ABSTRACT
Highly pathogenic avian influenza (HPAI) H5N1 is an ongoing global health concern due to its severe sporadic outbreaks in Asia, Africa and Europe, which poses a potential pandemic threat. The development of safe and cost-effective vaccine candidates for HPAI is considered the best strategy for managing the disease and addressing the pandemic preparedness. The most potential vaccine candidate is the antigenic determinant of influenza A virus, hemagglutinin (HA). The present research was aimed at developing optimized expression in Nicotiana benthamiana and protein purification process for HA from the Malaysian isolate of H5N1 as a vaccine antigen for HPAI H5N1. Expression of HA from the Malaysian isolate of HPAI in N. benthamiana was confirmed, and more soluble protein was expressed as truncated HA, the HA1 domain over the entire ectodomain of HA. Two different purification processes were evaluated for efficiency in terms of purity and yield. Due to the reduced yield, protein degradation and length of the 3-column purification process, the 2-column method was chosen for target purification. Purified HA1 was found immunogenic in mice inducing H5 HA-specific IgG and a hemagglutination inhibition antibody. This paper offers an alternative production system of a vaccine candidate against a locally circulating HPAI, which has a regional significance.
Introduction
Intermittent outbreaks of influenza A and its spread in animals and transmission to humans have been drawing the attention of the world media. Outbreaks of H5N1 influenza in 2003 with a high mortality rate of roughly 60%Citation1 and of H1N1 influenza in 2009 with over 18,000 deathsCitation2 have posed unpredictable threats to the global populations. Ongoing circulation of H5N1 in poultry, especially in Asia, is of immense concern for the potential pandemic of highly pathogenic avian influenza (HPAI), H5N1 strain.
According to a phylogenetic analysis based on the hemagglutinin (HA) gene of H5N1 avian influenza (AI) strain,Citation3 Vietnam, Thailand and Malaysia (VTM) virus isolates (2003–2005) were grouped into a sublineage (Clade 1), whereas viruses isolated from 2003 onward in Indonesia formed another independent sublineage (Clade 2), suggesting the regional maintenance of certain sublineages in the poultry population. Since cross-clade protection against H5N1 viruses remains questionable, the wide antigenic diversity with a high mutation rate of HA circulating in poultry and wild birds means no one can envisage which H5N1 subtype will cause the next potential pandemic. This significantly challenges the current practice of reliance on a single or limited vaccine candidate(s) for pandemic preparedness. Therefore, it is believed that the development of a cost-effective vaccine candidate against an HPAI strain with a regional importance offers a significant tool to provide a higher level of immunoprotection against AI for the VTM provinces. A Malaysian isolate, A/chicken/Malaysia/5744/2004 (H5N1), belongs to the VTM sublineage and has been demonstrated to be highly pathogenic in chicken.Citation4 Its HA was only used to develop a DNA-based vaccineCitation5 and has not been expressed as a recombinant protein in any heterologous hosts including plants.
The use of plants as biofactories for recombinant protein production has gained great attention due to their pronounced advantages over other expression systems. Plant systems offer opportunities to produce vaccine antigens relatively fast using straightforward, cost-effective procedures, are highly scalable, and do not harbor mammalian pathogens.Citation6-10 In addition, plant cells are capable of performing eukaryotic post-translational modifications of target proteins, including N-linked glycosylation, which are substantially similar to those found in mammalian cells.Citation11 The transient expression approach is now the most extensively used and allows for the production of large quantities of target proteins within a short time frame,Citation12 which is particularly important in the case of epidemics.
Over the last decade, a growing number of candidate vaccines produced in plant systems have reached clinical or advanced preclinical stages of development (reviewed in refs.Citation13,14), including hepatitis B surface antigen,Citation15,16 plague F1-V fusion antigen,Citation17,18 anthrax protective antigen,Citation19,20 malaria Pfs25 and Pfs230 antigens,Citation21-23 and influenza HA.Citation24-28 Furthermore, a veterinary vaccine against Newcastle disease virus in poultry, produced in transgenic tobacco plant cell suspension by Dow AgroSciences LLC (Indianapolis, IN) and approved by the U.S. Department of Agriculture Center for Veterinary Biologics,Citation29 also shows the potential of vaccine development in planta.
Protein extraction and purification procedures can account for the largest percentage of the production cost; therefore, the development of simple, low-cost and reliable systems for purifying target proteins is ultimately important. Different purification approaches have been critically reviewed by Ward and Swiatek,Citation30 and column chromatography is widely used in plant molecular pharming. In this study, we engineered, expressed and purified HA of the abovementioned Malaysian isolate in N. benthamiana using a plant virus-based transient expression system,Citation31 and evaluated its immunogenicity in mice.
