ABSTRACT
Big data analysis has become a common way to extract information from complex and large datasets among most scientific domains. This approach is now used to study large cohorts of patients in medicine. This work is a review of publications that have used artificial intelligence and advanced machine learning techniques to study physio pathogenesis-based treatments in pSS. A systematic literature review retrieved all articles reporting on the use of advanced statistical analysis applied to the study of systemic autoimmune diseases (SADs) over the last decade. An automatic bibliography screening method has been developed to perform this task. The program called BIBOT was designed to fetch and analyze articles from the pubmed database using a list of keywords and Natural Language Processing approaches. The evolution of trends in statistical approaches, sizes of cohorts and number of publications over this period were also computed in the process. In all, 44077 abstracts were screened and 1017 publications were analyzed. The mean number of selected articles was 101.0 (S.D. 19.16) by year, but increased significantly over the time (from 74 articles in 2008 to 138 in 2017). Among them only 12 focused on pSS but none of them emphasized on the aspect of pathogenesis-based treatments. To conclude, medicine progressively enters the era of big data analysis and artificial intelligence, but these approaches are not yet used to describe pSS-specific pathogenesis-based treatment. Nevertheless, large multicentre studies are investigating this aspect with advanced algorithmic tools on large cohorts of SADs patients.
Introduction
According to the National Institute of Health (NIH),Citation1 systemic autoimmune diseases (SADs) affect 23.5 million people in United States (prevalence of 8%). The proportion of primary Sjogren's syndrome (pSS) among patients affected by systemic autoimmune diseases (SADs) remains debated, but prevalence of the disease is estimated around 0.05%.Citation2 pSS usually affects the salivary and lacrymal glands and results in severe dryness of mucosal surfaces, mainly in mouth and eyes.Citation3 pSS predominantly affects middle-aged women (with 16:1 sex ratioCitation4) and can lead to lymphoma.Citation5 The clinical characterization of this disease is actually based on several factors such as the detection of autoantibodies in serum and histological analysis of biopsied salivary gland tissue.Citation6 Clinical outcomes such as systemic activity scoring (ESSDAI) and patient-reported scoring (ESSPRI) are driving research but researchers are currently working on new outcomes. Treatment decisions are based on the initial evaluation of symptoms and extraglandular manifestations but also risk of mortality. Indeed, Sjogren's syndrom is classically associated to an increased risk of lymphoma but also to an overall elevated standardized mortality ratio with most common causes of death respiratory,Citation5, Citation7-Citation9 although overall survival of patients with pSS was not different from that of the general population in the US.Citation2 Even though knowledge of pSS has progressed substantially over the last years, it remains a complex disease, used as a study case for the development of pathogenesis-based treatments, which is a very active field of research.Citation10 Three Europeans studies (Big data project which collected previously obtained cohorts; HarmonicSS, an H2020 project aimed to the harmonization of data collected; PRECISEADS, a European IMI aimed to molecularly reclassify systemic autoimmune diseases including pSS) have started to investigate unprecedented quantity of data generated on large cohorts of SADs patients with the aim to improve the accuracy of diagnosis and find new therapeutic targets.
Although over the last decades technologies of signal acquisition in biology have evolved to produce an increasing amount of data, approaches such as Next generation sequencing (NGS), Flow cell cytometry or quantitative PCR have led to an explosion of the possible number of variables used to describe a patient. The development of these technologies and the reduction of their cost over the years make it possible to use these approaches on large cohorts of patients. They also lead to the generation of huge datasets where a significant number of patients are described by a large number of heterogeneous variables: genetic expression, cellular profile, metabolites detection and so on.
Extracting knowledge from these extremely large and often unstructured datasets is a very complex task, since the information they contain may not be analyzable by means of classical statistical methods. Over the past few years, new approaches based on artificial intelligence and advanced machine learning techniques have been used to analyze this kind of “big data”, especially in a clinical context, including the study of SAD.Citation11 This led to an accelerated growth in the number of publications in scientific and medical journals.
