPA vaccine, which promoted a Gag-specific cellular immune response with broad and long-lasting protection.
Previously, some cytokines and immune-stimulation regulators had been developed as vaccine adjuvants. For example, several cytokines and chemokines, including IL-12, IL-28, GM-CSF, and IL-15, have demonstrated their ability to augment DNA vaccine potency.Citation18-23 Recombinant DNA vaccine adjuvants have included fusion constructs composed of the tumor necrosis factor (TNF) superfamily ligands, such as 4-1 BBL, OX40L, RANKL, LIGHT, CD70, and BAFF, that enhanced immune responses to a HIV-1 Gag DNA vaccine.Citation24 Mice immunized with a DNA plasmid expressing a LAMP/Gag chimera showed strong, broad cellular and humoral immune responses.Citation25-29 In our study, CyPA as genetic adjuvant augmented Gag-specific cellular immunity. The cellular immune response by CyPA was approximately 2–2.5 fold greater than that in the Gag only group, similar to the response observed with cytokine-based adjuvants.
We speculate that the underlying mechanism of CyPA adjuvant activation is the interaction of CyPA with Gag protein, which provokes downstream innate immunity signaling. Dendritic cells (DCs) induce protective immune responses through recognition of pathogen-conserved elements; however, they do not recognize HIV-1 as a dangerous invader. In fact, DCs encourage HIV-1 to infect neighboring CD4+ T cell by a trans enhancement process.Citation30 The absence of DC activation by HIV-1 is in part a consequence of this relatively poor ability to infect these cells, which is imposed by the restriction factor SAMHD1.Citation31-33 When HIV-1 is complemented with the Vpx protein, which overcomes the SAMHD1 restriction, DCs are readily infected.Citation34 Earlier studies have shown that Citation14 infected DCs recognize HIV-1 viral protein particles through an intracellular CyPA and IRF3 pathway that initiates the innate immune response to HIV-1, induces virus-specific T cell immunity, and promotes naïve multifunctional CD4+ T cell proliferation. Because CyPA is secreted in response to cellular stress, we investigated whether CyPA could shuttle Gag out of the cell and more efficiently target to dendritic cells. However, when Gag alone and Gag with CyPA were transferred into mammalian cells, Gag was no longer found in the supernatants of co-transfected cells (data not shown). According to these reports and observations, we determined that the mechanism underlying the CyPA adjuvant effect was CyPA/Gag bound complex initiation of downstream anti-viral pathways. Therefore, interaction between the newly synthesized Gag and the cellular CyPA was necessary to promote HIV-1 vaccine effectiveness and immunogenicity. Hence, the CyPA exogenously delivered by the DNA vaccine allowed the CyPA protein to accumulate in the cytoplasm and prompted its binding with Gag antigen.
A previous study showedCitation12 that 3 site mutations (H54Q, R55A, and F60A) in CyPA could dissociate the specific interactions with Gag protein. Therefore, it is logical to assume that mutations at these 3 sites might also interrupt the interactions. Surprisingly, Gag-3mut (H54Q, R55A, and F60A 3 sites mutant simultaneously) did not reduce Gag-specific cellular immunity; rather, it increased the strength, breadth, and duration of cellular immunity. Unfortunately, we could not identify whether these mutations at these 3 site in CyPA changed the spatial conformation of the protein. To verify the relationship between the CyPA mutant and its adjuvant activity, we incorporation CyPA with the single-site mutations H54Q and F60A in DNA chimeric vaccines. Interestingly, the single-site CyPA mutants reduced the Gag cellular immune response significantly, which was the hypothesized response based on previous research. This result, combined with the in vivo results of administering CyPA and Gag separately into 2 different legs, has given us confidence in concluding that CyPA plays a role in Gag antigen-specific stimulation of immune cells and pathways through Gag/CyPA interactions in vivo. However, we still need to validate the physical interaction between CyPA mutants and Gag protein in another study, either by molecular docking experiments to predicate this interaction or by glutathione S-transferase (GST)-pull down assays or protein-protein interaction technologies. Finally, we need to determine the correlation between the adjuvant effect and CyPA-Gag interaction.