Results
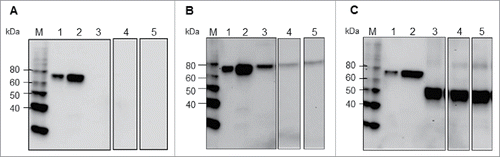
HA-MY and HA1-MY expression
HA sequences with or without codon optimization were cloned into the pGR-D4 vector, resulting in the constructs designated as pGR-D4::HA-MYNATIVE or pGR-D4::HA-MYOPT, respectively. The constructs were transformed into agrobacterium and then introduced into N. benthamiana as described in Materials and Methods. Expression of the HA protein in plants using each of these constructs was analyzed by Western blotting using an anti-4xHis antibody at 4 to 8 d post infiltration (dpi). The HA protein expression was not detected in plants infiltrated with agrobacterium containing pGR-D4::HA-MYnative (), while plants infiltrated with agrobacterium containing pGR-D4::HA-MYopt expressed detectable HA (), confirming the importance of codon optimization to the expression host. However, less protein was detected in total soluble protein (TSP) and TSP with 0.5% Triton X-100 (TST) preparations of the plant extract compare with plant total protein (PTP). Protein expression was also evaluated in plants infiltrated with pGR-D4 harboring truncated HA (HA1 domain) with codon optimization, pGR-D4::HA1-MYopt. The HA1 protein was detected as a ∼50 kDa band in all fractions of plant extract in a similar manner, suggesting that HA1 was expressed in plants as a soluble protein up to 8 dpi (). Considering higher expression and better solubility of HA1-MY over HA-MY in plants, further experiments were conducted for HA1-MY as a target protein.
Purification and characterization of recombinant HA1-MY
HA1-MY was first purified using a 3-column chromatography process, including immobilized metal affinity chromatography (IMAC; Ni-charge resin column) followed by hydrophobic interaction chromatography (HIC; Butyl column) and then by anion exchange chromatography (AEC; Q column). Target protein was detected in the elution (E) fraction from IMAC and flow through (FT) fractions from both Butyl and Q columns (). The ∼75 kDa major contaminant protein (arrow in the figure) was found in E and FT fractions of IMAC and Butyl columns, respectively (), and this major contaminant was removed during the Q column process as the ∼75 kDa protein band was not observed in the FT fraction while remained in the E fraction of Q column (). There was no residual target protein in the E fraction of Q column (). Ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) is one of the most abundant proteins in plants and acts as the major contaminant when purifying recombinant proteins expressed in plants.Citation32 In this study, we also evaluated the removal of RuBisCO during the purification process by Western blotting using an anti-RuBisCO antibody. With the Butyl column process, RuBisCO was successfully removed and no detectable band was observed on Western blot, whereas TSP and the E fraction of IMAC contained RuBisCO (). However, a loss of target protein was observed between the Butyl and Q column processes with a more than 50% decrease in the band intensity in Western blot analysis ().
Figure 2. Three-column purification process analyzed by SDS-PAGE and Western blot. (A) Coomassie-stained SDS-PAGE gel, (B) Western blot analysis using an anti-4xHis antibody, and (C) Western blot analysis using an anti-RuBisCO antibody. M: Molecular weight marker, Lane 1: E fraction from IMAC, Lane 2: FT fraction from Butyl column, Lane 3: FT fraction from Q column, Lane 4: E fraction from Q column, Lane 5: TSP.
Development of a 2-column purification process
To improve the yield throughout the purification process by decreasing the loss of target, the Butyl column process was omitted and the Q column process was applied immediately after IMAC with buffer exchange. Based on the SDS-PAGE with Coomassie blue staining, the major contaminant protein at ∼75 kDa in the E fraction of IMAC was removed by the Q column process without the Butyl column process (). Western blot analysis using an anti-4xHis antibody revealed that the HA1-MY protein was recovered from Q column without significant loss of protein, although the residual HA1-MY protein was detected in the E fraction of Q column (). RuBisCO was detected abundantly in the E fraction of IMAC and Q column, but not in the target fraction, FT of Q column ().
Figure 3. Two-column purification process analyzed by SDS-PAGE and Western blot. (A) Coomassie-stained SDS-PAGE gel, (B) Western blot analysis using an anti-4xHis antibody, and (C) Western blot analysis using an anti-RuBisCO antibody. M: Molecular weight marker, Lane 1: E fraction from IMAC, Lane 2: FT fraction from Q column, Lane 3: E fraction from Q column.
Analysis of glycosylation of HA1-MY
Although the putative molecular size of HA1-MY is 37 kDa, the target HA1-MY protein was detected as smear bands with molecular weight of around 45–55 kDa in both SDS-PAGE and Western blot (), suggesting that HA1-MY was expressed in plants with different degrees of glycosylation. To analyze glycosylation of the HA1-MY protein, purified HA1-MY was enzymatically treated with Peptide-N-Glycosidase F (PNGase F). The resulting products were detected by Coomassie staining as smeared 45–55 kDa, a major ∼40 kDa and a ∼35 kDa bands (). The 35 kDa protein, which was not initially seen in either SDS-PAGE gel or Western blot analysis and appeared in SDS-PAGE only after treatment with PNGase F, presumably is the PNGase F enzyme with a predicted molecular size of 36 kDA (). The deglycosylated ∼40 kDa protein was detected by Western blot analysis (), suggesting that HA1-MY underwent host glycosylation during expression in plants. Smear bands around 45–55 kDa were still observed after PNGase F treatment, but with less intensity. These could be residual glycosylated HA1-MY (due to insufficient PNGase F treatment) and HA1-MY that degraded during the enzyme treatment (a band just below the ∼40 kDa main band).