Since 2008, the National Center for Biotechnology and Information (NCBI) has released an Application Programming Interface (API), called E-utility and designed to access the contents of publications and their relative information stored in the Medline database. With this API came the possibility to perform automatic driven search on this database.
The objective of this work was to report on all publications that have used big data analysis and machine learning approaches to study pysio pathogenesis-based treatments in pSS. To this end, we performed a systematic literature review using a semi automatic approach.
Results
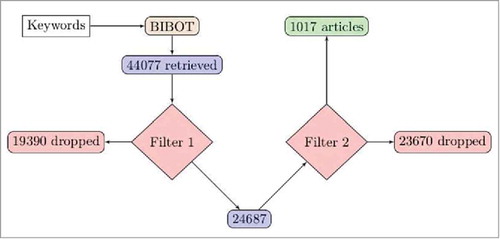
Altogether, 44077 abstracts were found in the NCBI using a combination of queries generated by BIBOT.Citation12-Citation14 Among them, 19390 abstracts failed to pass the first filter and 23670 were dropped by the second filter, leading to a final selection of 1017 abstracts. Among this selection 12 articles were related to the use of machine learning techniques for the classification of Sjögren patients, 885 were about the study of systemic lupus erythematosus patients, 173 were about the analysis of rheumatoid arthritis patients and 844 were about the use of machine learning to explore other autoimmune diseases. Over the last decade, the number of publications concerning the use of advanced statistical techniques applied to the exploration and classification of autoimmune diseases found by BIBOT has globally increased, and almost doubled from 2008 (74 articles retrieved) to 2017 (138 articles retrieved). The mean number of selected articles over this period of time is 101.0 (S.D. 19.16) by year, 93 of the selected articles were published in 2009, 97 in 2010, 77 in 2011, 85 in 2012, 106 in 2013, 123 in 2014, 113 in 2015 and 104 in 2016. The distribution of publications among the countries is unequal, with 412 publications from the United States, 313 publications from England and 292 publications dispatched between Australia, Brazil, Canada, China, Croatia, Egypt, France, Germany, Greece, India, Iran, Ireland, Israel, Italy, Japan, Netherlands, New Zealand, Poland, Puerto Rico, Romania, Saudi Arabia, Scotland, South Korea, Spain, Switzerland, Taiwan, Thailand, and Turkey.
General trends in publications
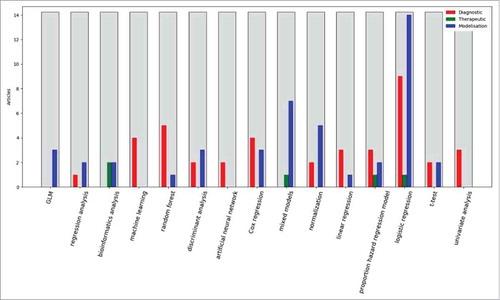
The 1017 selected articles were automatically classified into four categories: “Diagnostic” for the articles that focus on the diagnosis procedure for autoimmune diseases, “Therapeutic” for the articles that focus on therapy aspects, “Modelization” for the articles that emphasize on the study of the mechanistic aspect of SADS and “Unclassified” for the articles that failed to be automatically classified. 249 articles were categorized in the “Diagnostic” category, 91 in the “Therapeutic” category, 661 in the “Modelization” category and 16 in the “Unclassified” category. Among the 249 articles classified in the “Diagnostic” category, three deal with Sjögren's syndrome (1.2%). No article concerns pSS in the “Therapeutic” category and 9 concern pSS in the “Modelisation” category (1.4%). describes in more details these four categories.
Table 1. Distribution of publications among different topics.
Referenced statistical approaches
The different statistical approaches used in the selected articles were automatically extracted from the abstract when possible. The results are shown in , in which the different techniques were categorized in three classes: the machine learning techniques (advanced data mining techniques such as artificial neural networks and support vector machines), the regression techniques (classical regression techniques such as linear and logistic regressions) and other techniques (chi-square, Sudent t-test, etc.). In the “Diagnostic” category we found 33.3% of machine learning techniques, 38.9% of regression techniques and 27.8% of other techniques. In the “Modelization” category we found 42.2% of machine learning techniques, 37.8% of regression techniques and 20.0% of other techniques. Finally, in the “Therapeutic” category we found 20% of machine learning techniques, 40% of regression techniques and 40% of other techniques. When we analyzed the distribution of techniques without categorization we found 53.8% of machine learning techniques, 27% of regression techniques and 19.2% of other techniques.