To breadth the breadth and duration of the vaccine-induced immune responses, we utilized an ELISPOT mapping approach to identify novel epitopes and survey dynamic changes in cellular immunity after immunization. Finally, we showed that co-vaccination with CyPA could promote and enhance recognition of novel epitopes that were located in the C-terminus of Gag antigen. After an analysis of the protein alignment, we found the reactive peptides were not located in the Gag/CyPA binding loop region. This suggested that the broad cellular immunity we observed was not limited to this interaction site. From long-term monitoring, we found that cellular immunity induced by the dual expression cassette (Gag/CyPA chimeric vaccine) was stronger than that induced by the single and pre-mixed regimens. Up to 5 months after the last immunization, a sustained, high level of Gag-specific cellular immunity resulted from the dual cassette vaccine. This response was 2-fold higher than that seen in the Gag only and Gag pre-mixed with CyPA groups by ELISPOT assay. The Gag/CyPA dual-expression vaccine showed a trend toward greater immunogenicity than Gag+CyPA, but this difference was not significant. Based on this data, we speculate that the Gag and CyPA gene inserted into a single plasmid for co-expression of these genes will promote specific binding and subsequent innate signaling. The long-term immunity shown to be induced by the dual expression cassette suggests advantages of a shorter vaccination schedule, reduced number of shots, and optimized prime-booster potential, as well as decreased manufacturing costs, with a maximized cellular immune response.
Since this study was based on a Gag-CyPA specific interaction, Gag antigen sequence specificity is a potential restricting factor in the application of this strategy. In this study, we used HIV-1 subtype B/C strain CN54; however, it is unknown whether CyPA could serve as an adjuvant with other HIV-1 subtypes. The binding region (PVHAGPIPP) of Gag is a Pro-rich loop, and the critical amino acids AGPI are located in the Gag N-terminus. An amino acid alignment of HIV-1 types showed that the frequency of these 4 Pro residues in the binding region was 99.4%, 100%, 68.8%, and 98.8%, respectively. Moreover, the critical loop (AGPI) was also conserved; the frequency of each amino acid was 98.8%, 100%, 100%, and 84.4%, respectively. According to these results, the adjuvant effect of CyPA would not be restricted to B/C subtypes when co-administrated with Gag antigen, and should extend to most HIV-1 subtypes. Finally, it will be important to determine whether the enhancing effect of CyPA is additive or synergistic with other adjuvants such as IL-12 and GM-CSF. Gag-specific immunity could be increased when the adjuvants are combined.
CyPA can serve as a Gag antigen-specific genetic adjuvant in DNA vaccine formulations. The potential to increase Gag vaccine efficacy and duration of protection by co-formulation with other antigens in a polyvalent vaccine should be explored.
Materials and Methods
DNA vaccine construction
Single cassette plasmid
The eukaryotic expression vector pDRVI4.0, constructed by the Division of Research on Virology and Immunology (National Center for AIDS/STD Control and Prevention, China CDC), was used as the parental plasmid for constructing the expression vectors. The pDRVI4.0-Gag plasmid that encodes the Gag protein of HIV-1 strain CN54 and pDRVI4.0-EGFP that encodes green fluorescent protein were constructed previously by our laboratory.
Mouse CyPA (mCyPA) cDNA (GeneBank Ref. ID: NM_008907.1) was cloned from bone marrow-derived mature DCs (BMDC) according to a previously described approach.Citation35 Two primers were used for PCR amplification, 5´-AAAACTGCAGATGGTCAACCCCACCGTGTTCTTC-3′ (sense) and 5´-GCTCTAGATTAGAGCTGTCCACAGTCGGAAATG-3′ (antisense); and the reaction protocol comprised the following steps: 98°C for 10 min; 30 cycles of 98°C for 30 s, 59°C for 30 s, and 72°C for 1 min; and 72°C for 10 min. mCyPA was cloned into pDRVI4.0 using the PstI and XbaI restriction endonuclease sites.
The human CyPA (hCyPA) gene (GeneBank Ref. ID NM_021130.3) was synthesized by Invitrogen Corp., and was cloned into pDRVI4.0 using the PstI and XbaI restriction endonuclease sites.
Dual cassette chimeric plasmid
To generate the dual expression cassette, the hCyPA cassette containing the CMV promoter, SV40 enhancer, hCyPA gene, and BGH-pA signal elements was amplified by PCR from the parental pDRVI4.0-hCyPA plasmid, and the fragment was cloned into pDRVI4.0-Gag using the EcoRI restriction site. The primers used for hCyPA cassette amplification were 5´-TATTACTGTGACGTTGAATTCTGGTTGC-3′ (sense) and 5´-CCG GAATTCAGATCTGGATCCAACGTCGGTACC-3′ (antisense). The protocol employed for PCR amplification comprised the following steps: 94°C for 5 min; 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 2 min; and 72°C for 10 min.