Figure 4. Deglycosylation of HA1-MY treated with PNGase F. (A) Coomassie-stained SDS-PAGE gel and (B) Western blot analysis using an anti-A/Vietnam/1194/04 antibody. M: Molecular weight marker, Lane 1: Non-treated HA1-MY, Lane 2: Mock-treated HA1-MY without PNGase F, Lane 3: PNGase F-treated HA1-MY.
Immunogenicity of HA1-MY in mice
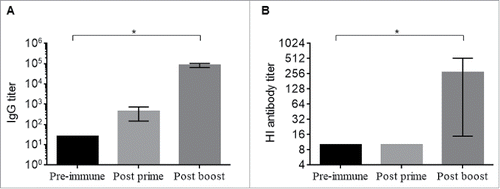
Immunogenicity of HA1-MY was determined by measuring anti-H5 IgG and hemagglutination inhibition (HI) antibody responses in sera from immunized mice. After the prime administration, anti-H5 IgG responses were slightly increased compare with pre-immunization, and these responses were significantly increased after the second administration of HA1-MY (p < 0.05, Kruskal-Wallis test, ). In addition, although the serum HI antibody was not detectable after the prime administration of HA1-MY, serum HI antibody responses were elicited after the second immunization, and the responses were significantly higher than those of pre-immune sera (p < 0.05, Kruskal-Wallis test, ).
Discussion
The present study was aimed at establishing efficient methods for plant transient expression and protein purification of HA from the Malaysian isolate of HPAI H5N1 virus strain, which would serve as the first step toward developing a plant-based recombinant subunit vaccine candidate in the future. We first targeted both the entire ectodomain of HA and the HA1 domain located at N-terminus of the HA molecule before the furin cleavage site, including globular receptor-binding domain.Citation33 HA is the most antigenic surface glycoprotein of influenza viruses, and several candidate subunit vaccines based on the ectodomain of HA produced in plants have been shown to induce the production of virus-neutralizing antibodies in mice34,35 and humansCitation26 and to protect ferrets against the virus challenge.Citation36,37 The HA1 domain contains almost all the antigenic and neutralization sites that have been characterized as conformational epitopes.Citation38,39 Hence, this domain is believed to be associated with neutralizing epitopes and ultimately, inducing an effective protective antibody response.Citation40,41
There are limited reports demonstrating production of the HA1 domain in planta. Although attempts of expression of (i) 37 kDa HA1 (amino acids 1–330), (ii) 34 kDa HA1 (amino acids 1–277) lacking a C-terminal fragment including a cleavage site and sequences that may confer poor solubility, and (iii) 27 kDa HA1 (amino acids 68–277) with an N-terminal deletion were reported,Citation42 the 37 kDa domain did not express in this previous study. The predicted 27 kDa domain only showed minimal expression with a 35 kDa product, whereas for the predicted 34 kDa domain, a 37 kDa product and a diffusely migrating 45 kDa product were observed.Citation42 In contrast, we have shown that the HA1 domain (i.e. the 37 kDa product in ref. Citation42) can be expressed at a high level. These differences might be due to the differences in the HA amino acid sequence among influenza strains used, since in the same plant expression system, the HA ectodomain from different H5N1 strains expressed as soluble protein.Citation35,37 Further investigation might support the hypothesis, and depending on the targeted influenza strain, either the HA ectodomain or the HA1 domain can be utilized to efficiently produce HA antigens as influenza vaccine candidates.
In the process of developing the HA1-MY purification method, 3-column purification was originally applied where IMAC chromatography was followed by Butyl column chromatography (HIC) and then by Q column chromatography (AEC). Given a high amount of impurities, the HIC and AEC steps were performed in the FT mode with the aim of efficient binding of major contaminants to the columns.Citation43-45 The least hydrophobic media (Butyl) and salt concentration as low as 1M NaCl were used, so that target protein would be fractionated in FT and not bind to the column. HIC was applied as the second step because of its ability to remove the most significant contaminant, RuBisCO,Citation46 which has the similar size to HA1-MY. RuBisCO was not detected in FT of Butyl column, confirming its binding to the column. AEC was then applied to remove another major contaminant with a molecular size of around 75 kDa, since HA1-MY (pI 8.6) has a positive charge on its surface and would not bind to the column at pH 7.5. The results demonstrated successful separation of target protein from RuBisCO using Butyl column and from the 75 kDa contaminant using Q column.
While the 3-column purification process is effective, it is time-consuming and a low recovery of target was observed from column to column, especially with column re-use, as reported elsewhere.Citation47,48 In order to reduce the process time and to improve the target yield, a 2-column purification process was evaluated for its efficiency and yield. IMAC eluent was dialyzed to remove excessive NaCl and applied immediately onto a Q column, without the Butyl column chromatography step. Based on Q column chromatogram profiles (data not shown), products of interest were observed in the 2 adjacent peaks in FT of Q column, suggesting heterogeneity of the target protein. Protein microheterogeneity may arise during translational or post-translational processing in eukaryotic systems, likely here due to differential glycosylation, deamidation or oxidation of amino acids or proteolytic cleavage by endogenous peptidases. In addition, the charge of specific amino groups may be reversed by acylation or carbamylation.Citation30 On the other hand, RuBisCO was found predominantly in the E fraction of Q column demonstrating that AEC (Q column) is comparable to HIC (Butyl column) in terms of RuBisCO removal from HA1-MY preparation ().