Trends evolution
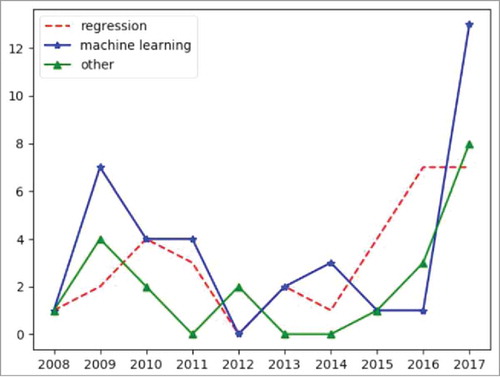
The dataset sizes have significantly increased over the last decade. For each article, BIBOT tries to automatically extract the number of patients considered, and use it to compute the mean and median sizes of cohorts by year. These results are based on 115 parsed abstracts. A linear regression on the mean values give a coefficient of 8224, which means a great increase of the cohorts sizes. However, the median size shows a lower increase from 144 in 2008 to 212.5 in 2017. Altogether, 177303 patients were associated to the “Diagnostic” category, 96115 patients to the “Modelisation” category and 1336 to the “Therapeutic” category. shows the evolution of the trends in publications through analysis of the apparition of specific keywords referencing to statistical approaches in abstracts over the last decade. We observed fluctuant values over this period of time although there seems to be a global increase of the terms associated to machine learning in the last years.
Study of pSS using big data approaches
The 12 articles reporting on pSS among the 1017 retained by the algorithm are presented in . We observed that 50% of this selection (6 articles) were published in 2017 and only 16% (2 articles) were published before 2015. The main objectives of these articles were to estimate the prevalence of pSSCitation15-Citation17 (and to this purpose use large cohorts of patients), to search new biomarkers to increase diagnosis accuracyCitation18-Citation22 and to focus on genetic aspects of the diseaseCitation23-Citation25 (mechanisms, susceptibility factor, and so on). Only one article emphasized on the pathogenesis aspect of the diseaseCitation26 but without focusing on the aspect of pathogenesis-based treatments.
Table 2. Selected articles reporting on pSS.
Discussion
This study has highlighted some important trends of publications over the past ten years, concerning the use of machine learning techniques and big data related approaches applied to the study of autoimmune diseases, with an emphasis on pSS.
Firstly, an important number of articles is available on this subject: the literature automatic search selected 1017 publications for further analysis. Secondly, we observe a global increase of the sizes of the cohorts during the last decade in the analyzed publications. Finally, the apparition of certain statistical analysis methods was monitored in the selected abstracts and we detected a global increase of the terms associated to machine learning approaches over time. These last two facts seem to be correlated, larger cohorts imply larger datasets and the necessity to use more “big data” suited algorithms to perform robust analysis.
The augmentation of the use of machine learning techniques in publications instead of simple correlation analysis might also occur because these techniques can be used to find indirect and non – linear relations between the components of complex and only partially known mechanisms involved in the apparition of diseases and in treatment responses. There are not always simple and direct relations to show in a complex disease such as pSS, whose systemic is only partially known and where the clinical symptoms are the indirect results of these underlying mechanisms.
Selected articles were automatically dispatched in four categories based on their abstract (see ). BIBOT succeeds to classify the majority of articles, with only 16 articles left in the “Unclassified” category. We identified some disparities between categories; the number of articles (and so the number of patients and techniques used) seems to be significantly lower in the “Therapeutic” category. Until now the analyzed studies seem to emphasize on the diagnosis and mechanistic aspects of the SADs using artificial intelligence techniques to identify new biomarkers, and not on physio pathogenesis-based treatments.