Gag/CyPA mutant plasmid
Mutants were created from the pDRVI4.0-Gag/hCyPA parental plasmid, and according to a classic site-mutation protocol.Citation12 The plasmid pDRVI4.0-Gag/hCyPA was used as a template to create the following missense mutations in the hydrophobic pocket of CyPA, which contained the critical HIV binding domain (PVHAGPIAP). The H54Q/R55A/F60A mutation primer was 5´-GTTATAAGGGTTCCTGCTTTCAGGCAATTATTCCAGGGGCTATGTGTCAGGGTGG-3´, (underlining denotes the mutation sites). The protocol employed for PCR amplification comprised: 95°C for 30 s; 18 cycles of 95°C for 30 s, 55°C for 1min, and 68°C for 4.5 min; and 72°C for 10 min. The PCR products were added to 1µl of Dpn1 (10U/µl) to digest template DNA at 37°C for 1 h, and the resulting mutation plasmids were pDRVI4.0-Gag/hCyPA-3mut, pDRVI4.0-Gag/hCyPA-H54Q, and pDRVI4.0-Gag/hCyPA-F60A.
All plasmids were produced by transforming HB101 Escherichia coli (Life Technologies) and were purified using an endotoxin-free column (Qiagen).
Analysis of protein expression and interaction
Cell culture and transfections
The expression of each plasmid was verified by transfection into 293T cells with Lipofectamine 2000 according to the manufacturer's protocol (Life Technologies). Cells were harvested at 48 h post transfection and analyzed by the following assay.
Western blot analysis
Harvested cell extracts were prepared by lysis in RIPA buffer for 5 min on ice. Protein lysates (100 μg) were fractionated by 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), transferred to polyvinylidene difluoride (PVDF) membranes, and incubated with rabbit anti-HIV Gag antibody (dilution 1:10000) or mouse anti-Human CyPA antibody (final concentration of 2 μg/ml) (Abcam), followed by secondary antibodies (rabbit or mouse peroxidase conjugates). Protein expression was detected using a Substrate ECL system (Thermo Fisher Scientific).
Studies with mice
Ethics statement
All experiments were conducted in accordance with the guidelines of the Laboratory Animal Center of China CDC and the National Center for AIDS/STD Control and Prevention. All procedures for animal use and care were approved by the National Center for AIDS/STD Control and Prevention Institutional Committee on Laboratory Animals.
Immunization strategy
Groups of 6-week-old female BALB/C mice (Vital River Laboratories) were immunized 3 times at 2-week interval. shows the immunization timeline and strategies. One hundred micrograms of each plasmid was dissolved in a final volume of 100 μl of PBS (pH 7.2) and was injected intramuscularly. Because of the different sizes of pDRVI4.0-Gag (4412 bp) and pDRVI4.0-Gag/hCyPA (6004 bp), these plasmids needed to be normalized to same number of gene copies as 50 μg pDRVI4.0-Gag in the immunization regimen. We used 68 μg of pDRVI4.0-Gag/hCyPA and 69 μg of pDRVI4.0-Gag/EGFP in this scheme (). This calculation was based on the assumption that the average weight of a base pair (bp) is 650 Da. Using Avogadro's number, 6.022 × 10ˆ23 molecules/mol, the number of molecules of the template per gram were calculated: mol/g × molecules/mol = molecules/g. The formula used was number of copies = (amount × 6.022 × 10ˆ23) / (length × 10ˆ9 × 650).
HIV-1 Gag-specific IFN-γ ELISPOT assay
The ELISPOT HIV-1 Gag-specific T cell response assay was carried out according to the protocol provided by the manufacturer (BD ELISPOT Mouse IFN-γ ELISPOT). Briefly, 96-well plates were coated with 10 μg/ml of anti-mouse IFN-γ in sterile PBS and incubated at 4°C overnight. The plates were washed with phosphate buffered saline, and blocked by RPMI-1640 medium containing 10% fetal bovine serum at room temperature for 2 h. Splenocytes were seeded into wells at a concentration of 5×10ˆ5 cells/well with 100 μl of Gag peptide (at a final concentration of 5 μg/peptide/ml) (peptide sequence: AAMQILKDTINEEAA) or peptide pools (shown in the Table S1 mapping strategy) and maintained in a humidified 5% CO2 incubator at 37°C for 24 h. After incubation, the ELISPOT plates were developed according to the kit instructions. Finally, the spot-forming cells were quantified with a Bioreader-4000 automated ELISPOT reader (BioSys) and normalized to 10ˆ6 splenocytes.
