ABSTRACT
HPV16 persistent infection is a well-known condition that precedes human cancer development. High risk HPV E5 proteins cooperate with E6/E7 oncogenes to promote hyper-proliferation of infected cells leading to possible cancer progression. Thus, presence of E5 viral transcripts could be a key marker of active infection and, in turn, a target of immunotherapy. Purpose of the study is to detect E5 transcripts in clinical samples and to explore the activity of novel anti-HPV16 E5 DNA vaccines. HPV transcripts were detected by PCR with specific primers encompassing the splice-donor sites of E5 transcript. For E5-based immunotherapies, 2 E5-based versions of DNA vaccines carrying whole E5 gene or a synthetic multiepitope gene were improved by fusion to sequence of PVX coat protein. These vaccines were challenged with a new luminescent animal model based on C3-Luc cell line. E5 transcripts were detected in clinical samples of women with HPV positive low-grade SIL, demonstrating the validity of our test. In C3 pre-clinical mouse model, vaccine candidates were able to induce a strong cellular immunity as indicated by ELISPOT assays. In addition, E5-CP vaccines elicited strong anti-tumor effects as showed by decreased tumor growth monitored by animal imaging. The tumor growth inhibition was comparable to those obtained with anti-E7 DNA vaccines. In conclusion, detection of E5 transcripts in clinical samples indicates that E5 is a possible target of immunotherapy. Data from pre-clinical model demonstrate that E5 genetic immunization is feasible, efficacious and could be utilized in clinical trials.
Introduction
E5 protein of high risk HPV can be considered an oncogene acting in the first stage of carcinogenesis.Citation1 The localization of HPV E5 to the endoplasmic reticulum suggests its activity may be related to the trafficking of cytoplasmic membrane proteins through this cellular compartment, in particular of growth factor receptors and of molecules involved in immune control. There are multiple documented intracellular binding targets for 16E5 such as some members of EGF receptor family,Citation2-3 the 16-kDa subunit of the vacuolar H+-ATPase,Citation4-5 the heavy chain of HLA type I,Citation6 calnexin,Citation7 the zinc transporter ZnT-1, the EVER1/ EVER2 transmembrane channel-like proteins,Citation8-9 the nuclear import receptor family member KNβ3,Citation10 BAP31, and A4.Citation11-12 However the role of HPV-16 E5 in carcinogenesis seems to be limited to the early stages of cervical carcinogenesis because the E5 gene is frequently deleted when the HPV genome is integrated during malignant progression.Citation13-15 Nevertheless, the expression of HPV-16 E5 as detected by immunohistochemistry, was reported in approximately 80, 90 and 60% of HPV 16-infected LSILs, high-grade SILs and cervical carcinomas, respectively.Citation13 Furthermore, data from E5 mRNA transcripts in 2 other clinical studies confirmed the presence of E5 in LSIL and HSIL. To further confirm these data and to highlight the possible role of E5 as key marker of early stage of infection/transformation and, in turn, as target of immunotherapy, a test for E5 transcripts in clinical samples and novel anti-HPV16 E5 DNA vaccines have been developed in the present study. Therefore, targeting E5 which is frequently expressed in earlier stages of malignant transformation may be a rational approach for preventing premalignant lesions from progressing into invasive cervical cancers.Citation16
Results
Presence of E5 transcripts in clinical samples
RT-real time-PCR with the specially designed primers for an E5 specific transcript was performed in a number of clinical samples including LSIL, HSIL and ASCUS. Seventy samples were analyzed and in 68 of them total RNA was successful extracted and utilized for reverse transcription into cDNA, followed by real-time PCR with the specific primers. Results in showed that specific E5 transcripts were detectable in all lesions with highest positivity (60%) in LSIL samples, indicating a possible biological activity of E5 products.
Table 1. Specific E5 mRNA in clinical samples.
Construction of E5 vaccines
In previous studies 2 DNA vaccines against E5 oncogene of HPV16 were prepared: pCI-E5H16 pCI-E5Multi. First DNA plasmid vaccine contained full E5 gene of HPV16 whereas the other construct was made by cloning in the same plasmid 2 previous described coding sequences for immune epitopes, in duplicate.Citation17-18 By this way, this E5Multi was showed to be more immunogenic with less, if any, oncogenic activity. In order to improve the activity of these E5 vaccines, E5 and E5Multi sequences were subcloned and fused to capsid protein of plant virus PVX that we already demonstrated to increase activity of antigens in DNA vaccine formulations.Citation19
Immunological response to E5 vaccines
The new E5 vaccines, pVAX-E5CP and pVAX-E5MultiCP, were delivered i.m. to C57BL/6 mice at 7 day interval with a scheme of a prime and 3 boosts. Same scheme was adopted for the administration of anti E6 (pVAX-E6CP) and anti E7 (pVAX-E7CP) vaccines in order to confront E5 vaccine activity with that of already tested vaccines. One week after last boost, peripheral blood samples were collected, animal sacrificed and splenocytes collected.
