ABSTRACT
It has recently become apparent that it is possible to characterize productively recombined, T-cell receptor (TcR) gene segments in tumor exome files, which presumably include representations of the DNA of other cells in the microenvironment. Similar characterizations have been done for TcR recombinations in tumor specimen RNASeq files. While exome files have been used to characterize immunoglobulin gene segment recombinations for tumors closely related to B-cells, immunoglobulin recombinations have yet to be characterized for putative microenvironment cells for solid tumors. Here we report a novel scripted algorithm that detects productive and unproductive immunoglobulin recombinations in both B-cell related tumor exome files and in solid tumor exome files, with the most important result being the relatively high level B-cell infiltrate in breast cancer. This analysis has the potential of streamlining and dramatically augmenting the knowledge base regarding B-cell infiltrates into solid tumors; and leading to antibody reagents directed against tumor antigens and tissue resident, infectious pathogens.
Introduction
Immune cell infiltrates into solid tumors are thought to have both positive and negative effects, for example as indicated in a recent report whereby B-cell signatures were associated with a positive outcome for a number of cancers but a negative outcome for renal cell carcinoma.Citation1 Another report has indicated increased gastric cancer survival correlates with a B-cell infiltrate,Citation2 but plasma cell infiltration of ovarian cancers was associated with the opposite result.Citation3 We recently noted the negative impact of B-cells in an artificial micro-environment on tumor cell apoptosis.Citation4
To develop more precise methods of immunoscoring, potentially leading to more accurate methods of linking a B-cell related immunoscore to tumor outcome or to a specific therapy, we and others have developed genomics-based immunoscoring approaches.Citation5-7 Recently, the linkage of TcR and MHCII expression has been established for particular tumor specimens using RNASeq files, and TcR recombinations have been studied in tumor exome files. In all of these cases, there has been the presumption that results represent non-tumor cells in the tumor microenvironment that leave RNA expression and genetic recombination signatures in the bulk preparations used for transcriptome and exome (WXS) generation. Here we report the development and application of a scripted algorithm for the V(D)J recombinations of the immunoglobulin loci, representing the first case of detection of these recombinations in solid tumor WXS files.
Results
To test the viability of detecting recombined immunoglobulin V(D)J sequences, using the novel algorithm developed for this study, we made use of Diffuse Large B-cell lymphoma (DLBL) TCGA WXS files, previously demonstrated by others to contain reads representing these recombinations.Citation8 We verified the extensive recovery of IGH, IGL, and IGK reads from the DBLC files ().
Table 1. Summary of results from the initial test application of the immunoglobulin recombination search algorithm applied to TCGA DLBL files: Unique recombinations detected.
We next searched TCGA WXS files representing only primary tumors, for a variety of solid tumor types, for both productive and unproductive recombinations (). Results indicated that such recombinations were detectable in a subset of the files representing all of the solid tumors examined. To be certain immunoglobulin recombinations could be detected in another source of WXS files, besides the TCGA WXS files, we searched WXS files representing BLCA patients at the Moffitt Cancer Center. This search revealed a total of 5 productive and unproductive recombinations among 16 WXS files ().
Table 2. Summary of results from application of the immunoglobulin recombination search algorithm applied to TCGA WXS files: Unique recombinations detected.
Table 3. Summary of results from application of the immunoglobulin recombination search algorithm applied to Moffitt Cancer Center bladder cancer patient exomes: Unique recombinations detected.
IGH recombinations were relatively rare. IGK recombinations were most common, and the TCGA BRCA dataset indicated the highest level of detectable immunoglobulin recombinations among the solid tumors WXS files examined (; ). The BRCA data set not only represented the highest number of total recombinations, but also the largest number of TCGA barcodes (samples) that revealed the presence of immunoglobulin V(D)J recombinations ().
Examination of individual reads indicated a variety in the use of distinct V and J gene segment for all 3 immunoglobulin genes (). Examples of N region nucleotides are indicated in .
Figure 1. Total number of productive and unproductive Ig recombinations detected for the indicated cancer datasets, with each data set represented by 100 barcodes. BRCA represents the largest number of Ig recombinations, but the only the comparison of BRCA and KIRP represents a statistical significance, with a p value < 0.02 (t-test).