HA1-MY purified by the 2-column purification process migrated in SDS-PAGE as heterogeneous proteins in the 45–55 kDa range rather than as the predicted 38.69 kDa protein, indicating various levels of HA1-MY glycosylation. PNGase F treatment resulted in the appearance of one major protein of about 40 kDa, which is close to the predicted size (), demonstrating that several intermediate glycoforms were produced in the plant system during expression. Similar findings were obtained with yeast-expressed N1 neuraminidase (NA),Citation49 where after PNGase F treatment, the diffuse 72 kDa N1 NA protein reduced to 45 and 60 kDa species that were recognized by an anti-NA antibody. PNGase F cleaves N-glycans from glycoproteins, except when the (1–3) core is fucosylated in the Golgi body. Protein retention in the endoplasmic reticulum (ER) mediated by the KDEL signal might not be completely achieved because plant cells can express proteins in an unsynchronized manner. Therefore, some proteins have occasionally been transported across the ER to the Golgi body before being retrieved back to the ER.Citation50 This may result in a minor subpopulation of proteins exhibiting plant-specific glycosylation such as 1,3-Fucose linked to the Asparagine-linked N-acetylglucosamine that is resistant to PNGase F cleavage.Citation51
Purified HA1-MY was evaluated for immunogenicity in mice and demonstrated the ability to induce both anti-H5 IgG and HI antibody responses after 2 immunizations. These results indicate that the plant-produced HA1 domain of HA from the Malaysian HPAI H5N1 isolate, A/chicken/Malaysia/5744/2004 (H5N1), is immunogenic in mice inducing protection-related antibody responses. However, direct comparison of HA-MY versus HA1-MY in terms of immunogenicity was not performed in this study and further investigation would provide immunogenicity profile of the ectodomain and HA1 domain of HA from the Malaysian HPAI strain.
In summary, we have engineered the HA1 domain of regionally circulating HPAI H5N1 strain, expressed it in a plant transient expression system, and successfully purified the target protein. Plant-produced HA1-MY contained several glycoforms and demonstrated its immunogenicity in mice. The results of this study suggest feasibility of cost-effective production of vaccine candidates against regionally significant HPAI strains that would be affordable in developing countries.
Materials and methods
Cloning and expression of the HA sequence using a plant viral vector
The HA gene sequence of a Malaysian HPAI virus strain A/chicken/Malaysia/5744/2004 (H5N1) encoding amino acids 17–532 (AGH30707) was synthesized with or without codon optimization for expression in N. benthamiana (Blue Heron, USA). The HA gene sequence encoding amino acids 17–345 (the HA1 domain of HA) was synthesized with codon optimization. For construction of the expression cassette, the pathogenesis-related protein 1a (PR-1a) signal peptide sequence, originated from Nicotiana tabacum (NCBI accession number: BAA14220; amino acids 1–30) was incorporated at the N-terminus of either native HA, codon-optimized HA or codon-optimized HA1. A hexa-histidine (6xHis) tag and the ER retention signal peptide (KDEL) were added at the C-terminus. Each resulting gene cassette was sub-cloned into the tobacco mosaic virus-based pGR-D4 launch vector.Citation31 Constructs were designated as pGR-D4::HA-MYnative, pGR-D4::HA-MYopt or pGR-D4::HA1-MYopt. The constructed recombinant vectors were transformed into Agrobacterium tumefaciens GV 3101 strain and then introduced into N. benthamiana plants by vacuum infiltration as described elsewhere.Citation31,37 Expressed HA proteins using pGR-D4::HA-MYnative, pGR-D4::HA-MYopt or pGR-D4::HA1-MYopt were designated as HA-MYnative, HA-MY or HA1-MY, respectively.
Western blot analysis of HA-MY and HA1-MY expression
Each target protein accumulating in plants was extracted in the buffer (50 mM Na2HPO4, 500 mM NaCl, 20 mM Imidazole, pH 7.5) and analyzed using SDS-PAGE and Western blot as PTP, TSP, TST and chromatographically purified fractions. Briefly, protein samples separated by 10% SDS-PAGE were transferred onto polyvinylidenedifluoride (PVDF) membranes using a mini trans-blot cell system (Biorad, Hercules, CA). For target detection, primary mouse anti-4xHis antibody (Qiagen, USA), primary sheep anti-A/Vietnam/1194/04 (NIBSC, UK) polyclonal antisera, and horseradish peroxidase-conjugated secondary goat anti-mouse (Qiagen, USA) and donkey anti-sheep (Jackson Immuno Research, UK) antibodies were used. Plant-produced HA of influenza virus strain A/Indonesia/05/05 (HA Indonesia) was used as a control H5 HA protein.Citation37 SuperSignal® West Pico Chemiluminescent substrate solution (Pierce, Rockford, IL) was used for signal development. Images were captured using GeneGnome5 instrument (Syngene, Frederick, MD).