In peripheral lymphocytes, flow cytometry assay was performed in order to define CD8+ and CD4+ T cell response to specific peptides after vaccination.Citation20 No difference was detected in CD8+ T cell response (data not shown) whereas an higher CD4+ T cell response was detected with E5 vaccines respect to E6 or E7 vaccines. This high response was particularly detected with pVAX-E5CP vaccine ().
Figure 1. Immune responses to anti-E5 DNA vaccines. A. Flow cytometry assay for CD4+ circulating lymphocytes. Blood samples were collected one week after last boost and flow cytometry performed as in Methods. E5, E5M, E6, and E7 groups are mice vaccinated with pVAX-E5CP, pVAX-E5MultiCP, pVAX-E6CP, or pVAX-E7CP DNA preparations, respectively. Data are from pooled blood samples from each group. pVAX mice are control group vaccinated with empty vector. B ELISPOT assay. Pooled splenocytes from all animals in the group were analyzed for the presence of Interferon-γ- secreting cells upon stimulation with specific peptides, as described in Methods. Columns represent data ± SD of triplicate wells.
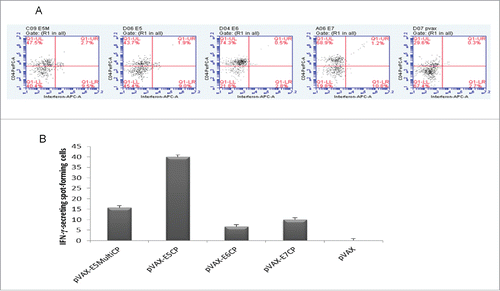
Collected splenocytes were utilized for ELISPOT assay. Interferon-γ-secreting splenocytes were detected with all vaccine preparations but pVAX-E5CP and pVAX-E5MultiCP induced stronger immune response with 4-fold increase by pVAX-E5CP respect to pVAX-E7CP ().
Anticancer activity of E5 vaccines
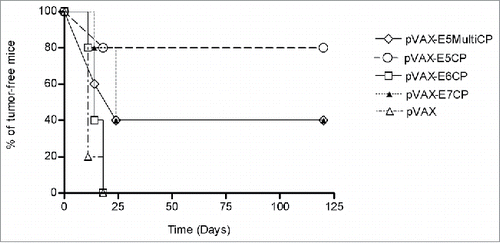
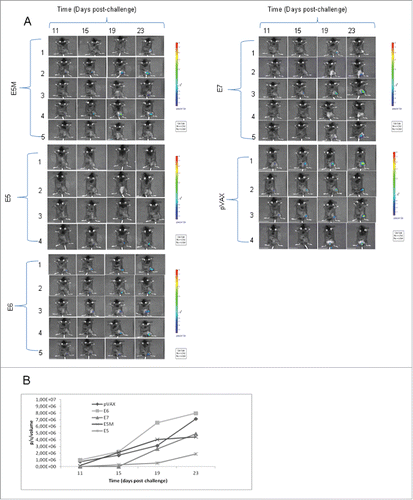
Therapeutic activity of new E5 vaccines was assessed in a special designed mouse model, the C3 cell line. This already well known model was engineered to render cells and developed tumors luminescent by infection with lentivirus expressing luciferase gene (). In this model 3 d after challenge with 5×105 C3-Luc cells, DNA vaccines were delivered i.m. After seven days a boost dose was injected i.m. Tumor growth was monitored each 4 d by imaging as shown in . In control group (pVAX) all animals developed luminescent tumors whereas in vaccinated mice by pVAX-E5CP, pVAX-E5MultiCP or pVAX-E7CP a number of mice did not developed any tumor. The intensity of luminescent signal was recorded and quantified (). The intensity of the signal is directly linked to the tumor burden. After 23 days, the lowest level of luminescent signal was recorded with pVAX-E5CP vaccine while pVAX-E5MultiCP gave same result as for pVAX-E7CP, indicating a decreased rate of tumor growth. In this C3-Luc model, pVAX-E6CP was unable to reduce tumor burden. The same experiment was repeated with same results (data not shown). Kaplan-Meier analysis of experiments as time-to tumor development indicated a slight delay in tumor onset for mice affected by tumor and treated with pVAX-E5CP, pVAX-E5MultiCP or pVAX-E7CP. On the contrary, a dramatic statistically significant (p < 0,0075) effect was seen on tumor inhibition. Indeed, 80% of pVAX-E5CP-treated mice were totally tumor free respect to 40% of pVAX-E5-MultiCP or pVAX-E7CP-treated mice ().