This study has some limitations: it was not trivial to perform a literature research that aims at retrieving articles with an emphasis on SADs and advanced statistical approaches, two independent domains. Furthermore, much of the information processed by our program came from the abstracts, which do not always contain all the needed information for further analysis. However, we selected an important number of articles, enough to detect trends in publications even if the complete analysis is made only on a fraction of the retrieved publications.
The partial automating of this systematic literature review by means of a program allowed us to filter a lot of candidate articles. As the number of publications on the subject increases over the years (from 74 in 2008 to 138 in 2017) and will probably continue to increase, it might be more and more difficult to retrieve manually all the relevant articles. This new kind of bibliographical approach could prove itself very relevant in the near future.
In conclusion, SADs studies provide an increasing number of patients and thus larger datasets to analyze. The study of autoimmune diseases has entered the era of big data analysis and faces this new challenge with an increasing use of suitable algorithmic tools, such as machine learning approaches.
Material and methods
A semi automatic approach: BIBOT
We have developed a program, called BIBOT, which stands for “bibliography bot”, in order to explore the use of artificial intelligence and advanced machine learning techniques to analyze datasets related to a given application subject. To this end, BIBOT uses natural language processing (NLP) approaches to parse the content of abstracts of large number of publications; NLP is an emerging field of machine learning, which aims at capturing the meaning of sentences and texts written in a natural language (English), such as scientific articles.
BIBOT is written in python 2.7 language, and is designed to interact with the Medline database through the NCBI API, in order to retrieve abstracts, articles and meta-data about theses articles (year of publication, author list, journal of publication, language, conflict of interest statement, etc.). The source code is available on a Github repository.Citation12 The program uses the Bio.Entrez packageCitation13 and the natural language tool kit (nltk) package.Citation14
Data collection
A systematic literature review using BIBOT was performed in January 2018. The search aimed at retrieving all articles published within the past 10 years, and reporting research results on autoimmune diseases with an emphasis on pSS, big data and the use of advanced machine learning techniques to extract information from large cohorts of patients. Only original articles published in English after 2007 were considered.
The Medline database was explored, using the NCBI API from the Bio.Entrez package for python. The lists of references were also automatically added to the selected articles. The requests used by the program to investigate the MedLine database were automatically generated from a list of keywords used as an input for the program. Each combination of at least two keywords is used to generate a query, which leads to 2n-n-1 generated queries, where n is the number of keywords provided to the program.
Article selection
Candidate abstracts were found by the program with a list of 9 keywords: «big data», «artificial intelligence», «machine learning», «autoimmunity», «Sjogren», «modelisation», «diagnosis», «therapeutic», and «mechanisms». BIBOT then evaluated each candidate article with a first filter based on the year of publication (necessarily after 2007) and the language used in the article (necessarily in English). Finally, the program performed a second filter on the remaining articles by means of a text analysis of the abstract using natural language processing approaches. Each sentence from the abstract was segmented into a list of words and symbols, this list was then analyzed by the program to identify the main subjects of the article. Once those topics are identified they are compared to two validation lists of keywords, one containing machine learning terms, and the other autoimmunity terms. Only articles which match at least one element from each of the two validation lists were selected. For example, this last sentence of an abstract: “ Nevertheless, large multicentre studies are investigating this aspect with advanced algorithmic tools on large cohorts of SADs patients.” would be segmented into a list of words and expression among which we could find “advanced algorithmic tools”, and “SADs patients”, two subjects related to machine learning and autoimmune diseases respectively. In this case the article matches one element from each one of the two validations lists and therefore passes the second filter. A flow chart () describes the search and selection process.
Data analysis
The articles which were automatically selected based on their abstract were obtained in full text. General elements were automatically collected for each article, including year of publication, country of origin of the data, conflict of interest. The contents of the selected articles were then analyzed by the authors and the extracted data were entered into a spreadsheet file.
The number of articles published over time was analyzed using univariate linear regression. The evolution of the global frequency of appearance of terms referencing to statistical analysis approaches in the abstracts of publications over the last decade was computed by our program, and is presented in the «results» section. The evolution of the sizes of datasets over the last decade were given by the number of patients constituting the cohorts used for the research presented in each article, and were retrieved automatically by BIBOT from the abstract, when the information was available.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Financial support or other benefits from commercial sources
none
Significance & Innovation
Big data analysis has become a common way to extract information from complex and large datasets.