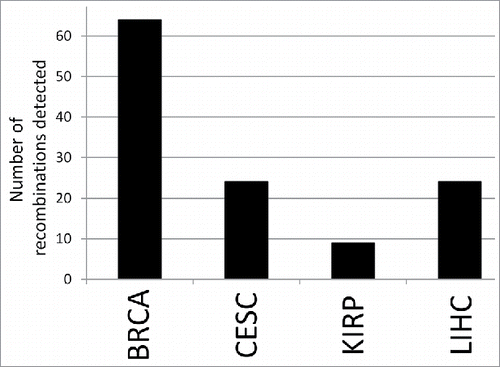
Figure 2. Number of barcodes for the indicated datasets, out of a total of 100 barcodes each, representing either a productive or unproductive Ig gene recombination. BRCA represents the highest number, however, only the difference between BRCA and KIRP represents a statistically significant difference, with a p value < 0.044 (t-test).
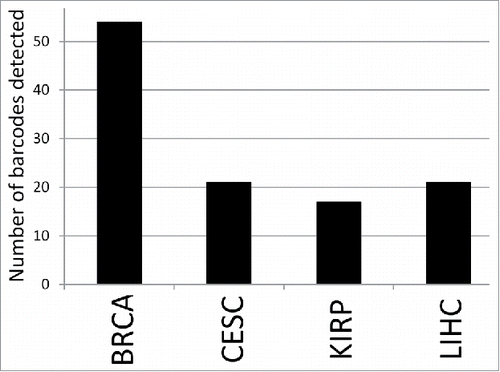
Figure 3. Example immunoglobulin recombinations with V-, J-, and N-region nucleotides indicated. The recombinations indicated are taken from the Moffitt BLCA reads, , bottom 3 barcodes, in order in this figure as they appear in .

Table 4. Summary of results from application of the immunoglobulin recombination search algorithm applied to TCGA WXS files: Barcodes with immunoglobulin recombinations.
Table 5. Example IGH recombinations detected in TCGA WXS files. (All VJ usage in this table was verified using the web tool at https://www.ncbi.nlm.nih.gov/igblast/). Reads indicated by nucleotide number for TCGA samples to meet publication standards.
Table 6. Example IGK recombinations detected in TCGA WXS files.
Table 7. Example IGL recombinations detected in TCGA WXS files.
Discussion
The above results indicate for the first time the opportunity to detect both productive and unproductive immunoglobulin V(D)J recombinations with solid tumor specimen exome files. This raises the question of whether these recombinations represent the adventitious collection of B-cells during the preparation of the tumor specimen or whether the V(D)J recombinations represent detection of B-cells that could have an impact on the development and prognosis of the tumor? Similar results have been obtained for algorithms designed to detect TcR V(D)J recombinations, and in those cases, the initial evidence is indicating that the TcR V(D)J recombinations do indeed represent T-cells that could have an impact on tumor development and prognosis (manuscript submitted). In the case of this study, there is the hint the search algorithm does identify B-cells that are an effective part of the tumor micro-environment: the algorithm leads to a much more robust identification of immunoglobulin V(D)J recombinations in DLBL, as would be expected given the likely B-cell clonality of the tumor source for the WXS file ().
The current, scripted algorithm for identifying the recombined Ig V(D)J regions represents a relatively modest yield, particularly in the case of some cancer datasets. Further work will address computational methods to improve the yield, assuming the upper limit has not be reached due to the limit on the number of tumors with a B-cell infiltrate. Such a number (or percentage) of tumors will likely only be established after processing very large databases, particularly if the numbers of tumors or patients with B-cell infiltrates is small.