Figure 2. Luminescent C3 cell model. Luminescent C3-Luc cells were obtained as described in Methods. 5×105 C3-Luc cells were injected s.c. into a C57BL/6 mouse. An example of imaging results is shown at 11 d after inoculation. The intensity of bioluminescence was color- or gray-coded and the used scale is reported.
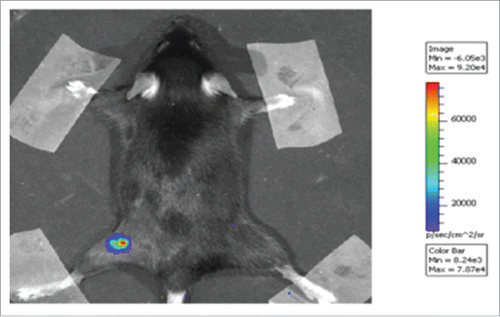
Figure 3. Anticancer activity of E5 vaccines. E5, E5M, E6, E7 and pVAX groups are mice vaccinated with pVAX-E5CP, pVAX-E5MultiCP, pVAX-E6CP, pVAX-E7CP, or empty pVAX DNA preparations, respectively. A. Imaging of mice challenged with C3-Luc cells at the reported time interval. Mice were vaccinated with the different DNA preparations at 3 and 7 d post cell inoculum. The intensity of bioluminescence was color- or gray-coded for imaging purposes; the used scale is reported for each mouse group. B. Luminescent signal quantification of pooled data from animals developing luminescent tumors. The intensity of luminescent signal was recorded as in Methods and expressed as photon/second/cm2/steradian (p/s/cm2/sr). Higher signal intensity represents higher tumor mass.
Discussion
HPV 16 E5 gene can transform human cells but its role has been suggested mostly in early stage of cervical carcinogenesis because E5 gene is frequently deleted during HPV integration in cancer development.Citation1
However, a subset of HPV16-positive invasive cervical carcinomas maintains viral DNA only as episomesCitation21 and besides the integrated monomeric forms, head-to-tail concatemers of full-length HPV genomes flanked by truncated copies also exist.Citation22
Finally, the E5 protein was identified by mass spectrometry in CaSki cells giving the definitive proof that HPV-16 E5 does exist and it may contribute to the malignant phenotype of some cervical cancers, even in cells exclusively containing an integrated HPV genome.Citation23 Thus, E5 may play a role in carcinogenesis and must be present in early stages. Expression of HPV-16 E5 was detected by immunohistochemistry, in LSILs, HSILs and cervical carcinomas.Citation24 These data were confirmed in other studies utilizing RT real time PCR on mRNA from LSILs and HSILs, respectively.Citation25-26 However, all HPV16 transcripts are polycistronic and the resulting early primary transcripts have 3 exons and 2 introns, which undergo alternative RNA splicing.
Each intron of the early transcripts could be spliced out through utilization of 3 alternative splices within the 2 introns lead to the production of at least 14 species of mRNA transcripts with various coding potential.Citation27 Thus, it is hard to ascertain the coding potential for E5 alone. Recently, a transcript with coding capacity for E5 alone was identified in clinical samples and W12 cell line.Citation28 Other reports indicate that other bicistronic transcripts from another promoter can be responsible for E5 transcription.Citation29 In our study by designing forward primers encompassing splice/donor site, a specific E5 transcript was detected avoiding any possible DNA contamination. This E5 transcript was expressed in the majority of LSIL confirming literature data and indicating us that a specific immunotherapy can be developed against this oncogene.
Eradication of HPV-induced tumors in mice by anti-E5 vaccinationCitation30 was already reported and a substantial improvement of previous reported DNA vaccines was achieved in this work. By fusing E5 or E5Multi gene to CP gene of PVXCitation19 new vaccines showed a stronger anticancer activity with more that 80% of animal cured by pVAX-E5CP. This result is particularly interesting because C3 model is more resistant than widely used TC-1 to specific immunotherapy.Citation31 Indeed, anti E7 vaccine was less effective in this model in respect to TC-1 model. This anticancer activity of E5 vaccines is linked to specific immune response and in particular to a higher E5-induced activity in CD4+ circulating lymphocytes. Inducing CD4+ T helper cell immune responses is one of the best strategies to enhance vaccine potency. CD4+ T helper cells are known to play an important role in the generation of CD8+ T cell immune responses as well as memory T cell responses (for a review see ref. Citation32). In addition, our study was also designed to define antitumoral improvement of E5 vaccines. This improvement was fully achieved. Indeed, previous vaccines made with same E5 or E5Multi sequences were able only to decrease tumor burdenCitation20 whereas new CP fused vaccines were able to cure same C3 tumors. Future development of this therapeutic strategy could be the intra-tumor administration of DNA vaccine as well as heterologous (DNA/protein) prime-boost schedules to be applied together with chemo-radiotherapy.