The mean number of articles using big data in autoimmune diseases was 101.0 (S.D. 19.16) by year and increase significantly over the time (from 74 articles in 2008 to 138 in 2017).
Only 12 articles focused on pSS but none of them emphasized on the aspect of physio pathogenesis-based treatments.
References
- Brandt JE, Priori R, Valesini G, Fairweather D. Sex differences in Sjögren's syndrome: a comprehensive review of immune mechanisms. Biol Sex Differ. 2015;6:19. doi:10.1186/s13293-015-0037-7. PMID:26535108.
- Maciel G, Crowson CS, Matteson EL, Cornec D. Prevalence of primary sjögren's syndrome: Sjögren may have indeed been right. Arthritis Care Res. 2017;69(10):1612–6. doi:10.1002/acr.23173.
- Park YS, Gauna AE. Mouse models of primary Sjögren's syndrome. Curr Pharm Des. 2015;21(18): 2350–64. doi:10.2174/1381612821666150316120024. PMID:25777752.
- Fairweather D, Petri M, Coronado MJ, Cooper LT. Autoimmune heart disease: role of sex hormones and autoantibodies in disease pathogenesis. Expert Rev Clin Immunol. 2012;8(3):269–84. doi:10.1586/eci.12.10. PMID:22390491.
- Brito-Zerón P, Kostov B, Fraile G, Caravia-Durán D, Maure B, Rascón FJ, et al. SS Study Group GEAS-SEMI.Characterization and risk estimate of cancer in patients with primary Sjögren syndrome. J Hematol Oncol. 2017;10(1):90. doi:10.1186/s13045-017-0464-5. PMID:28416003.
- Horvath IF, Szanto A, Papp G, Zeher M. Clinical Course, Prognosis, and Cause of Death in Primary Sjögren's Syndrome. Journal of Immunology Research. 2014;2014:647507. doi:10.1155/2014/647507. PMID:24963499.
- Kim HJ, Kim KH, Hann HJ, Han S, Kim Y, Lee SH, Kim DS, Ahn HS. Incidence, mortality, and causes of death in physician-diagnosed primary Sjögren's syndrome in Korea: A nationwide, population-based study. Semin Arthritis Rheum. 2017 Oct;47(2):222–7. doi:10.1016/j.semarthrit.2017.03.004.
- Tobón GJ, Saraux A, Gottenberg JE, Quartuccio L, Fabris M, Seror R, Devauchelle-Pensec V, Morel J, Rist S, Mariette X, et al. Role of Fms-like tyrosine kinase 3 ligand as a potential biologic marker of lymphoma in primary Sjögren's syndrome. Arthritis Rheum. 2013 Dec;65(12):3218–27 doi:10.1002/art.38129.
- Fragkioudaki S, Mavragani CP, Moutsopoulos HM. Predicting the risk for lymphoma development in Sjogren syndrome: An easy tool for clinical use. Medicine (Baltimore). 2016 Jun;95(25):e3766. doi:10.1097/MD.0000000000003766.
- Saraux A, Pers JO, Devauchelle-Pensec V. Treatment of primary sjö̈gren syndrome. Nat Rev Rheumatol. 2016;12(8):456–71. doi:10.1038/nrrheum.2016.100. PMID:27411907.
- Ostmeyer J, Christley S, Rounds WH, Toby I, Greenberg BM, Monson NL, Cowell LG. Statistical classifiers for diagnosing disease from immune repertoires: a case study using multiple sclerosis. BMC Bioinformatics. 2017;18:401. doi:10.1186/s12859-017-1814-6. PMID:28882107.
- Foulquier N. BIBOT: Bibliography bot. GitHub; 2018.
- Kans J. Entrez Direct: E-utilities on the UNIX Command Line. US: National Center for Biotechnology Information; 2013.
- Bird S, Klein E, Loper E. Natural Language Processing with Python. Sebastopol, California: O'Reilly Media; 2009.