The HIV-1 Gag peptide mapping pool is described in Table S1. A total of 121 single peptides covered the full-length Gag protein. Each peptide was a 15-mer with an overlapping 11 amino acids. The subsequent procedures were performed as described above.
Enzyme-linked immunosorbent assay for antibody response
Flat-bottom plates (96-well, Costar) were coated with purified recombinant p55 protein at a concentration of 0.5 μg/ml in carbonate buffer at 4°C overnight. The protein was expressed in 293F cells and adjusted to 95% purity. Plates were washed 5 times with phosphate buffered saline with Tween20 and blocked with 3% BSA in PBST at 37°C for 1 h. After five washings, 100 μ1 of serially diluted mouse serum was added to wells in duplicate. After 2 h of incubation at 37°C, the plates were washed 5 times with PBST and then incubated for 1 h with 1:10000 diluted HRP-labeled anti-mouse IgG antibody (Merck) at 37°C. After the final wash, plates were developed with 100 μl per well of fresh Tetramethylbenzidine (TMB) substrate (Sigma-Aldrich) for 15 min at room temperature. The reactions were stopped by adding 25 μl of 2 M H2SO4, and then the plates were read at OD 450 nm.
Statistical analysis
A one-way ANOVA analysis was used to compare experimental groups, and this was followed by non-pairwise multiple comparisons using a Newman-Keuls test. In all figures, p values are labeled by asterisks for p < 0.05 (*), p < 0.01(**), p < 0.001 (***), and p < 0.0001 (****). All statistical calculations were computed using Prism 6.0 Software (GraphPad Inc..).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
KHVI_A_1082692.docx
Download MS Word (117.7 KB)Funding
This work was supported by National Major projects for Infectious Diseases Control and Prevention (2012ZX10001008); National Natural Science Foundation of China (81020108030); State Key Laboratory for Infectious Disease Prevention and Control Development Grant (2011SKLID303); Young Scholar Scientific Research Foundation of China CDC (2012A105).
References
- Riviere Y, McChesney MB, Porrot F, Tanneau-Salvadori F, Sansonetti P, Lopez O, Pialoux G, Feuillie V, Mollereau M, Chamaret S, et al. Gag-specific cytotoxic responses to HIV type 1 are associated with a decreased risk of progression to AIDS-related complex or AIDS. AIDS Res Hum Retroviruses 1995; 11:903-7; PMID:7492437; http://dx.doi.org/10.1089/aid.1995.11.903
- Pontesilli O, Klein MR, Kerkhof-Garde SR, Pakker NG, de Wolf F, Schuitemaker H, Miedema F. Longitudinal analysis of human immunodeficiency virus type 1-specific cytotoxic T lymphocyte responses: a predominant gag-specific response is associated with nonprogressive infection. J Infect Dis 1998; 178:1008-18; PMID:9806028; http://dx.doi.org/10.1086/515659
- Rosenberg ES, Billingsley JM, Caliendo AM, Boswell SL, Sax PE, Kalams SA, Walker BD. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science 1997; 278:1447-50; PMID:9367954; http://dx.doi.org/10.1126/science.278.5342.1447
- Ranasinghe S, Flanders M, Cutler S, Soghoian DZ, Ghebremichael M, Davis I, Lindqvist M, Pereyra F, Walker BD, Heckerman D, et al. HIV-specific CD4 T cell responses to different viral proteins have discordant associations with viral load and clinical outcome. J Virol 2012; 86:277-83; PMID:22031937; http://dx.doi.org/10.1128/JVI.05577-11
- Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med 2007; 13:46-53; PMID:17173051; http://dx.doi.org/10.1038/nm1520
- Rosenberg ES, Walker BD. HIV type 1-specific helper T cells: a critical host defense. AIDS Res Hum Retroviruses 1998; 14 Suppl 2:S143-7; PMID:9672231