In conclusion, results from this study indicate that E5 is expressed in early stage carcinogenesis and can be a target of immunotherapy. The efficacy of E5 vaccines fused to CP in eradicating HPV-associated tumor in a mouse model suggests that these vaccine preparations can be used in human immunotherapy. Finally, the neutralization (immunological or chemical) of E5 or of E5-induced signaling pathways could be advantageous particularly in HPV infections and pre-cancer lesions where medical treatments are still lacking.
Materials and methods
Samples
The study included 70 fresh specimens collected in PreservCyt (Cytyc Corp., Rome, Italy). Cytological diagnosis was: LSIL (59%), HSIL (18%) and ASCUS (23%). All samples were collected for diagnostic purposes and studied in accordance with national ethical principles. The investigation protocol was approved by the review board of our Institute.
Design a specific primers set for E5 transcripts and RT-Real time PCR
RNA was extracted from sample by the RNeasy Plus Mini kit (QIAGEN, Milan, Italy), according to the manufacturer's instructions. Total RNA pre-treated with deoxyribonuclease I (DNase I, Amplification grade, Invitrogen, Milan, Italy) was retro-transcribed into cDNA for 1 h at 42°C using a random hexamer primer kit as described by the manufacturer (GeneAmp RNA PCR kit Applied Biosystem, Foster City, CA USA). The synthesized cDNA underwent real time-PCR with 2× Kapa SYBR Fast qPCR Master Mix (KAPA biosystems, Milano, Italy). Briefly, the amplification of E5 and β-Actin sequences was performed in a 20 µl volume containing, 10 μM of each primer and 20 ng of cDNA, using the following protocol for 40 cycles: 3 sec denaturation at 95°C, annealing and extension at 60°C for 30 sec, with the initial denaturation at 95°C for 3 min. The E5 primers were designed by Beacon Design software (BioRad, Milan, Italy) straddling the splicing site (880–3358) for forward primer (5′-GCGACGTGAGAGCAACG-3′) and within E4 region for reverse primer (5′-AGGGGTTTCCGGTGTCTGG-3′).
Plasmids
To construct the pVAX-E5CP, pVAX-E5MultiCP, pVAX-E7CP and pVAX-E6CP expression plasmids, the coding sequence of the E5, E5MultiCitation20 mutated E633 and mutated E719 were fused to Potato Virus X coat protein gene (CP) and amplified by PCR, according to standard procedures. The PCR products and plasmid pVAX1 vector (Invitrogen, Milan, Italy) were digested with Not I and Hind III, and religated by T4 ligase. Then, recombinant plasmids were transfected into Escherichia coli strain TOP10 (Invitrogen, Milan, Italy) by electroporation. Plasmids for vaccine preparations were purified by double CsCl gradient to obtain endotoxin-free products.
Generation of C3 luminescent cells
C3 tumor cells (a gift of C.J.M. Melief, Leiden, Netherlands) are B6 embryonic mouse cells transformed by HPV 16 genome and EJ ras.Citation34 The C3 cells are syngeneic to C57BL/6 mice and were cultured in RPMI-1640 with 10% fetal bovine serum (Life Technology, Milan, Italy). C3 cells were infected with a lentivirus containing the firefly luciferase gene to generate C3-Luc. Briefly, the luciferase coding sequence isolated from the pGL-3 vector (Promega, Milan, Italy) was cloned into the lentiviral vector FG9 behind the CMV-LTR and UBiC promoters. This vector was co-transfected using calcium phosphate with packaging constructs pRSVREV, pMDLg/ pRRE, and the VSV-G expression plasmid pHCMVG into HEK-293T cells to generate luciferase expressing lentivirus (Lenti-luc), according to standard procedures. C3 cells were then infected with Lenti-luc virus mixed with polybrene (4μg/mL medium). Blasticidin-selected C3-Luc cells were analyzed by luciferase assay to detect the highest luminescent clones as described by Brasier et al.Citation35 For mouse inoculation, C3-Luc cells were harvested by trypsinization, washed twice, and resuspended in saline solution.