Chromatographic purification of recombinant HA1-MY
Purification of recombinant HA1-MY was conducted using 3 chromatographic methods, including the Ni-Sepharose-based IMAC (GE Healthcare), Butyl Sepharose-based High Performance (HP) HIC (GE Healthcare) and Quaternary amine (Q)-based AEC (TOSOH). At 7 dpi, infiltrated leaf tissues were harvested and frozen at −80°C. Tissues were subsequently thawed, homogenized and extracted in 3 volumes of IMAC binding buffer (50 mM Na2HPO4, 500 mM NaCl, 20 mM Imidazole, pH 7.5). The extract was then centrifuged at 3,000 × g for 20 min at 4°C and the supernatant was filtered through cheesecloth. The filtrate was then centrifuged at 40,000 × g for 20 min at 4°C, and the supernatant was filtered through a 0.2 µm filter. Recombinant HA1 was subsequently purified using IMAC. For 3-column purification process, the E fraction of IMAC was passed through both Butyl (HIC) and Q (AEC) columns. For 2-column purification process, the E fraction of IMAC was dialyzed using a 10 kDa MWCO dialysis cassette to reduce NaCl concentration to 50 mM. The dialysis cassettes were put into 1L start buffer for AEC and stirred slowly with a magnetic stirrer at 4°C for 2–3 hr. The buffer was changed at least twice and samples were retrieved from the cassettes. Both HIC and AEC were performed using an AKTA™ purifier 10 controlled by the Unicorn software (Unicorn Software Solutions, Bowling Green, KY).
Deglycosylation of purified HA1-MY by PNGase F treatment
Deglycosylation of target molecules was conducted using the reaction buffer of Enzymatic CarboRelease Kit (QA-Bio, USA) and PNGase F (New England Biolabs, UK). Protein samples were individually mixed with 50 mM Sodium phosphate (pH 7.0) and 5% (v/v) denaturing solution followed by boiling for 5 min. The denatured protein samples were then chilled on ice followed by adding 1% Triton X-100 and 750 U PNGase F in the total reaction volume of 50 µL. These samples were incubated at 37°C overnight.
Immunogenicity of HA1-MY in mice
Six-week-old female Balb/c mice were immunized intramuscularly with 30 µg of HA1-MY in the presence of 10 µg of Quil A (Accurate Chemical, NY) on study days 0 and 21. Serum samples were collected prior to the prime immunization (Pre-immune) and 2 weeks after the prime (Post prime) and booster (Post boost) immunizations. Collected serum samples were evaluated for anti-H5 HA IgG responses by ELISA using inactivated A/Vietnam/1194/04 (H5N1, clade 1) as a coating antigen and for HI antibody responses in the HI assay using A/Vietnam/1194/04 virus (NIBRG-14, MIBSC, UK) and horse erythrocytes as described previously.Citation37
Abbreviations
| AEC | = | anion exchange chromatography |
| AI | = | avian influenza |
| dpi | = | days post infiltration |
| E | = | elution |
| ELISA | = | enzyme-linked immunosorbent assay |
| ER | = | endoplasmic reticulum |
| FT | = | flow through |
| HA | = | hemagglutinin |
| HI | = | hemagglutination inhibition |
| HIC | = | hydrophobic interaction chromatography |
| HPAI | = | highly pathogenic avian influenza |
| IMAC | = | immobilized metal affinity chromatography |
| kDa | = | kilodalton |
| KDEL | = | endoplasmic reticulum retention signal peptide |
| mM | = | millimolar |
| MWCO | = | molecular weight cut off |
| MY | = | Malaysia |
| PNGase F | = | peptide-N-Glycosidase F |
| PTP | = | plant total protein |
| PVDF | = | polyvinylidenedifluoride |
| TSP | = | total soluble protein |
| TST | = | TSP with 0.5% Triton X-100 |
| RuBisCO | = | ribulose-1,5-bisphosphate carboxylase/oxygenase |
| SDS-PAGE | = | sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| U | = | unit |
| VTM | = | Vietnam Thailand and Malaysia |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Dr. Natasha Kushnir (Fraunhofer USA - Center for Molecular Biotechnology) for editorial assistance.
Funding
The work was partly supported by the Ministry of Agriculture and Agro-based Industry (MOA), Malaysia under the Science Fund (05–02–12-SF1007).