- Lichterfeld M, Kaufmann DE, Yu XG, Mui SK, Addo MM, Johnston MN, Cohen D, Robbins GK, Pae E, Alter G, et al. Loss of HIV-1-specific CD8+ T cell proliferation after acute HIV-1 infection and restoration by vaccine-induced HIV-1-specific CD4+ T cells. J Exp Med 2004; 200:701-12; PMID:15381726; http://dx.doi.org/10.1084/jem.20041270
- Geldmacher C, Currier JR, Herrmann E, Haule A, Kuta E, McCutchan F, Njovu L, Geis S, Hoffmann O, Maboko L, et al. CD8 T-cell recognition of multiple epitopes within specific Gag regions is associated with maintenance of a low steady-state viremia in human immunodeficiency virus type 1-seropositive patients. J Virol 2007; 81:2440-8; PMID:17182686; http://dx.doi.org/10.1128/JVI.01847-06
- Franke EK, Yuan HE, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 1994; 372:359-62; PMID:7969494; http://dx.doi.org/10.1038/372359a0
- Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh CT, Sodroski J, Gottlinger HG. Functional association of cyclophilin A with HIV-1 virions. Nature 1994; 372:363-5; PMID:7969495; http://dx.doi.org/10.1038/372363a0
- Gamble TR, Vajdos FF, Yoo S, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 1996; 87:1285-94; PMID:8980234; http://dx.doi.org/10.1016/S0092-8674(00)81823-1
- Braaten D, Ansari H, Luban J. The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J Virol 1997; 71:2107-13; PMID:9032343
- Zhao Y, Chen Y, Schutkowski M, Fischer G, Ke H. Cyclophilin A complexed with a fragment of HIV-1 gag protein: insights into HIV-1 infectious activity. Structure 1997; 5:139-46; PMID:9016720; http://dx.doi.org/10.1016/S0969-2126(97)00172-X
- Manel N, Hogstad B, Wang Y, Levy DE, Unutmaz D, Littman DR. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature 2010; 467:214-7; PMID:20829794; http://dx.doi.org/10.1038/nature09337
- Priddy FH, Brown D, Kublin J, Monahan K, Wright DP, Lalezari J, Santiago S, Marmor M, Lally M, Novak RM, et al. Safety and immunogenicity of a replication-incompetent adenovirus type 5 HIV-1 clade B gag/pol/nef vaccine in healthy adults. Clin Infect Dis 2008; 46:1769-81; PMID:18433307; http://dx.doi.org/10.1086/587993
- Catanzaro AT, Koup RA, Roederer M, Bailer RT, Enama ME, Moodie Z, Gu L, Martin JE, Novik L, Chakrabarti BK, et al. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 candidate vaccine delivered by a replication-defective recombinant adenovirus vector. J Infect Dis 2006; 194:1638-49; PMID:17109335; http://dx.doi.org/10.1086/509258
- McElrath MJ, De Rosa SC, Moodie Z, Dubey S, Kierstead L, Janes H, Defawe OD, Carter DK, Hural J, Akondy R, et al. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet 2008; 372:1894-905; PMID:19012957; http://dx.doi.org/10.1016/S0140-6736(08)61592-5
- Yu H, Tawab-Amiri A, Dzutsev A, Sabatino M, Aleman K, Yarchoan R, Terabe M, Sui Y, Berzofsky JA. IL-15 ex vivo overcomes CD4+ T cell deficiency for the induction of human antigen-specific CD8+ T cell responses. J leukoc Biol 2011; 90:205-14; PMID:21474552; http://dx.doi.org/10.1189/jlb.1010579
- Zhu Q, Egelston C, Gagnon S, Sui Y, Belyakov IM, Klinman DM, Berzofsky JA. Using 3 TLR ligands as a combination adjuvant induces qualitative changes in T cell responses needed for antiviral protection in mice. J Clin Investig 2010; 120:607-16; PMID:20101095; http://dx.doi.org/10.1172/JCI39293
- Terabe M, Tagaya Y, Zhu Q, Granger L, Roederer M, Waldmann TA, Berzofsky JA. IL-15 expands unconventional CD8alphaalphaNK1.1+ T cells but not Valpha14Jalpha18+ NKT cells. J Immunol 2008; 180:7276-86; http://dx.doi.org/10.4049/jimmunol.180.11.7276
- Shedlock DJ, Talbott KT, Cress C, Ferraro B, Tuyishme S, Mallilankaraman K, Cisper NJ, Morrow MP, Wu SJ, Kawalekar OU, et al. A highly optimized DNA vaccine confers complete protective immunity against high-dose lethal lymphocytic choriomeningitis virus challenge. Vaccine 2011; 29:6755-62; PMID:21238574; http://dx.doi.org/10.1016/j.vaccine.2010.12.064