Animals and vaccination schedule
Animal study was approved by the Institutional Animal Care of the Regina Elena National Cancer Institute and by the Government Committee of National Minister of Health (85/2016-PR) and was conducted according to EU Directive 2010/63/EU for animal experiments. Mice were divided into 5 groups of 5 mice per group. Female 6–8 week C57BL/6 were injected i.m. with 100 μg of plasmid vaccine DNA suspended in 100μl sterile saline solution, whereas control mice received empty pVAX vector. All mice were vaccinated 4 times at weeks 0, 1, 2 and 3. One week after the final immunization, mice were sacrificed for blood collection and spleen removal for ELISPOT assay. In therapeutic setting, all mouse groups were challenged s.c. with 5 × 105 C3-Luc cells and, thereafter, treated with vaccines as in previous report.Citation20 The mice were monitored by imaging to detect tumor growth. All efforts were made to minimize the number of animals and their suffering. Indeed only 5 animals (the minimum to obtain statistical significance in our modelCitation36) per group was utilized and the experiment repeated only one time.
Imaging
The luciferase activity was utilized for the bioluminescent approach. The mice were anesthetized and 75 mg/kg of d-luciferin (Caliper, PerkinElmer company) was injected intraperitoneally. Ten minutes later, quantification of light emission was acquired for 5 min. Signal was detected using the IVIS Lumina II CCD camera system and analyzed with the Living Image 2.20 software package (Caliper Life Sciences, Milan, Italy). Photon emission was measured in specific regions of interest (ROIs). Data were expressed as photon/second/cm2/steradian (p/s/cm2/sr). The intensity of bioluminescence was color- or gray-coded for imaging purposes; the scale used in each experiment is reported in each figure.
IFN-gamma Enzyme-Linked Immuno-spot assay (ELISPOT)
Interferon-γ-secreting cells were detected by enzyme linked immunospot assay for IFN- γ -secreting cells (BDTM ELISPOT BD Biosciences PharMingen, San Diego, CA, USA). Briefly, the mice were sacrificed and a single-cell suspension of splenocytes were harvested from each group of vaccinated mice. 106 splenocytes were added to microtiter well that had been pre-coated overnight at 4°C with anti-mouse-IFN-γ antibody (5 μg/ml) along with IL-2 (50 units/ml; Sigma–Aldrich Italia, Milan, Italy). Stimulation was performed in triplicates at 37°C for 24–72 h, with the E5-, E6- or E7-specific peptidesCitation20 (synthesized by GenScript, Piscataway, USA). A mixture of phorbol myristate acetate and ionomycin was used to detect splenocyte responsiveness. An unrelated peptide was used as a negative control. The plates were incubated with a biotinylated anti-mouse IFN-γ antibody (2 μg/ml) for 4 h at room temperature. Streptavidin–HRP was then used for 1 h at room temperature, and the cell spots stained by addition of the filtered 3-amino-9-ethylcarbazole substrate, for 1–5 min. The reaction was terminated once the formation of discrete purple-colored spots was detected, each individual spot representing an individual analyte-secreting cell. Spots were counted under a dissecting microscope.
Detection of CD4+ and CD8+ T cells by flow cytometry
To determine CD4+ and CD8+ T cell response to vaccines, single-cell suspensions of peripheral blood lymphocytes (PBL) were prepared and examined by flow cytometry assay (Cytofix/Cytoperm™ Plus Kit With GolgiPlug™ -BD Biosciences San Diego, CA, USA) according to manufacturer instruction with PerCP-Cy™5.5 and FITC-labeled antibodies against CD4+ and CD8+ T cells, respectively, and APC-labeled antibodies for Interferon-γ (BD PharMingen™). Briefly, PBLs were isolated from heparinized blood samples, washed twice in PBS, and adjusted to a final concentration of 5 × 105 cells per ml and stimulated for 20 h with 10 µg/ml of E5, E6 or E7 specific peptides, already described in ELISPOT assay. Then, stimulated cells were harvested and stained for cell surface antigens: anti-CD4 monoclonal antibody and anti-CD8 monoclonal antibody. After incubation for 30 min at 4°C in the dark, labeled secondary antibodies were added. To enhance detection of cytokine-producing cells, GolgiPlug™ (BD PharMingen™) buffer containing brefeldin A was utilized to block intracellular transport processes, resulting in the accumulation of most cytokines in the Golgi complex. Subsequently, the cells were washed with PBS and resuspended in 500μL PBS, followed by flow cytometry analysis. Viable lymphocytes were gated on the basis of forward and side scatter characteristics, and 10 000 events were analyzed for positive staining with FITC. Data analysis was performed using BD FACSAria software.