References
- World Health Organization. Cumulative Number of Confirmed Human Cases of Avian Influenza A/(H5N1) Reported to WHO: 10 August 2012. http://www.who.int/influenza/human_animal_interface/EN_GIP_20120810CumulativeNumberH5N1cases.pdf, 2012
- World Health Organization. Pandemic (H1N1) 2009 – update 103, 4 June 2010. http://www.who.int/csr/don/2010_06_04/en/index.html, 2010
- Chen H, Smith GJD, Li KS, Wang J, Fan XH, Rayner JM, Vijaykrishna D, Zhang JX, Zhang LJ, Guo CT, et al. Establishment of multiple sublineages of H5N1 influenza virus in Asia: Implications for pandemic control. PNAS USA 2006; 103:2845-50; PMID:16473931; http://dx.doi.org/10.1073/pnas.0511120103
- Balasubramaniam VRMT, Hassan SS, Omar AR, Mohamed M, Noor SM, Mohamed R, Othman I. Cellular transcripts regulated during infections with Highly Pathogenic H5N1 Avian Influenza virus in 3 host systems. Virology J 2011; 8:196; http://dx.doi.org/10.1186/1743-422X-8-196
- Oveissi S, Omar AR, Yusoff K, Jahanshiri F, Hassan SS. DNA vaccine encoding avian influenza virus H5 and Esat-6 of Mycobacterium tuberculosis improved antibody responses against AIV in chickens. Comp Immunol Microbiol Infect Dis 2010; 33:491-503; PMID:19781778; http://dx.doi.org/10.1016/j.cimid.2009.08.004
- Basaran P, Rodríguez-Cerezo E. Plant molecular farming: opportunities and challenges. Crit Rev Biotechnol 2008; 28:153-72; PMID:18937106; http://dx.doi.org/10.1080/07388550802046624
- Tiwari S, Verma PC, Singh PK, Tuli R. Plants as bioreactors for the production of vaccine antigens. Biotechnol Adv 2009; 27:449-67; PMID:19356740; http://dx.doi.org/10.1016/j.biotechadv.2009.03.006
- Twyman RM, Schillberg S, Fischer R. The Production of Vaccines and Therapeutic Antibodies in Plants. In: Molecular Farming in Plants: Recent Advances and Future Prospects. Wang, A. and Ma, S, eds. Springer Science+Business Media B.V., The Netherlands. Chapter 7. 2012; pp. 145-58
- Xu J, Dolan MC, Medrano G, Cramer CL, Weathers PJ. Green factory: plants as bioproduction platforms for recombinant proteins. Biotechnol Adv 2012; 30:1171-84; PMID:21924345; http://dx.doi.org/10.1016/j.biotechadv.2011.08.020
- Stoger E, Fischer R, Moloney M, Ma JK. (2014) Plant molecular pharming for the treatment of chronic and infectious diseases. Annu Rev Plant Biol 2014; 65:743-68; PMID:24579993; http://dx.doi.org/10.1146/annurev-arplant-050213-035850
- Gomord V, Faye L. Posttranslational modification of therapeutic proteins in plants. Curr Opin Plant Biol 2004; 7:171-81; PMID:15003218; http://dx.doi.org/10.1016/j.pbi.2004.01.015
- Rybicki EP. Plant-made vaccines for humans and animals. Plant Biotechnol J 2010; 8:620-37; PMID:20233333; http://dx.doi.org/10.1111/j.1467-7652.2010.00507.x
- Yusibov V, Streatfield S, Kushnir N. Clinical development of plant-produced recombinant pharmaceuticals vaccines, antibodies and beyond. Human Vaccin 2011; 7:1-9; http://dx.doi.org/10.4161/hv.7.3.14207
- Yusibov V, Kushnir N, Streatfield SJ. Advances and challenges in the development and production of effective plant-based influenza vaccines. Expert Rev Vaccin 2015; 14:519-35; http://dx.doi.org/10.1586/14760584.2015.989988
- Sunil Kumar GB, Ganapathi TR, Srinivas L, Revathi CJ, Bapat VA. Expression of hepatitis B surface antigen in transgenic banana plants. Planta 2005a; 222:484-93; PMID:15918027; http://dx.doi.org/10.1007/s00425-005-1556-y
- Sunil Kumar GB, Ganapathi TR, Srinivas L, Revathi CJ, Bapat VA. Secretion of hepatitis B surface antigen in transformed tobacco cell suspension cultures. Biotechnol Lett 2005b; 27:927-32; PMID:16091888; http://dx.doi.org/10.1007/s10529-005-7185-5
- Arlen PA, Singleton M, Adamovicz JJ, Ding Y, Davoodi-Semiromi A, Daniell H. Effective plague vaccination via oral delivery of plant cells expressing F1-V antigens in chloroplasts. Inf Immun 2008; 76:3640-50; http://dx.doi.org/10.1128/IAI.00050-08
- Chichester JA, Musiychuk K, Farrance CE, Mett V, Lyons J, Mett V, Yusibov V. A single component two-valent LcrV-F1 vaccine protects non-human primates against pneumonic plague. Vaccine 2009; 27:3471-4; PMID:19200825; http://dx.doi.org/10.1016/j.vaccine.2009.01.050
- Chichester JA, Musiychuk K, de la Rosa P, Horsey A, Stevenson N, Ugulava N, Rabindran S, Palmer GA, Mett V, Yusibov V. Immunogenicity of a subunit vaccine against Bacillus anthracis. Vaccine 2007; 25:3111-4; PMID:17280756; http://dx.doi.org/10.1016/j.vaccine.2007.01.068