The opportunity to detect immunoglobulin V(D)J recombinations in tumor specimen WXS files affords the possibility of highly efficient, data-mining paradigms for the association of V(D)J usage with a number of cancer related parameters, owing to the very large number of tumor WXS files available. For example, it will be possible to determine whether B-cell infiltrates into the tumor micro-environment have an impact on prognosis, through minimization of apoptosisCitation4 or other tumor-promoting or tumor-destroying mechanisms; or whether there is an association of V and J usage with HLA types, as has been postulated for TcR V and J usageCitation9; or whether certain cancer mutations correlate with the detection of immunoglobulin recombinations or V and J usage. In addition, more recent studies have indicated that tumors with higher levels of mutations are more immunogenic, possibly due to a greater availability of neoantigens, based on T-cell responses.Citation10-12 A similar question could be asked regarding the detection of immunoglobulin recombinations. In short, all of the above approaches have the possibility of the addressing the issue raised in the above paragraph, is there a prognostic value or detectable impact of the B-cells, represented by WXS detectable recombinations, for the tumor. Indeed the answer to such a question could be related to whether the tumor-resident B-cells have productive or unproductive recombinations, allowing an understanding of the role of B-cells that are having effects on the tumor that are distinct from producing a BcR with antigen binding capacity. Such antigen-independent effects could include cytokine release or disruption of the local tissue architecture. However, it should be kept in mind that the detection of an unproductive recombination at this stage in the methodology cannot rule out the possibility that the second allele has undergone a productive recombination. Finally, a positive answer to the question, are the tumor resident B-cells indicated by the above analysis specifically associated with tumor development, would provide a potential guide in identifying immunoglobulin recombinations, and hypermutation results, for immunoglobulin molecules that could be used in therapy, not only for cancer but for infectious diseases as well. For example, tissue resident B-cells at sites of infectious pathology, such as in cases of flesh eating, or Group A Streptomyces infections, could facilitate the identification and development of therapeutic Ig molecules.
Methods
A list of V and J region sequences for human IGH, IGK, and IGL were obtained from the IMGT Repertoire (http://www.imgt.org/IMGTrepertoire/LocusGenes/#h1_36) (supporting online material, SOM). Whole exome sequence (WXS) samples for the cancer data sets originated from the Cancer Genome Atlas (TCGA) consortium (http://cancergenome.nih.gov/). The files were downloaded from Cancer Genomics Hub (CGHub) hosted by UC Santa Cruz (currently hosted by NCI GDC). In addition, WXS files for bladder cancer specimens were obtained from Moffitt Cancer Center, as described in ref.Citation5
The “SearchV” bash scripts for IGH, IGK, and IGL were executed on the WXS files. The SearchV bash scripts are modified versions of the “FindV2” bash script described in ref.Citation5 The SearchV scripts used SAMtools v1.31 to view the sequence for IGH, IGK, and IGL regions in the WXS files. The sequence region locations were obtained for assembly GRCh37.p13 from the NCBI Gene website (http://www.ncbi.nlm.nih.gov/gene/). The SearchV script used 10 base pairs to represent the V region 3′ ends, with the 10 base pair segment beginning at −8 from the 3′ end of the V for IgK; and at −5 from the 3′ end of the V regions for IgH and IgL. The differences were due to technical convenience. In all cases, the 10 base segment V region segment then goes toward the 5′ end. The use of segments several nucleotides away from the 3′ end is to account for N-region diversity. (The V regions sequences for all 3 genes were obtained from the IMGT web site indicated above). The SearchV script then used the Linux tool “grep” to search for the list of V sequences in a region approximately 6 million base pairs on either side of the location of the gene sequences. A list of reads from each WXS containing a known V region were outputted into individual tab-delimited text (TSV) files. A second bash script, SearchJ, modified from ref.,Citation5 was used to search for an exact match, to portions of the known J regions, in the list of reads; and the found J regions were stored in separate TSV files. The productive or unproductive status of the all the matched reads for all samples was determined through the International Immunogenetics Information System's website (IMGT/V-QUEST) and recorded into a single TSV file containing information regarding the matched reads as well as their corresponding known human immunoglobulin nucleotide sequences. This process was automated by a PHP script that submits each read to the IMGT website for processing using the cURL library and places the output into a TSV file (PHP v7.0.6, cURL v7.43.0). Manual analysis of the output for each immunoglobulin type was done using Microsoft Excel 2016. Versions of the above code, the list of V's and J's used, and example outputs, are in the SOM.
The above described pipeline can be parallelized, but in serial, a single threaded execution of the scripts for IGH, IGK, and IGL are CPU-limited and takes approximately 24 hours using an Intel Xeon E5649 @ 2.53GHz for 100 WXS files. The RAM usage is typically less than 500 MBs.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary files
Download PDF (700.2 KB)Acknowledgments
Authors would like to acknowledge the extensive assistance of Dr. Tony Green of USF research computing and the Moffitt Cancer Center functional genomics and tissue core facilities.