Statistical analysis
Statistical analyses were performed using one-way ANOVA parametric test and Bonferroni analysis of variance, using the GraphPad Prism 5 software (www.graphpad.com). Kaplan–Meier analysis was performed as time-to-tumor development, and Log-Rank test was used to determine the significance among Kaplan–Meier curves. The statistical significance was set as p < 0.05.
Abbreviations
| ASCUS | = | Atypical squamous cells of undetermined significance |
| BAP31 | = | B-cell-associated protein 31 |
| CP | = | coat protein |
| CMV-LTR | = | cytomegalovirus long-terminal repeat |
| ELISPOT | = | enzyme-linked immuno spot |
| FITC | = | fluorescein isothiocyanate |
| HSIL | = | high grade squamous intraepithelial lesion |
| h | = | hours |
| HEK-293T | = | human embryonic kidney 293T |
| HLA | = | human leucocyte antigen |
| HPV | = | Human papillomavirus; Interferon |
| i.m | = | intramuscularly |
| LSIL | = | low grade squamous intraepithelial lesion |
| PerCP-Cy 5.5 | = | peridinin-chlorophyll-protein Complex: CY5.5 Conjugate |
| PBL | = | peripheral blood lymphocytes |
| PBS | = | phosphate buffered saline |
| RT | = | reverse transcription |
| PCR | = | polymerase chain reaction |
| s.c. | = | sub-cutaneous |
| UBiC | = | ubiquitin-C promoter |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
Authors thank all the staff of IRE animal house for mouse stabling and maintenance.
Funding
Work partially supported by AIRC IG 12916; Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq/PVE 2014, 401305/2014); Fundação de Amparo a Ciência e Tecnologia do Estado de Pernambuco (FACEPE/PRONEM APQ-0562–2.02/14).
References
- Venuti A, Paolini F, Nasir L, Corteggio A, Roperto S, Campo MS, Borzacchiello G. Papillomavirus E5: the smallest oncoprotein with many functions. Mol Cancer 2011; 10:140. Review; PMID:22078316; http://dx.doi.org/10.1186/1476-4598-10-140
- Crusius K, Auvinen E, Steuer B, Gaissert H, Alonso A: The human papillomavirus type 16 E5-protein modulates ligand-dependent activation of the EGF receptor family in the human epithelial cell line HaCaT. Exp Cell Res 1998; 241:76-83; PMID:9633515; http://dx.doi.org/10.1006/excr.1998.4024
- Paolini F, Curzio G, Melucci E, Terrenato I, Antoniani B, Carosi M, Mottolese M, Vici P, Mariani L, Venuti A. Human papillomavirus 16 E2 interacts with neuregulin receptor degradation protein 1 affecting ErbB-3 expression in vitro and in clinical samples of cervical lesions. Eur J Cancer 2016; 58:52-61; PMID:26963794; http://dx.doi.org/10.1016/j.ejca.2016.02.001
- Venuti A, Salani D, Poggiali F, Manni V, Bagnato A. The E5 oncoprotein of human papillomavirus type 16 enhances endothelin-1-induced keratinocyte growth. Virology 1998; 248:1-5; PMID:9705249; http://dx.doi.org/10.1006/viro.1998.9227
- Regan JA, Laimins LA. Bap31 is a novel target of the human papillomavirus E5 protein. J Virol 2008; 82:10042-51; PMID:18684816; http://dx.doi.org/10.1128/JVI.01240-08
- Oh JM, Lee YI, Song YS, Kim WH, Juhnn YS. Human papillomavirus E5 protein induces expression of the EP4 subtype of prostaglandin E2 receptor in cyclic AMP response element-dependent pathways in cervical cancer cells. Carcinogenesis 2009; 30:141-49; PMID:18849297; http://dx.doi.org/10.1093/carcin/bgn236
- Burkhardt A, Willingham M, Gay C, Jeang KT, Schlegel R. The E5 oncoprotein of bovine papillomavirus is oriented asymmetrically in Golgi and plasma membranes. Virology 1989; 170:334-9; PMID:2541554; http://dx.doi.org/10.1016/0042-6822(89)90391-7
- Ronco LV, Karpova AY, Vidal M, Howley PM. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev 1998; 12:2061-72; PMID:9649509; http://dx.doi.org/10.1101/gad.12.13.2061