- Chichester JA, Manceva SD, Rhee A, Coffin MV, Musiychuk K, Mett V, Shamloul M, Norikane J, Streatfield SJ, Yusibov V. A plant-produced protective antigen vaccine confers protection in rabbits against a lethal aerosolized challenge with Bacillus anthracis Ames spores. Hum Vaccin Immunother 2013; 9:544-52; PMID:23324615; http://dx.doi.org/10.4161/hv.23233
- Farrance CE, Rhee A, Jones RM, Musiychuk K, Shamloul M, Sharma S, Mett V, Chichester JA, Streatfield SJ, Roeffen W. A plant-produced Pfs230 vaccine candidate blocks transmission of Plasmodium falciparum. Clin Vaccine Immunol 2011; 18:1351-7; PMID:21715576; http://dx.doi.org/10.1128/CVI.05105-11
- Jones RM, Chichester JA, Mett V, Jaje J, Tottey S, Manceva S, Casta LJ, Gibbs SK, Musiychuk K, Shamloul M. A plant-produced Pfs25 VLP malaria vaccine candidate induces persistent transmission blocking antibodies against Plasmodium falciparum in immunized mice. PLoS One 2013; 8:e79538; PMID:24260245; http://dx.doi.org/10.1371/journal.pone.0079538
- Jones RM, Chichester JA, Manceva S, Gibbs SK, Musiychuk K, Shamloul M, Norikane J, Streatfield SJ, van de Vegte-Bolmer M, Roeffen W. A novel plant-produced Pfs25 fusion subunit vaccine induces long-lasting transmission blocking antibody responses. Hum Vaccin Immunother 2015; 11:124-32; PMID:25483525; http://dx.doi.org/10.4161/hv.34366
- Landry N, Ward BJ, Trépanier S, Montomoli E, Dargis M, Lapini G, Vézina LP. Preclinical and clinical development of plant-made virus-like particle vaccine against avian H5N1 influenza. PLoS One 2010; 5:e15559; PMID:21203523; http://dx.doi.org/10.1371/journal.pone.0015559
- Chichester JA, Jones RM, Green BJ, Stow M, Miao F, Moonsammy G, Streatfield SJ Yusibov V. Safety and immunogenicity of a plant-produced recombinant hemagglutinin-based influenza vaccine (HAI-05) derived from A/Indonesia/05/2005 (H5N1) influenza virus: a phase 1 randomized, double-blind, placebo-controlled, dose-escalation study in healthy adults. Viruses 2012; 4:3227-44; PMID:23202523; http://dx.doi.org/10.3390/v4113227
- Cummings JF, Guerrero ML, Moon JE, Waterman P, Nielsen RK, Jefferson S, Gross FL, Hancock K, Katz JM, Yusibov V and Fraunhofer USA Center for Molecular Biotechnology Study Group. Safety and immunogenicity of a plant-produced recombinant monomer hemagglutinin-based influenza vaccine derived from influenza A (H1N1)pdm09 virus: a Phase 1 dose-escalation study in healthy adults. Vaccine 2014; 32:2251-9; PMID:24126211; http://dx.doi.org/10.1016/j.vaccine.2013.10.017
- Landry N, Pillet S, Favre D, Poulin JF, Trépanier S, Yassine-Diab B, Ward BJ. Influenza virus-like particle vaccines made in Nicotiana benthamiana elicit durable, poly-functional and cross-reactive T cell responses to influenza HA antigens. Clin Immunol 2014; 154:164-77; PMID:25128897; http://dx.doi.org/10.1016/j.clim.2014.08.003
- Ward BJ, Landry N, Trépanier S, Mercier G, Dargis, M, Couture M, D'Aoust MA, Vézina LP. Human antibody response to N-glycans present on plant-made influenza virus-like particle (VLP) vaccines. Vaccine 2014; 32:6098-106; PMID:25240757; http://dx.doi.org/10.1016/j.vaccine.2014.08.079
- The Poultry Site. USDA Issues License For Plant Cell Produced Newcastle Disease Vaccine For Chickens. 2 February 2006. http://www.thepoultrysite.com/poultrynews/8949/usda-issues-license-for-plant-cell-produced-newcastle-disease-vaccine-for-chickens/, 2006
- Ward WW, Swiatek G. Protein purification. Curr Anal Chem 2009; 5:1-21; http://dx.doi.org/10.2174/157341109787846171
- Musiychuk K, Stephenson N, Bi H, Farrance CE, Orozovic G, Brodelius M, Brodelius P, Horsey A, Ugulava N, Shamloul AM, et al. A launch vector for the production of vaccine antigens in plants. Influenza Other Respir Viruses 2007; 1:19-25; PMID:19453476; http://dx.doi.org/10.1111/j.1750-2659.2006.00005.x
- Buyel JF, Twyman RM, Fischer R. Extraction and downstream processing of plant-derived recombinant proteins. Biotechnol Adv 2015; 33(6 Pt 1):902-13; PMID:25922318; http://dx.doi.org/10.1016/j.biotechadv.2015.04.010
- Hong M, Lee PS, Hoffman RM, Zhu X, Krause JC, Laursen NS, Yoon SI, Song L, Tussey L, Crowe JE Jr, et al. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J Virol 2013; 87:12471-80; PMID:24027321; http://dx.doi.org/10.1128/JVI.01388-13
- Shoji Y, Chichester JA, Bi H, Musiychuk K, de la Rosa P, Goldschmidt L, Horsey A, Ugulava N, Palmer GA, Mett V, et al. Plant-expressed HA as a seasonal influenza vaccine candidate. Vaccine 2008; 26:2930-4; PMID:18440103; http://dx.doi.org/10.1016/j.vaccine.2008.03.045