Funding
Authors would like to acknowledge the the financial contributions of the Anna Valentine Fund and the taxpayers of the State of Florida.
References
- Iglesia MD, Parker JS, Hoadley KA, Serody JS, Perou CM, Vincent BG. Genomic analysis of immune cell infiltrates across 11 tumor types. J Natl Cancer Inst 2016; 108(11); PMID:27335052; http://dx.doi.org/10.1093/jnci/djw144
- Hennequin A, Derangere V, Boidot R, Apetoh L, Vincent J, Orry D, Fraisse J, Causeret S, Martin F, Arnould L, et al. Tumor infiltration by Tbet+ effector T cells and CD20+ B cells is associated with survival in gastric cancer patients. Oncoimmunol 2016; 5(2):e1054598; PMID:27057426; http://dx.doi.org/10.1080/2162402X.2015.1054598
- Lundgren S, Berntsson J, Nodin B, Micke P, Jirstrom K. Prognostic impact of tumour-associated B cells and plasma cells in epithelial ovarian cancer. J Ovarian Res 2016; 9:21; PMID:27048364; http://dx.doi.org/10.1186/s13048-016-0232-0
- Szekeres K, Koul R, Mauro J, Lloyd M, Johnson J, Blanck G. An Oct-1-based, feed-forward mechanism of apoptosis inhibited by co-culture with Raji B-cells: towards a model of the cancer cell/B-cell microenvironment. Exp Mol Pathol 2014; 97(3):585-9; PMID:25236570; http://dx.doi.org/10.1016/j.yexmp.2014.09.010
- Gill TR, Samy MD, Butler SN, Mauro JA, Sexton WJ, Blanck G. Detection of Productively Rearranged TcR-α V-J Sequences in TCGA Exome Files: Implications for Tumor Immunoscoring and Recovery of Antitumor T-cells. Cancer Inform 2016; 15:23-8; PMID:26966347
- Brown SD, Raeburn LA, Holt RA. Profiling tissue-resident T cell repertoires by RNA sequencing. Genome Med 2015; 7(1):125; PMID:26620832; http://dx.doi.org/10.1186/s13073-015-0248-x
- Butler SN, Blanck G. Immunoscoring by correlating MHC class II and TCR expression: high level immune functions represented by the KIRP dataset of TCGA. Cell Tissue Res 2016; 363(2):491-6; PMID:26293619; http://dx.doi.org/10.1007/s00441-015-2261-1
- Paciello G, Acquaviva A, Pighi C, Ferrarini A, Macii E, Zamo A, Ficarra E. VDJSeq-Solver: in silico V(D)J recombination detection tool. PloS One 2015; 10(3):e0118192; PMID:25799103; http://dx.doi.org/10.1371/journal.pone.0118192
- Klarenbeek PL, Doorenspleet ME, Esveldt RE, van Schaik BD, Lardy N, van Kampen AH, Tak PP, Plenge RM, Baas F, de Bakker PI, et al. Somatic variation of T-Cell receptor genes strongly associate with HLA class restriction. PloS One 2015; 10(10):e0140815; PMID:26517366; http://dx.doi.org/10.1371/journal.pone.0140815
- Maletzki C, Schmidt F, Dirks WG, Schmitt M, Linnebacher M. Frameshift-derived neoantigens constitute immunotherapeutic targets for patients with microsatellite-instable haematological malignancies: frameshift peptides for treating MSI+ blood cancers. Eur J Cancer 2013; 49(11):2587-95; PMID:23561850; http://dx.doi.org/10.1016/j.ejca.2013.02.035
- Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Eng J Med 2014; 371(23):2189-99; PMID:25409260; http://dx.doi.org/10.1056/NEJMoa1406498
- Westdorp H, Fennemann FL, Weren RD, Bisseling TM, Ligtenberg MJ, Figdor CG, Schreibelt G, Hoogerbrugge N, Wimmers F, de Vries IJ. Opportunities for immunotherapy in microsatellite instable colorectal cancer. Cancer Immunol Immunother 2016; 65(10):1249-59; PMID:27060000