- Obalek S, Favre M, Szymanczyk J, Misiewicz J, Jablonska S, Orth G. Human papillomavirus (HPV) types specific of epidermodysplasia verruciformis detected in warts induced by HPV3 or HPV3-related types in immunosuppressed patients. J Investig Dermatol 1992; 98:936-41 ; PMID:1317396; http://dx.doi.org/10.1111/1523-1747.ep12460892
- Hu L, Plafker K, Vorozhko V. Human papillomavirus E5 induces bi-nucleated cell formation by cell-cell fusion. Virology 2009; 384:125-34; PMID:19041112; http://dx.doi.org/10.1016/j.virol.2008.10.011
- Hu L, Ceresa BP. Characterization of the plasma membrane localization and orientation of HPV16 E5 for cell-cell fusion. Virology 2009; 393:135-43 ; PMID:19712955; http://dx.doi.org/10.1016/j.virol.2009.07.034
- Yang DH, Wildeman AG, Sharom FJ. Overexpression, purification, and structural analysis of the hydrophobic E5 protein from human papillomavirus type 16. Protein Expr Purif 2003; 30:1-10; PMID:12821315; http://dx.doi.org/10.1016/S1046-5928(03)00049-4
- Valle FG, Banks L. The human papillomavirus (HPV)-6 and HPV-16 E5 proteins co-operate with HPV-16 E7 in the transformation of primary rodent cells. J Gen Virol 1995; 76:1239-45; PMID:7730808; http://dx.doi.org/10.1099/0022-1317-76-5-1239
- Genther Williams SM, Disbrow GL, Schlegel R, Lee D, Threadgill DW, Lambert PF. Requirement of epidermal growth factor receptor for hyperplasia induced by E5, a high-risk human papillomavirus oncogene. Cancer Research 2005; 65:6534-42; PMID:16061632; http://dx.doi.org/10.1158/0008-5472.CAN-05-0083
- Ashrafi GH, Pitts JD, Faccini AM, McLean P, O'Brien V, Finbow ME, Campo S. Binding of bovine papillomavirus type 4 E8 to ductin (16K proteolipid), down-regulation of gap junction intercellular communication and full cell transformation are independent events. J Gen Virol 2000; 81:689-94; PMID:10675405; http://dx.doi.org/10.1099/0022-1317-81-3-689
- Grindlay GJ, Campo MS, O'Brien V. Transactivation of the cyclin A promoter by bovine papillomavirus type 4 E5 protein. Virus Res 2005; 108:29-38; PMID:15681052; http://dx.doi.org/10.1016/j.virusres.2004.07.010
- Liu D-W, Yang Y-C, Lin H-F, Lin M-F, Cheng Y-W, Chu C-C, Tsao YP, Chen SL. Cytotoxic T-lymphocyte responses to human papillomavirus type 16 E5 and E7 proteins and HLA-A*0201-restricted T-cell peptides in cervical cancer patients. J Virol 2007; 81:2869-79; PMID:17202211; http://dx.doi.org/10.1128/JVI.02256-06
- Chen Y-F, Lin C-W, Tsao Y-P, Chen S-L. Cytotoxic-T-lymphocyte human papillomavirus type 16 E5 peptide with CpG-oligodeoxynucleotide can eliminate tumor growth in C57BL/6 mice. J Virol 2004; 78:1333-43; PMID:14722288; http://dx.doi.org/10.1128/JVI.78.3.1333-1343.2004
- Massa S, Simeone P, Muller A, Benvenuto E, Venuti A, Franconi R. Antitumor activity of DNA vaccines based on the human papillomavirus-16 E7 protein genetically fused to a plant virus coat protein. Hum Gene Ther 2008; 19:354-64; PMID:18439124; http://dx.doi.org/10.1089/hum.2007.122
- Cordeiro MN, Paolini F, Massa S, Curzio G, Illiano E, Duarte Silva AJ, et al. Anti-tumor effects of genetic vaccines against HPV major oncogenes. Hum Vaccin Immunother 2015; 11:45-52; PMID:25483514; http://dx.doi.org/10.4161/hv.34303
- Gray E, Pett MR, Ward D, Winder DM, Stanley MA, Roberts I, Scarpini CG, Coleman N. In vitro progression of human papillomavirus 16 episome-associated cervical neoplasia displays fundamental similarities to integrant-associated carcinogenesis. Cancer Res 2010; 70:4081-91; PMID:20442284; http://dx.doi.org/10.1158/0008-5472.CAN-09-3335
- Xu B, Chotewutmontri S, Wolf S, Klos U, Schmitz M, Dürst M, Schwarz E. Multiplex identification of Human Papillomavirus 16 DNA integration sites in cervical carcinomas. PLoS One 2013; 8:e66693; PMID:23824673; http://dx.doi.org/10.1371/journal.pone.0066693