- Shoji Y, Farrance CE, Bi H, Shamloul M, Green B, Manceva S, Rhee A, Ugulava N, Roy G, Musiychuk K, et al. Immunogenicity of hemagglutinin from A/Bar-headed Goose/Qinghai/1A/05 and A/Anhui/1/05 strains of H5N1 influenza viruses produced in Nicotiana benthamiana plants. Vaccine 2009a; 27:3467-70; PMID:19200814; http://dx.doi.org/10.1016/j.vaccine.2009.01.051
- Mett V, Musiychuk K, Bi H, Farrance CE, Horsey A, Ugulava N, Shoji Y, de la Rosa P, Palmer GA, Rabindran S, et al. A plant-produced influenza subunit vaccine protects ferrets against virus challenge. Influenza Other Respir Viruses 2008; 2:33-40; PMID:19453491; http://dx.doi.org/10.1111/j.1750-2659.2008.00037.x
- Shoji Y, Bi H, Musiychuk K, Rhee A, Horsey A, Roy G, Green B, Shamloul M, Farrance CE, Taggart B, et al. Plant-derived hemagglutinin protects ferrets against challenge infection with the A/Indonesia/05/05 strain of avian influenza. Vaccine 2009b; 27:1087-92; PMID:19100806; http://dx.doi.org/10.1016/j.vaccine.2008.11.108
- Suzuki Y, Nei M. Origin and Evolution of Influenza Virus Hemagglutinin Genes. Mol Biol Evol 2002; 19:501-9; PMID:11919291; http://dx.doi.org/10.1093/oxfordjournals.molbev.a004105
- Wiley DC, Skehel JJ. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Ann Rev Biochem 1987; 56:365-94; PMID:3304138; http://dx.doi.org/10.1146/annurev.bi.56.070187.002053
- Chiu FF, Venkatesan N, Wua CR, Chou AH, Chen HW, Lian SP, Liu SJ, Huang CC, Lian WC, Chong P, et al. Immunological study of HA1 domain of hemagglutinin of influenza H5N1 virus. Biochem Biophys Res Commun 2009; 383:27-31; PMID:19324009; http://dx.doi.org/10.1016/j.bbrc.2009.03.106
- Khurana S, Verma S, Verma N, Crevar CJ, Carter DM, Manischewitz J, King LR, Ross TM, Golding H. Bacterial HA1 vaccine against pandemic H5N1 influenza virus: Evidence of oligomerization, hemagglutination, and cross-protective immunity in ferrets. J Virol 2011a; 85:1246-56; PMID:21084473; http://dx.doi.org/10.1128/JVI.02107-10
- Spitsin S, Andrianov V, Pogrebnyak N, Smirnov Y, Borisjuk N, Portocarrero C, Veguilla V, Koprowski H, Golovkin M. Immunological assessment of plant derived avian flu H5/HA1 variants. Vaccine 2009; 27:1289-92; PMID:19162113; http://dx.doi.org/10.1016/j.vaccine.2008.12.050
- Bendandi M, Marillonnet S, Kandzia R, Thieme F, Nickstadt A, Herz S, Frode R, Inoges S, Lopez-Diaz de Cerio A, Soria E. et al. Rapid, high-yield production in plants of individualized idiotype vaccines for non-Hodgkin's lymphoma. Ann Oncol 2010; 21:2420-7; PMID:20494963; http://dx.doi.org/10.1093/annonc/mdq256
- Lai H, Engle M, Fuchs A, Keller T, Johnson S, Gorlatov S, Diamond MS, Chen Q. Monoclonal antibody produced in plants efficient treats West Nile virus infection in mice. PNAS USA 2010; 107:2419-24; PMID:20133644; http://dx.doi.org/10.1073/pnas.0914503107
- Wilken LR, Nikolov ZL. Recovery and purification of plant-made recombinant proteins. Biotechnol Adv 2012; 30:419-33; PMID:21843625; http://dx.doi.org/10.1016/j.biotechadv.2011.07.020
- Holler CJ. Purification of an acidic recombinant protein from transgenic tobacco (master thesis). Master Thesis, Virginia Polytechnic Institute and State University, Blacksburg, VA; 2007
- Drossard J. Downstream processing of plant-derived recombinant therapeutic proteins. In Fischer R, Schillberg S (Eds.), Molecular farming: Plant-made pharmaceuticals and technical proteins. 2004. Wiley-VCH Verlag GmbH & Co., Germany
- Valdes R, Gomez L, Padilla S, Brito J, Reyes B, Alvarez T, Mendoza O, Herrera O, Ferro W, Pujol M, et al. Large-scale purification of an antibody directed against hepatitis B surface antigen from transgenic tobacco plants. Biochem Biophys Res Commun 2003; 308:94-100; PMID:12890485; http://dx.doi.org/10.1016/S0006-291X(03)01335-4
- Yongkiettrakul S, Boonyapakron K, Jongkaewwattana A, Wanitchang A, Leartsakulpanich U, Chitnumsub P, Eurwilaichitr L, Yuthavong Y. Avian influenza A/H5N1 neuraminidase expressed in yeast with a functional head domain. J Virol Methods 2009; 156:44-51; PMID:19038288; http://dx.doi.org/10.1016/j.jviromet.2008.10.025
- Pagny S, Cabanes-Macheteau M, Gillikin JW, Leborgne-Castel N, Lerouge P, Boston RS, Faye L, Gomord V. Protein recycling from the Golgi apparatus to the endoplasmic reticulum in plants and its minor contribution to calreticulin retention. Plant Cell 2000; 12:739-56; PMID:10810147; http://dx.doi.org/10.1105/tpc.12.5.739
- Tretter V, Altmann F, Marz L. Peptide-N4-(N-acetyl-β-glucosaminyl) asparagine amidase F cannot release glycans with fucose attached α 1→3 to the asparagine-linked N-acetylglucosamine residue. Eur J Biochem 1991; 299:647-52; http://dx.doi.org/10.1111/j.1432-1033.1991.tb16166.x