- Liu FC, Kuo CF, See LC, Tsai HI, Yu HP. Familial aggregation of myasthenia gravis in affected families: a population-based study. Clinical Epidemiology. 2017;9:527–535. doi:10.2147/CLEP.S146617.
- Brito-Zerón P, Acar-Denizli N, Zeher M, Rasmussen A, Seror R, Theander E, Li X, Baldini C, Gottenberg JE, Danda D, et al. Influence of geolocation and ethnicity on the phenotypic expression of primary sj ̈ogren's syndrome at diagnosis in 8310 patients: a cross-sectional study from the big data sjogren project consortium. Ann Rheum Dis. 2017;76(6):1042–50. doi:10.1136/annrheumdis-2016-209952. PMID:27899373.
- Ramos-Casals M, Brito-Zerón P, Kostov B, Sisó-Almirall A, Bosch X, Buss D, Trilla A, Stone JH, Khamashta MA, Shoenfeld Y. Google-driven search for big data in autoimmune geoepidemiology: analysis of 394,827 patients with systemic autoimmune diseases. Autoimmun Rev. 2015;14(8):670–9. doi:10.1016/j.autrev.2015.03.008. PMID:25842074.
- Sun HY, Lv AK, Yao H. Relationship of miRNA-146a to primary sjögren's syndrome and to systemic lupus erythematosus: a meta-analysis. Rheumatol Int. 2017;37(8):1311–6. doi:10.1007/s00296-017-3756-8. PMID:28573480.
- Tayob N, Do KA, Feng Z. Unbiased estimation of biomarker panel performance when combining training and testing data in a group sequential design. Biometrics. 2016;72(3):888–96. doi:10.1111/biom.12480. PMID:26845527.
- Hobson P, Lovell BC, Percannella G, Vento M, Wiliem A. Benchmarking human epithelial type 2 interphase cells classification methods on a very large dataset. Artif Intell Med. 2015;65(3):239–50. doi:10.1016/j.artmed.2015.08.001. PMID:26303104.
- Alevizos I, Illei GG. Micrornas in sjögren's syndrome as a prototypic autoimmune disease. Autoimmun Rev. 2010;9(9):618–21. doi:10.1016/j.autrev.2010.05.009. PMID:20457282.
- Huang ZC, Shi YY, Cai B, Wang LL, Wu YK, Ying BW, Feng WH, Hu CJ, Li YZ. Promising diagnostic model for systemic lupus erythematosus using proteomic fingerprint technology. Sichuan Da Xue Xue Bao Yi Xue Ban. 2009;40(3):499–503. PMID:19627014.
- Fang YF, Chen YF, Chung TT, See LC, Yu KH, Luo SF, Kuo CF, Lai JH. Hydroxychloroquine and risk of cancer in patients with primary sjögren syndrome: propensity score matched landmark analysis. Oncotarget. 2017;8(46):80461–71. doi:10.18632/oncotarget.19057. PMID:29113317.
- Qu S, Du Y, Chang S, Guo L, Fang K, Li Y, Zhang F, Zhang K, Wang J. Common variants near ikzf1 are associated with primary sj ̈ogren's syndrome in han chinese. PLoS One. 2017;12(5):e0177320. doi:10.1371/journal.pone.0177320. PMID:28552951.
- Liu M, Wu X, Liu X, He J, Su Y, Guo J, Li Z. Contribution of dendritic cell immunoreceptor (dcir) polymorphisms in susceptibility of systemic lupus erythematosus and primary sjogren's syndrome. Hum Immunol. 2015;76(11):808–11. doi:10.1016/j.humimm.2015.09.040. PMID:26429306.
- Wang-Renault SF, Boudaoud S, Nocturne G, Roche E, Sigrist N, Daviaud C, Bugge Tinggaard A, Renault V, Deleuze JF, Mariette X, et al. Deregulation of microrna expression in purified t and b lymphocytes from patients with primary sjögren's syndrome. Ann Rheum Dis. 2017;77(1):133–40. doi:10.1136/annrheumdis-2017-211417. PMID:28916716.