- Sahab Z, Sudarshan SR, Liu X, Zhang Y, Kirilyuk A, Kamonjoh CM, et al. Quantitative measurement of human papillomavirus type 16 E5 oncoprotein levels in epithelial cell lines by mass spectrometry. J Virol 2012; 86:9465-73; PMID:22740411; http://dx.doi.org/10.1128/JVI.01032-12
- Chang JL, Tsao YP, Liu DW, Huang SJ, Lee WH, Chen SL. The expression of HPV- 16 E5 protein in squamous neoplastic changes in the uterine cervix. J Biomed Sci 2001; 8:206-13; PMID:11287752; http://dx.doi.org/10.1007/BF02256414
- Lorenzon L, Mazzetta F, Venuti A, Frega A, Torrisi MR, French D. In vivo HPV 16 E5 mRNA: expression pattern in patients with squamous intra-epithelial lesions of the cervix. J Clin Virol 2011; 52:79-83; PMID:21767984; http://dx.doi.org/10.1016/j.jcv.2011.06.007
- French D, Belleudi F, Mauro MV, Mazzetta F, Raffa S, Fabiano V, Frega A, Torrisi MR. Expression of HPV16 E5 down-modulates the TGFbeta signaling pathway. Mol Cancer 2013; 12:38; PMID:23651589; http://dx.doi.org/10.1186/1476-4598-12-38
- Zheng ZM, Baker CC. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci 2006; 11:2286-302. Review; PMID:16720315; http://dx.doi.org/10.2741/1971
- Chen J, Xue Y, Poidinger M, Lim T, Chew SH, Pang CL, Abastado JP, Thierry F. Mapping of HPV transcripts in four human cervical lesions using RNAseq suggests quantitative rearrangements during carcinogenic progression. Virology 2014; 462-463:14-24; PMID:25092457; http://dx.doi.org/10.1016/j.virol.2014.05.026
- Kho EY, Wang HK, Banerjee NS, Broker TR, Chow LT. HPV-18 E6 mutants reveal p53 modulation of viral DNA amplification in organotypic cultures. Proc Natl Acad Sci U S A 2013; 110:7542-9; PMID:23572574; http://dx.doi.org/10.1073/pnas.1304855110
- Liu DW, Tsao YP, Hsieh CH, Hsieh JT, Kung JT, Chiang CL, Huang SJ, Chen SL. Induction of CD8 T cells by vaccination with recombinant adenovirus expressing Human Papillomavirus Type 16 E5 Gene Reduces Tumor Growth. J Virol 2000; 74:9083-89; PMID:10982354; http://dx.doi.org/10.1128/JVI.74.19.9083-9089.2000
- Accardi L, Paolini F, Mandarino A, Percario Z, Di Bonito P, Di Carlo V, Affabris E, Giorgi C, Amici C, Venuti A. In vivo antitumor effect of an intracellular single-chain antibody fragment against the E7 oncoprotein of human papillomavirus 16. Int J Cancer 2014; 134:2742-7; PMID:24226851; http://dx.doi.org/10.1002/ijc.28604
- Bedoui S, Heath WR, Mueller SN. CD4(+) T-cell help amplifies innate signals for primary CD8(+) T-cell immunity. Immunol Rev 2016; 272:52-64; PMID:27319342; http://dx.doi.org/10.1111/imr.12426
- Ristriani T, Fournane S, Orfanoudakis G, Travé G, Masson M. A single-codon mutation converts HPV16 E6 oncoprotein into a potential tumor suppressor, which induces p53-dependent senescence of HPV-positive HeLa cervical cancer cells. Oncogene 2009; 28:762-72; PMID:19015633; http://dx.doi.org/10.1038/onc.2008.422
- Feltkamp MC, Smits HL, Vierboom MP, Minnaar RP, de Jongh BM, Drijfhout J W, et al. Vaccination with cytotoxic T lymphocyte epitope-containing peptide protects against a tumor induced by human papillomavirus type 16-transformed cells. Eur J Immunol 1993; 23:2242-49; PMID:7690326; http://dx.doi.org/10.1002/eji.1830230929
- Brasier AR, Tate JE, Habener JF. Optimized use of the firefly luciferase assay as a reporter gene in mammalian cell lines. Biotechniques 1989; 7:1116-22; PMID:2698191
- Venuti A, Massa S, Mett V, Vedova LD, Paolini F, Franconi R, Yusibov V. An E7-based therapeutic vaccine protects mice against HPV16 associated cancer. Vaccine 2009; 27:3395-7; PMID:19200826; http://dx.doi.org/10.1016/j.vaccine.2009.01.068