- Morrow MP, Pankhong P, Laddy DJ, Schoenly KA, Yan J, Cisper N, Weiner DB. Comparative ability of IL-12 and IL-28B to regulate Treg populations and enhance adaptive cellular immunity. Blood 2009; 113:5868-77; PMID:19304955; http://dx.doi.org/10.1182/blood-2008-11-190520
- Kutzler MA, Weiner DB. Developing DNA vaccines that call to dendritic cells. J Clin Investig 2004; 114:1241-4; PMID:15520855; http://dx.doi.org/10.1172/JCI23467
- Kanagavelu SK, Snarsky V, Termini JM, Gupta S, Barzee S, Wright JA, Khan WN, Kornbluth RS, Stone GW. Soluble multi-trimeric TNF superfamily ligand adjuvants enhance immune responses to a HIV-1 Gag DNA vaccine. Vaccine 2012; 30:691-702; PMID:22146759; http://dx.doi.org/10.1016/j.vaccine.2011.11.088
- Arruda LB, Sim D, Chikhlikar PR, Maciel M, Jr., Akasaki K, August JT, Marques ET. Dendritic cell-lysosomal-associated membrane protein (LAMP) and LAMP-1-HIV-1 gag chimeras have distinct cellular trafficking pathways and prime T and B cell responses to a diverse repertoire of epitopes. J Immunol 2006; 177:2265-75; http://dx.doi.org/10.4049/jimmunol.177.4.2265
- Chikhlikar P, Barros de Arruda L, Agrawal S, Byrne B, Guggino W, August JT, Marques ET, Jr. Inverted terminal repeat sequences of adeno-associated virus enhance the antibody and CD8(+) responses to a HIV-1 p55Gag/LAMP DNA vaccine chimera. Virology 2004; 323:220-32; PMID:15193918; http://dx.doi.org/10.1016/j.virol.2004.02.025
- Chikhlikar P, Barros de Arruda L, Maciel M, Silvera P, Lewis MG, August JT, Marques ET. DNA encoding an HIV-1 Gag/human lysosome-associated membrane protein-1 chimera elicits a broad cellular and humoral immune response in Rhesus macaques. PloS one 2006; 1:e135; PMID:17205139; http://dx.doi.org/10.1371/journal.pone.0000135
- de Arruda LB, Chikhlikar PR, August JT, Marques ET. DNA vaccine encoding human immunodeficiency virus-1 Gag, targeted to the major histocompatibility complex II compartment by lysosomal-associated membrane protein, elicits enhanced long-term memory response. Immunology 2004; 112:126-33; PMID:15129672; http://dx.doi.org/10.1111/j.1365-2567.2004.01823.x
- Marques ET, Jr., Chikhlikar P, de Arruda LB, Leao IC, Lu Y, Wong J, Chen JS, Byrne B, August JT. HIV-1 p55Gag encoded in the lysosome-associated membrane protein-1 as a DNA plasmid vaccine chimera is highly expressed, traffics to the major histocompatibility class II compartment, and elicits enhanced immune responses. J Biol Chem 2003; 278:37926-36; PMID:12824194; http://dx.doi.org/10.1074/jbc.M303336200
- Kwon DS, Gregorio G, Bitton N, Hendrickson WA, Littman DR. DC-SIGN-mediated internalization of HIV is required for trans-enhancement of T cell infection. Immunity 2002; 16:135-44; PMID:11825572; http://dx.doi.org/10.1016/S1074-7613(02)00259-5
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 2011; 474:658-61; PMID:21720370; http://dx.doi.org/10.1038/nature10195
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 2011; 474:654-7; PMID:21613998; http://dx.doi.org/10.1038/nature10117
- Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, et al. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet 2009; 41:829-32; PMID:19525956; http://dx.doi.org/10.1038/ng.373
- Goujon C, Jarrosson-Wuilleme L, Bernaud J, Rigal D, Darlix JL, Cimarelli A. With a little help from a friend: increasing HIV transduction of monocyte-derived dendritic cells with virion-like particles of SIV(MAC). Gene Therapy 2006; 13:991-4; PMID:16525481; http://dx.doi.org/10.1038/sj.gt.3302753
- Hou J, Liu Y, Shao Y. The MSHA Strain of Pseudomonas Aeruginosa Activated TLR Pathway and Enhanced HIV-1 DNA Vaccine Immunoreactivity. PloS one 2012; 7:e47724; PMID:23077664; http://dx.doi.org/10.1371/journal.pone.0047724
